Abstract
Programmed cell death (PCD) in plant is triggered by abiotic and biotic stress. Light-dependent PCD is unique to plants. Light-induced PCD also requires reactive oxygen species (ROS) and salicylic acid (SA). In this study, lesion simulating disease1 (LSD1) and elongated hypocotyl 5 (HY5) perform opposite roles to regulate excess red light (RL)-triggered PCD associated with ROS and SA production. Under RL, the lsd1 mutant released more ROS and SA and displayed a stronger cell death rate than the hy5 mutant. It was shown that active LSD1 converted into inactive form by changing the redox status of the plastoquinone pool, and HY5 interacted with phytochrome B (phyB) to promote PCD in response to RL. LSD1 inhibited the enhanced disease susceptibility 1 (EDS1) expression by upregulating SR1, whereas HY5 enhanced the enhanced EDS1 expression by binding to the G-box of the EDS1 promoter. This study suggested that LSD1 and HY5 antagonistically modulated EDS1-dependent ROS and SA signaling; thus, PCD was mediated in response to RL.
Keywords: Arabidopsis, excess red light, EDS1, HY5, LSD1, programmed cell death, ROS, SA
Introduction
Programmed cell death (PCD) is involved in plant growth and development. PCD is also closely related to defense during plant–pathogen interactions or acclimation in response to abiotic stress (Rusterucci et al., 2001; Gechev et al., 2006). The onset of PCD in plants is dependent on light, which is essential to the life cycle of plants. Excess light (EL) stress triggers the release of reactive oxygen species (ROS) and subsequently induces PCD in plants; red light (RL) is vital in this process (Asada, 1999; Mateo et al., 2004). ROS, which often interacts with salicylic acid (SA) and ethylene (ET), contributes to the progress of light-dependent PCD (Mazel and Levine, 2001; Muhlenbock et al., 2008; Li et al., 2013). RL-induced PCD is controlled partly by specific changes in redox status of the photosynthetic electron carrier chain, namely, plastoquinone (PQ) pool (Muhlenbock et al., 2008). Meanwhile, phytochrome B (phyB) is a major RL photoreceptor that positively regulates ROS production and cell death during de-etiolation response (Nagatani, 2004; Chen et al., 2013).
The phytochrome family (phyA–phyE) perceives red light (660 nm) and far-red (750 nm) light; this family also monitors light quality and intensity to regulate plant growth and development, such as seed germination, seedling de-etiolation, stem elongation, phototropism, stomata and chloroplast movement, shade avoidance, and flowering time (Franklin and Quail, 2010; Strasser et al., 2010). Phytochromes exist in two different photoreversible forms: RL-absorbing (Pr) and far-RL-absorbing (Pfr) forms. An inactive Pr is converted to an active Pfr; upon RL exposure, the active Pfr is translocated into the nucleus (Nagatani, 2004). Phytochrome-mediated light signals are dependent on SA signaling by regulating the expression of the enhanced disease susceptibility 1 (EDS1) gene (Xie et al., 2011). phyB serves as the upstream of elongated hypocotyl 5 (HY5) and participates in HY5 accumulation under RL (Osterlund et al., 2000). HY5, a basic leucine zipper (bZIP) transcription factor, is the first known positive regulator of light signaling under various wavelengths of light, including RL, far-RL, blue light, and UV-B (Oyama et al., 1997; Osterlund et al., 2000; Ulm et al., 2004). HY5 directly binds the G-box (CACGTG) DNA sequence of downstream genes, including light, ROS, and hormone-responsive genes (Lee et al., 2007; Shi et al., 2011; Chen et al., 2013; Abbas et al., 2014). HY5 positively regulates Pchlide synthesis, ROS production, and cell death in the light during seedling greening (Chen et al., 2013). However, limited information is known whether HY5 is involved in modulating ROS and SA signaling to promote PCD and enhance plant tolerance in response to RL.
Lesion simulating disease 1 (LSD1) encodes a small C2C2 zinc finger protein and acts as a negative regulator of PCD (Dietrich et al., 1997). LSD1 participates in controlling the redox signals generated from the PQ pool in the chloroplast and suppresses cell death under EL and RL conditions. LSD1 and EDS1 elicit opposite effects on ROS and SA-dependent PCD under UVC and cold stress (Muhlenbock et al., 2008; Huang et al., 2010; Wituszynska et al., 2013, 2015). The lsd1 mutant shows an increase in the EDS1 expression but displays a decrease in signal response (SR) family gene transcript levels in response to light stress (Wituszynska et al., 2013). These results suggest that the signal response 1 (SR1) protein combines with the DNA sequence (ACGCGT) of the EDS1 promoter to suppress gene expression; the SR1 protein is necessary to repress SA-dependent defense (Du et al., 2009). However, the EDS1 gene sequence contains a G-box domain, which is possibly combined by HY5. A possible mechanism is that LSD1 suppresses the EDS1 expression by upregulating SR1, but HY5 promotes the EDS1 expression under RL.
LSD1 activity requires three C2C2-type zinc finger domains, and LSD1 is also a redox-sensitive protein (Dietrich et al., 1997). Previous studies suggested that the zinc finger protein often functions as a redox sensor, and redox targets are thiols in Zn/S-coordination center (Junming et al., 2014). After oxidizing treatment is administered, the zinc finger protein releases zinc and subsequently converts into oxidized conformation (dimerization state) to change activity (Hwang et al., 2001; Tsao and Su, 2001; Derong et al., 2005; Ungureanu et al., 2012). RL likely causes the more oxidized status of the cell and then inhibits LSD1 activity by changing conformation, which does not interact with other proteins. Arabidopsis basic leucine zipper transcription factor 10 (AtbZIP10) and LSD1 regulate antagonistically oxidative stress-induced PCD (Kaminaka et al., 2006). Arabidopsis metacaspase 1 (AtMC1) interacts with LSD1 and then blocks AtMC1-dependent PCD (Coll et al., 2010). GSH-induced LITAF domain protein (GILP) and CATALASEs (CAT1, CAT2, and CAT3) interact with LSD1 to negatively regulate hypersensitive cell death (He et al., 2011; Li et al., 2013).
In this study, LSD1 and HY5 performed opposite roles in regulating RL-triggered PCD associated with ROS and SA production. Unexpectedly, LSD1 conformation and activity were controlled by the change in PQ pool under RL. However, phyB interacted with HY5 and contributed to RL response. Our results demonstrated that LSD1 and HY5 antagonistically modulated the EDS1-dependent ROS and SA signaling; thus, PCD was mediated in response to RL.
Materials and methods
Plant materials, growth, light conditions
Arabidopsis ecotype Columbia-0 (Col-0), lsd1-2 (Coll et al., 2010), hy5-215 (Osterlund et al., 2000), hy5-ox-YFP (Oravecz et al., 2006), phyb, phyB-ox-YFP (Wang et al., 2010; Zhao et al., 2014), eds1-2 (Wang et al., 2011) were sterilized and grown in soil culture with 16/8 h light/dark cycle (100 nm μmol photons m−2·s−1) and 54% relative humidity at 22°C. Four-week old plants were used for experiments. Arabidopsis rosettes were fully exposed to excess white light (EL, 1500 μmol m−2 s−1; 6 h) or (RL, 120 μmol photons m−2s−1; 6 h, 650 nm) supplied by light-emitting diode panels (Photon System Inst.). The above light conditions provided similar energy at the indicated spectral regions. Heat emission from the light source was insignificant. Plants were also sprayed with or exposed to a 30–40 mL drop of DCMU (8 μM) applied to the leaf 2–3 h before RL treatment as described (Friedman et al., 2012).
Trypan blue staining
Four-week leaves were exposed to RL for the indicated period of time before staining. Trypan blue staining was performed as described (Muhlenbock et al., 2008). After staining, all samples were mounted on slides and photographed with a stereo microscope at 6 and 15-fold magnification.
Electrolyte leakage analysis
Cell death was quantified by ion leakage from whole rosettes. Four-week-old plants treated with RL, DCMU, and DCMU + RL were harvested. If the leaves were big enough to be cored, leaf disks were removed (7 mm diameter), floated in water for 30 min and subsequently transferred to tubes containing 6 mL distilled water. Conductivity of the solution was detected with an Orion Conductivity Meter at the indicated time points. For each measurement, we used six leaf disks or 40–50 mg fresh weight, which equaled 4–6 leaves. The entire experiment was performed three times (Coll et al., 2010).
Fluorescence imaging of ROS and hydrogen peroxide quantification
ROS fluorescence determination was performed, which the leaves in the dark for 1 h with 10 μM 2′7′-dichlorofluorescin diacetate (H2DCFDA) and oberving the stain by the Zeiss LSM 510. Hydrogen peroxide was quantified using previous method (Muhlenbock et al., 2008).
SA quantifications
The total SA was extracted by previous method (Xing et al., 2013). The extracting total SA was detected by high performance liquid chtomatoraphy (HPLC) using fluorescence detectors: excitation wavelength is 294 nm and emission wavelength is 426 nm.
Determination of glutathione
Glutathione levels were detected through the 5, 5′-dithiobis (2-nitrobenzoic acid)-recycling enzymatic method. The extracted samples were divided in half for assay of total glutathione (GSH + GSSG) and for GSSG alone as described previously (Ge et al., 2007). The absorbance change at 412 nm was measured, and the glutathione concentration was evaluated by comparison with a standard calibration curve.
Catalase activity
CAT activity previously described and was measured spectrophotometrically by monitoring H2O2 decrease at 240 nm (Mateo et al., 2004).
RNA extraction and qRT-PCR analysis
Total RNAs were extracted from detached Arabidopsis leaves using Trizol according to the supplier's recommendations. The integrity of the total RNA extracts was tested by 1% agarose gel electrophoresis. RNA concentrations and purities were measured spectrophotometrically using OD260/OD280 and OD260/OD230 ratios. First-strand cDNA was synthesized with the SuperScript II First-Strand Synthesis System for qRT-PCR (Invitrogen). qRT-PCR was performed using the Roche Light Cycler TM 2.0 Real-time Detection System. The expression of target gene was normalized relative to the housekeeping gene ACTIN2 (Li et al., 2012). The primers are presented LSD1 (5′-TGTCAAACTACGAACCTTGTGC-3′ and 5′-TTGATCTGCGCAACCTGA-3′); HY5 (5′-ATGCAGGAACAAGCGACT-3′ and 5′-TCAAAGGCTTGCATCAGC-3′); EDS1(5′-CGAAGGGGACATAGATTGGA-3′and5′-ATGTACGGCCCTGTGTCTTC-3′); PAD4 (5′-TATGGTCGACGCTGCCATAC-3′ and 5′-ATAGAAGCCAAAGTGCGGTG-3′); NPR1 (5′-CCGCCGCTAAGAAGGAGAAA-3′and 5′-GCCAAAACAGTCACAACCGA-3′); ICS1 (Du et al., 2009); PAL(Xing et al., 2013) and PR1(Zhao et al., 2014). The results were calculated using the 2−ΔΔCT method. Preliminary statistics were performed using the Roche Light Cycler system and the significance of differences was analyzed in SPSS 15.0.
Protein extraction and western blot analysis
For protein extraction, 0.4 g treated 14-day-old seedlings were ground to powder in liquid nitrogen and dissolved in extraction buffer [50 mM Tris–HCl, pH 6.8, 50 mM DTT, 4% (w/v) SDS, 10% (v/v) glycerol, 1% (w/v) polyvinylpolypyrrolidone (PVPP), 5 mM PMSF]. After centrifugation at 12,000 g for 20 min twice at 4°C, the supernatants were transferred into new tubes, quickly stored at −80 °C until analyzed. The protein content was determined by the BCA Protein Assay Kit (TIANGEN, China). Extracts containing 40 mg protein were separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to nitrocellulose membranes. The membranes were blocked with TBST (10 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20) containing 5% non-fat milk for 1 h and then incubated with LSD1 antibody (Agrisera), HY5 antibody (Osterlund et al., 2000). After washing three times, the membranes were blocked with secondary antibody anti-rabbit IRDyeTM800 (Rockland Immunochemicals, Gilbertsville, PA, USA). Proteins were detected by using Odyssey two-color infrared imaging system (Li-Cor, Inc., Lincoln, NE, USA).
Co-immunoprecipitation assay
Co-immunoprecipitation assay was performed as described previously (Zhao et al., 2014) with some modifications. Total proteins were extracted from plants in an extraction buffer [50 mM Tris-HCl, pH 7.5–8.0, 100 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1 mM sodium or thovanadate, 50 mM sodium fluoride, 1 mM PMSF, containing protease inhibitor cocktail (Roche)]. Proteins extracts were inoculated with antibody for 4 h, and then protein A beads were added. After incubation overnight at 4°C, the beads were centrifuged and washed four times with a wash buffer (PBS, pH 7.4). The immunoprecipitated proteins were detected by SDS-PAGE gel with YFP antibody or LSD1 antibody (Agrisera). HY5 protein was detected with a rabbit polyclonal HY5 antibody (Osterlund et al., 2000) after immunoprecipitation with YFP and HY5 antibody (Zhao et al., 2014).
Chromatin immunoprecipitation (ChIP)
For the ChIP analysis, 1.5 g fresh leaves were crosslinked with 10 mL of 1% formaldehyde under vacuum infiltration conditions. ChIP assays were performed as described previously (Vani et al., 2007) with a minor modification of the three wash buffers: Low wash buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8), High Salt Wash buffer (500 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8), LiCl Wash Buffer (0.25 M LiCl, 1% v/v NP-40, 1% w/v sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8). The amount of each precipitated DNA fragment was determined by semi-quantitative PCR and qRT-PCR using to amplify the sequences with G-box regions contained in the EDS1 gene promoter (from −221 to −101) (5′-AAAACCGACACGTGGAAAGC-3′ and 5′-CCCCATCATGAGACCATTTCA-3′) and the sequences without G-box regions (from −688 to −571) (5′-CGAACGCAAAAACGGACCAG-3′ and 5′-CTTATCCGGAAAGTAAACCGGA-3′).
Statistical analysis
Data were subjected to analyzing by Duncan's multi- ple range tests (DMRTs; p < 0.05) and significance by analysis of variance (ANOVA) using SPSS12.0 software for Windows (Zhao et al., 2014).
Results
Excess light promotes programmed cell death regulated by LSD1 and HY5, red light is a major inducer
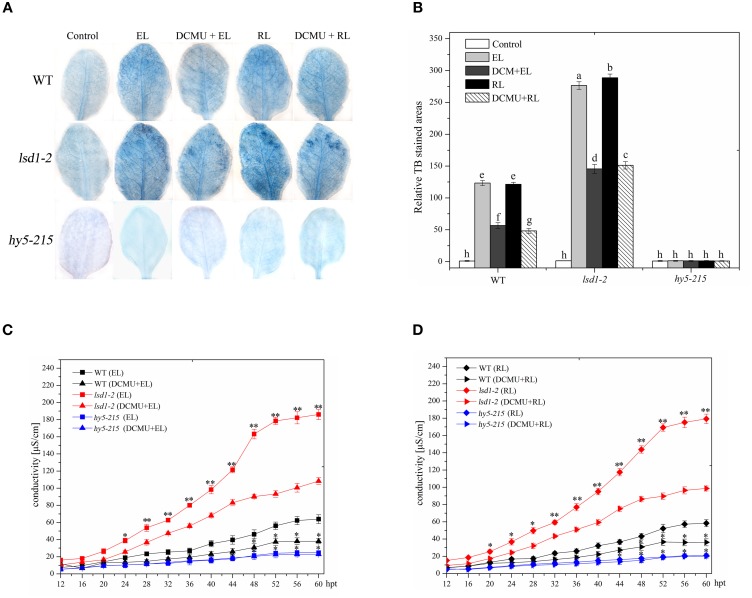
In Arabidopsis wild-type leaves, excess white light or excess RL caused PCD, as indicated by trypan blue staining and ion leakage detection results; by contrast, the extent of cell death in response to RL was similar to EL irradiation. The lsd1-2 mutant showed a more prominent increase in PCD than the WT under EL and RL treatments. However, a significant decrease in PCD was observed in the hy5-215 mutant (Figure 1). The redox state of the PQ pool was involved in regulating EL-induced PCD and light acclimatory responses (Muhlenbock et al., 2008). EL and RL maintain the redox state of PQ to a more reduced state to promote PCD, whereas DCMU inhibits this process and maintains the redox state of PQ to a more oxidized state (Vandewalle et al., 2007; Muhlenbock et al., 2008; Busch, 2014). The lsd1-2 mutant and WT treated with DCMU under ambient light for 4 h and exposed to EL or RL showed a significant decrease in PCD compared with that treated with RL alone; by contrast, the hy5-215 mutant leaves of the same treatment were slightly different (Figure 1). These results indicated that LSD1 and HY5 acted as negative and positive regulators, respectively, in EL-induced PCD; RL was a major inducer. The redox status of PQ is implicated in RL-dependent PCD.
Figure 1.
Plant PCD in response to RL is regulated by LSD1, HY5 and the redox status of PQ pool. (A) Trypan blue staining of four-week-old leaves of wild type(WT), lsd1-2 mutant, and hy5-215 mutant, which were exposed to normal light (control, 100 μmol photons m−2s−1), excess white light (EL, 1500 μmol m−2 s−1; 6 h) or excess red light (RL, 120 μmol m−2 s−1; 660–680 nm; 6 h) excess red light (RL; 120 μmol photons m−2s−1; 6 h), and were pre-sprayed 8 μM DCMU under normal light for 3 h and then exposed to EL or RL for 6 h (DCMU + RL). (B) Areas of TB-stained foliar tissues in WT, lsd1-2, and hy5-215 mutant as described in (A). Different letters indicate statistically significant differences between treatments (Duncan's multiple range test: P < 0.05). Values represent means ± SD of three independent experiments. (C,D) Ion leakage of WT, lsd1-2 mutant, and hy5-215 mutant.
RL-induced ROS and SA accumulations are controlled by LSD1 and HY5
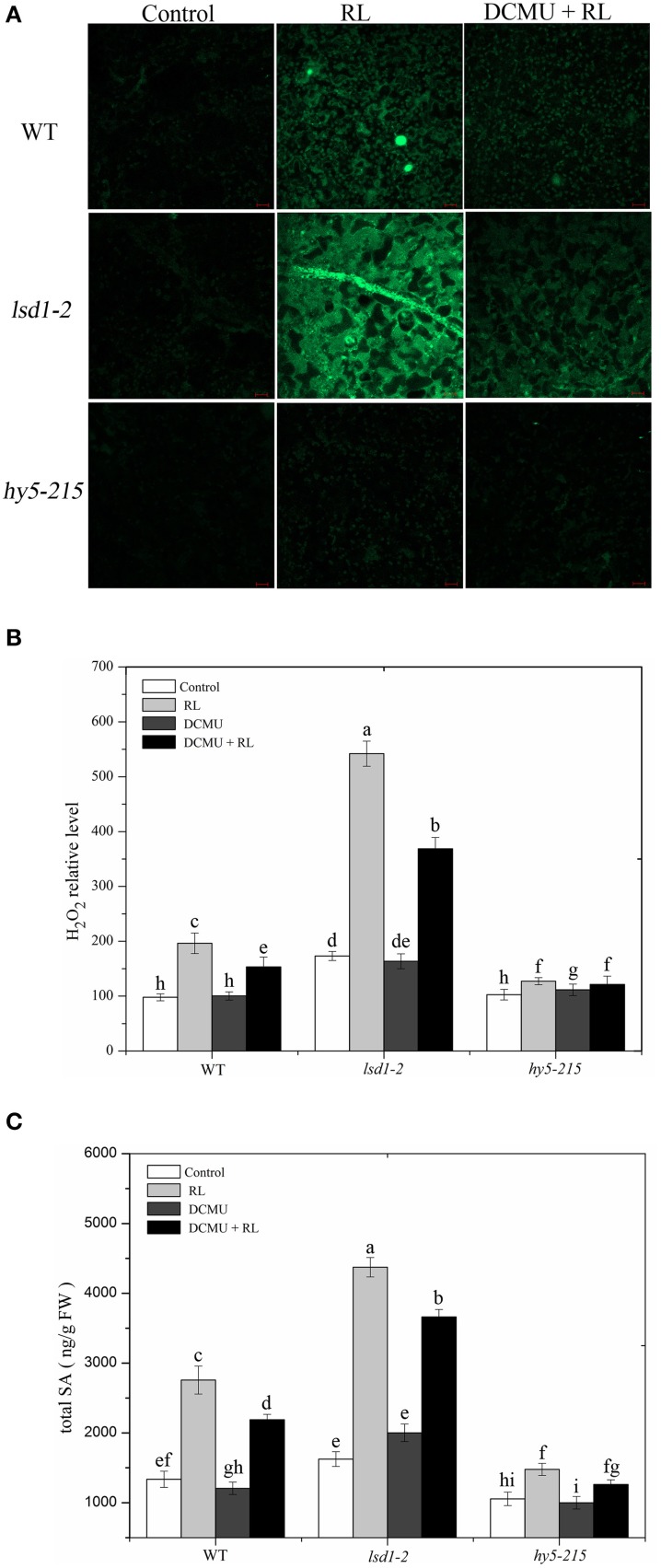
A previous study substantiated that EL-induced cell death is regulated by subsequent changes in cellular ROS and hormonal homeostasis, not by changes in ROS alone (Mackintosh et al., 1989). RL leads to ROS and SA accumulation, and therefore promotes PCD (Mateo et al., 2004; Muhlenbock et al., 2008; Szechynska-Hebda et al., 2010). H2O2 and SA concentrations in WT and two mutant (lsd1-2 and hy5-215) plants were measured in the present study. The exposure of WT to RL for 6 h resulted in a significant increase in H2O2 and SA concentrations (Figure 2). At the same time, H2O2 and SA concentrations were higher in the RL-treated lsd1-2 mutant than in WT; the production of H2O2 and SA of the hy5-215 mutant was prevented compared with that of WT (Figure 2). This result indicated that LSD1 could suppress the accumulation of H2O2 and SA. However, HY5 could promote accumulation in RL-induced PCD. H2O2 and SA concentrations were remarkably lower in the lsd1-2 mutant and WT pre-treated with DCMU and then treated with RL than in plants treated with RL alone; by contrast, H2O2 and SA concentrations in the hy5-215 mutant were weakly decreased. The results suggested that LSD1 and HY5 elicited an antagonistic effect on RL-induced ROS release, SA accumulation, and PCD, which were partially dependent on the redox status of the PQ pool (Figures 2A–C). Our results suggested that LSD1 and HY5 had antagonistic effect on RL-induced ROS burst, SA accumulation, and consequently PCD, which were partially dependent on the redox status of PQ pool.
Figure 2.
Plant cellular ROS/SA homeostasis modulated by LSD1, HY5, and the redox status of PQ pool under red light. (A) H2DCFDA fluorescence (green) indicates ROS in four-week-old leaves of WT, lsd1-2 mutant, and hy5-215 mutant, which were exposed to control, RL for 6 h, and DCMU + RL. (B) The cellular H2O2 levels by the quantitative measurement in four-week-old leaves of WT, lsd1-2 mutant, and hy5-215 mutant exposed to different treatment, including control, RL, DCMU, and DCMU +RL. (C) The cellular SA levels were detected in WT, lsd1-2 mutant, and hy5-215 mutant, the four-week old leaves were exposed to control, RL, DCMU, and DCMU+ RL. Data are means ± SD of three experiments. Different letters indicate statistically significant differences between treatments (Duncan's multiple range test: P < 0.05).
RL changes the cellular redox status and subsequently affects LSD1 conformation and activity
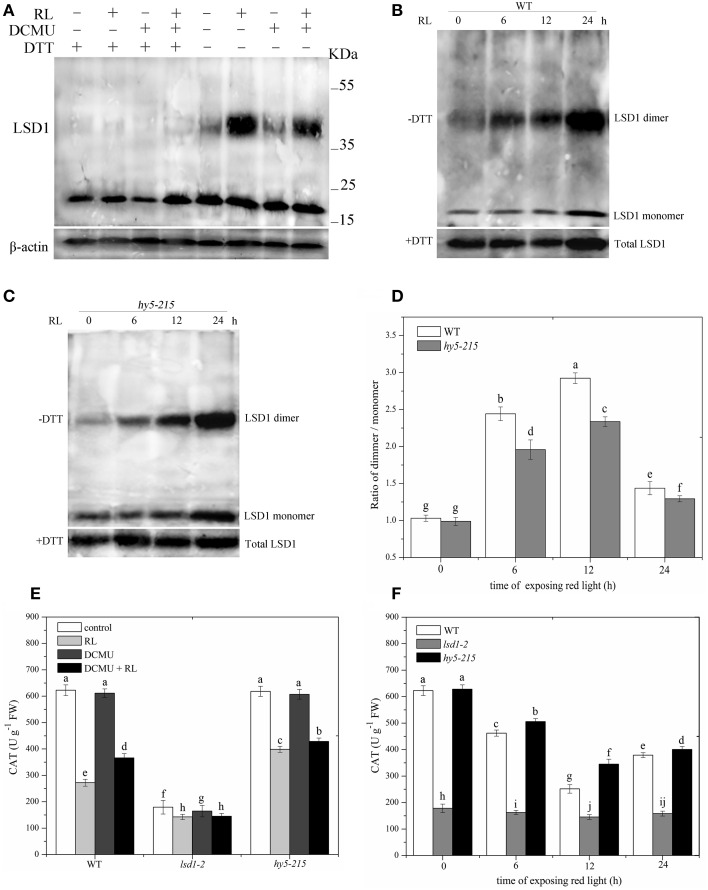
Many redox-regulated proteins, such as Hsp33 and SP1, use a cysteine-coordinating zinc center as a redox sensor. Upon exposure to oxidized condition, cysteines are quickly oxidized and zinc is released (Maret, 2006). LSD1 is a novel zinc finger protein that contains three zinc finger domains; this protein is also a redox-regulated protein. To determine whether RL can cause cellular redox state to change, we analyzed the content of reduced glutathione (GSH) and the ratio of reduced glutathione: total glutathione ([GSH]:[(GSH) + (GSSG)]) in RL-treated WT. The results showed that RL caused an increase in GSH levels within 6 h. GSH levels were maximal at 12 h after RL treatment was administered and decreased thereafter (Supplementary Figure 1A). A significant decrease in the redox status of the glutathione pool after 12 h indicated that oxidative stress was induced by RL irradiation (Supplementary Figure 1B). The GSH levels decreased when WT was pre-treated with DCMU and then exposed to RL for 12 h, but the (GSH):([GSH] + [GSSG]) ratio significantly increased compared with that of the 12 h RL treatment (Supplementary Figures 1C,D). The GSH levels and the (GSH):([GSH] + [GSSG]) ratio remained unchanged in the chloroplast, whereas the cytoplasmic (GSH):([GSH] + [GSSG]) ratio was altered and similar to the trend of the total cell (Supplementary Figure 1). The results demonstrated that RL disturbed the cellular PQ pool balance and subsequently resulted in a more oxidized environment in the cytoplasm.
The reducing agent DTT, included in the protein extraction solution, can modify protein conformation and function by reducing protein cysteine residues. Protein samples extracted with or without DTT were then subjected to immunoblot analysis in WT to investigate the specific conformation of the LSD1 protein. The addition of DTT to all samples reduced all of the LSD1 proteins to the monomeric form. Without DTT, the LSD1 protein was present at a higher molecular weight (dimer) in RL treatment but not in the control group (Figure 3A). Under RL, dimer and monomer of the LSD1 protein increased; by contrast, the ratio of dimer/monomer decreased. The dimer of LSD1 of plants subjected to DCMU pre-treatment and RL treatment was significantly decreased compared with those subjected to RL treatment alone (Figure 3A). To explore whether HY5 is involved in regulating the LSD1 conformation, we detected LSD1 conformation in the hy5-215 mutant without DTT. The results showed that the ratio of dimer/monomer of LSD1 in the hy5-215 mutant was slightly lower than that in WT after RL treatment was administered (Figures 3B–D). The RL-induced PQ pool change combined with HY5 modulated the LSD1 protein conformation change, and the redox status of the PQ pool played a major role.
Figure 3.
Conformation of LSD1 was changed by immunoprecipitation assay and LSD1 activity was indirectly measured under RL. (A) Total protein extracted with (+) or without (–) DTT (50 mM) in the extraction buffer from control, RL, DCMU, and DCMU + RL treated WT was subjected to immunoprecipitation analysis, both dimmer, and monomer LSD1 protein were detected. (B) Total protein was extracted from WT leaves at times after RL treatment and analyzed by an immunoprecipitation assay. (C) The LSD1 conformation was detected in hy5-215 mutant. (D) The ratio of LSD1 dimer/monomer was analyzed quantitatively. (E) The CAT activity was measured in WT, lsd1-2 mutant, and hy5-215 mutant, the four-week old leaves were exposed to control, RL, DCMU and DCMU+ RL for 6 h. (F) The CAT activity of WT, lsd1-2 mutant, and hy5-215 mutant was detected at times after RL treatment. Different letters indicate statistically significant differences between treatments (Duncan's multiple range test: P < 0.05). Values represent means ± SD of three independent replicates.
To analyze LSD1 activity, we indirectly examined CAT activity because the active LSD1 positively regulates CAT activity by interacting with CAT (Li et al., 2013). The CAT activity was lower in RL-treated WT than in the control group. The CAT activity of plants pre-treated with DCMU and exposed to RL was significantly enhanced compared with that of plants treated with RL alone. The lsd1-2 mutant also displayed the strong inhibition of the CAT activity, particularly in the RL treatment. However, the CAT activity showed a weak reduction in the hy5-215 mutant compared with that of WT (Figures 3E,F). The results showed that RL induced a more oxidized GSH pool, which mainly depended on the redox status of the PQ pool change, and caused the monomer of LSD1 to convert into dimer and suppress the LSD1 activity. HY5 weakly influenced the LSD1 conformation change and activity.
HY5 interacts with phyB in cell death induced by RL
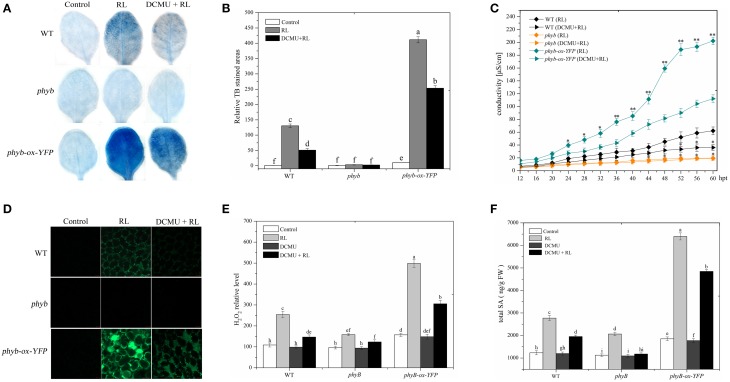
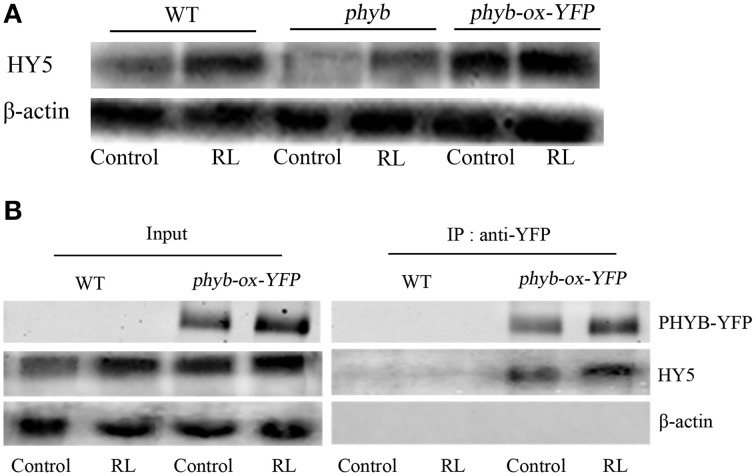
phyB is a specific RL-response factor, which positively modulates cell death and ROS signaling (Li et al., 2011; Chen et al., 2013). In this study, the phyB mutant significantly inhibited PCD and decreased ROS and SA concentrations; by contrast, phyB-ox-YFP significantly enhanced PCD and increased ROS and SA levels compared with WT exposed to RL (Figure 4). The phyB mutant and phyB-ox-YFP were also exposed to DCMU and RL. PCD and ROS/SA concentrations in phyB-ox-YFP decreased compared with those in phyB-ox-YFP subjected to RL treatment alone; by contrast, PCD and ROS/SA concentrations in the phyB mutant weakly decreased (Figure 4). It has also been found that phyB acted as upstream of HY5 and positively regulates HY5 (Osterlund et al., 2000). The mutation of phyB inhibited HY5 protein accumulation. However, the overexpressed phyB line increased the HY5 protein concentration compared with WT (Figure 5A). However, whether phyB promotes HY5 protein accumulation through the interaction of phyB with HY5 Co-immunoprecipitation assays were performed using proteins isolated from phyB-ox-YFP transgenic plants or WT to ensure that this interaction occurred in vivo. Figure 5B showed that the YFP antibody could immunoprecipitate the HY5 fusion protein, as detected by the HY5 antibody, under control and RL conditions. The result showed HY5 could interact with phyB under control and RL. The data suggested that phyB-mediated HY5 accumulation may rely on the interaction of phyB and HY5 under the RL-induced PCD.
Figure 4.
phyB is involved in regulating RL-induced PCD associated with ROS and SA production. (A) Trypan blue staining of four-week-old leaves of WT, phyB mutant, and phyB-ox-YFP under control, RL, and DCMU + RL treatment. (B) Areas of TB-stained foliar tissues in WT, phyB mutant, and phyB-ox-YFPas described in (A). (C) Relative ion leakage in WT, phyB mutant, and phyB-ox-YFP after RL or DCMU + RL. Asterisks indicate significant differences between the wild type and the mutants (Student's paired t-test: *P < 0.05, **P < 0.01). (D) H2DCFDA fluorescence (green) indicates ROS in leaves of WT, phyB mutant, and phyB-ox-YFP exposed to control, RL for 6 h, and DCMU + RL. (E) Measurement of ROS levels of WT, phyB mutant, and phyB-ox-YFP under control, RL, DCMU, and DCMU + RL conditions. (F) Detection of SA levels. Different letters indicate statistically significant differences between treatments (Duncan's multiple range test: P < 0.05). Values represent means ± SD of three independent replicates.
Figure 5.
HY5 interacts with phyB and promoted HY5 accumulation under RL. (A) The western blot analysis the HY5 of protein levels in WT, phyB mutant, and phyB-ox-YFP transgentic plant under untreated and RL treatment. (B) In vivo coimmunoprecipitation assay between phyB and HY5, WT and phyB-ox-YFP plant was exposed to control and RL. After precipitation with the anti-phyB-YFP antibody, proteins were immunoblotted with anti-HY5 or anti-YFP antibodies.
EDS1 is required for RL induced-cell death and its expression is modulated antagonistically by LSD1 and HY5
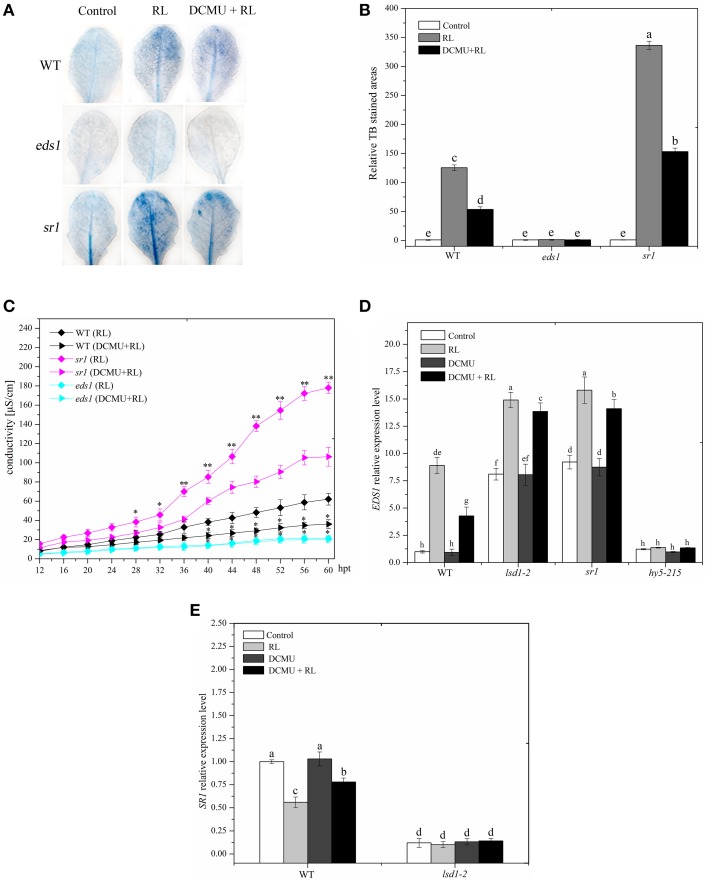
EDS1 is necessary to upregulate ROS and ET production in EL stress and caused the onset of cell death in the lsd1 mutant (Mateo et al., 2004). PCD was severely suppressed in the eds1 mutant compared with that in WT after RL treatment was administered (Figures 6A–C). The eds1 mutant also showed decreased ROS and SA concentrations compared with WT exposed to RL (Supplementary Figure 3). RL induced an increase in the EDS1 expression levels in WT (Figure 6C). The result indicated that EDS1 is required for RL-induced ROS and SA signaling and PCD.
Figure 6.
EDS1 is in response to RL-induced PCD and its expression is controlled by LSD1 and HY5. (A) Trypan blue staining of four-week-old leaves of WT, eds1 and sr1 mutant after control, RL, and DCMU + RL treatment. (B) Areas of TB-stained foliar tissues in WT, eds1 and sr1 mutant as described in (A). (C) Relative ion leakage was detected and showed the extent of cell death. Asterisks indicate significant differences between the wild type and the mutants (Student's paired t-test: *P < 0.05, **P < 0.01). (D) qRT-PCR showing the relative expression of EDS1 gene in WT, hy5-215,lsd1-2 and sr1 mutant exposed to control, RL, DCMU and DCMU + RL. (E) qRT-PCR determination of SR1 mRNA levels in WT and lsd1-2 mutant exposed to control, RL, DCMU, and DCMU + RL. Each data is the mean ± SD of three independent replicates. Different letters indicate statistically significant differences between treatments (Duncan's multiple range test: P < 0.05).
LSD1 negatively regulates EDS1-dependent PCD during EL (Muhlenbock et al., 2008). The EDS1 expression levels were increased in the lsd1-2 mutant compared with those in WT exposed to RL, indicating that LSD1 inhibited the EDS1 expression (Figure 6D). This previous study also showed that phyA and phyB could positively modulate the EDS1-dependent SA signaling by improving EDS1 expression (Xie et al., 2011). PhyB may promote the EDS1 expression by upregulating HY5. To determine whether HY5 positively regulates EDS1 expression, we performed qRT-PCR analysis of the hy5-215 mutant and WT after RL treatment was administered. Our results showed that the EDS1 expression levels strongly decreased in the hy5-215 mutant than in WT regardless of RL treatment (Figure 6D). These data demonstrated that HY5 could enhance the EDS1 expression. The EDS1 expression in plants pre-treated with DCMU and then treated with RL decreased compared with that in WT treated with RL only; however, no evident change occurred in lsd1-2 and hy5-215 mutants (Figure 6D). These results indicated that the PQ pool was also involved in the EDS1 expression in response to RL. Thus, LSD1 and HY5 antagonistically modulated the EDS1 expression during RL-induced cell death.
LSD1 inhibits EDS1 gene expression by contributing to SR1 expression
SR1 directly binds to promoter of EDS1 gene, and consequently suppressed EDS1 gene expression (Du et al., 2009). Recent study has showed that LSD1 positively regulates SR family expression (Wituszynska et al., 2013); SR1 prevents cell death, biotic defense responses, and RL response (Galon et al., 2008, 2010). The sr1 mutant displayed a strong increase in cell death and ROS/SA production compared with WT after RL treatment was administered (Figures 6A–C and Supplementary Figure 3). However, no difference was observed between sr1 and lsd1-2 mutants (Figures 1, 2, 6). TheEDS1 expression levels were enhanced in lsd1-2 and sr1 mutants compared with those in WT; by contrast, the EDS1 expression levels were similar between lsd1-2 and sr1 mutants regardless of RL treatment (Figure 6D). Cell death and ROS/SA generation were partly suppressed and EDS1 expression was decreased when the sr1 mutant was pre-treated with DCMU and then exposed to RL compared with that treated with RL alone (Figures 6A–C). Therefore, SR1 and LSD1 possibly prevented RL-induced PCD by inhibiting EDS1 expression.
LSD1 may be involved in regulating SR1 gene expression. SR1 expression levels decreased in the lsd1-2 mutant compared with WT. SR1 expression levels also decreased in WT exposed to RL than in the control group but were similar to the lsd1-2 mutant between the control group and the RL-treated group (Figure 6E). The SR1 expression also decreased in WT exposed to DCMU and then to RL compared with that exposed to RL alone. However, no difference was observed in the lsd1-2 mutant (Figure 6E). These results demonstrated that LSD1 inhibited the EDS1 expression by upregulating SR1 expression, but RL inhibited SR1 gene expression by inducing PQ pool to change.
HY5 directly binds the promoter of EDS1 gene to promote its expression
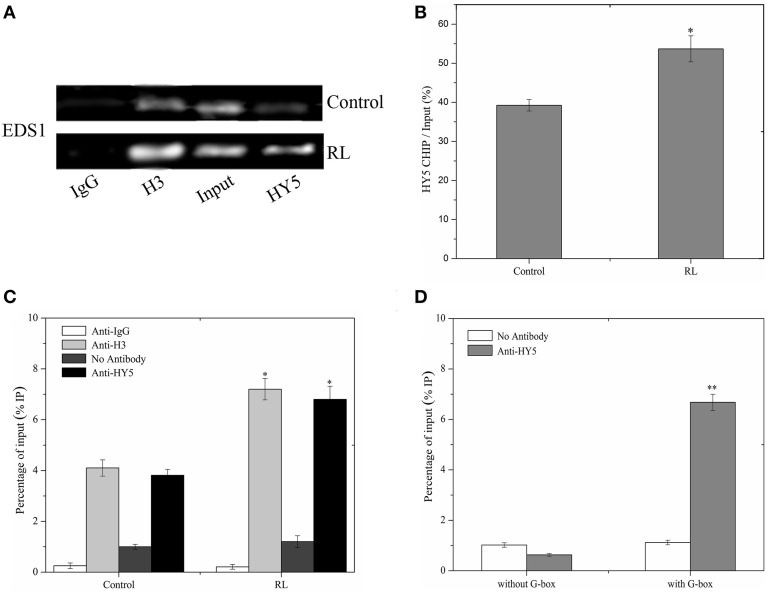
W This study showed that HY5 positively mediated the EDS1 transcript expression induced by RL (Figure 7C). HY5 acted as a positive transcription factor and directly bound a G-box (CACGTG) DNA sequence element in many light-regulated genes (Lee et al., 2007). It is interesting that promoter of EDS1 also contains G-box, which may be the binding region of HY5. To investigate whether HY5 could combine with the EDS1 promoter, a chromatin immunoprecipitation (ChIP) assay was performed followed by RT-PCR and analyses on hy5-ox-YFP transgenic plants with or without RL treatment. Our data showed that ChIP with either anti-acetyl-histone 3 or anti-HY5-YFP antibody detected the EDS1 promoter amplification products (Figure 7A, lane 2 and 4). However, almost no products were observed in ChIP with anti-rabbit IgG (Figure 7A, lane 1). As a control setup of ChIP efficiency, the input DNA samples without immunoprecipitation displayed a bright lane (Figure 7A, lane 3 and Figure 7B). ChIP–qPCR analysis also illustrated that HY5 could interact with G-box within the EDS1 promoter regardless of RL (Figure 7C). As shown in Figure 7D, HY5 strongly binds to the EDS1 promoter that encompasses the G-box motifs; by contrast, no interaction was observed between HY5 and EDS1 promoter without the G-box motifs. These results suggested that HY5 could bind to the EDS1 promoter and induce HY5 expression under RL.
Figure 7.
HY5 binds directly to the promoter of EDS1. (A) A ChIP assay was performed with specific antibodies for immunoprecipitation, and purified DNA samples were analyzed by RT-PCR assay. Immunoprecipitation was carried out with normal rabbit IgG (negative control, lane 1), anti-acetyl-histone H3 (positive control, lane 2), and anti-HY5-YFP serum (lane 4). As a control for detecting the efficiency of the ChIP assay, “input DNA” samples (lane3) without immunoprecipitation, were processed in parallel. (B) Quantitative analysis of EDS1 gene with anti HY5-YFP was shown in (B) with Image J software about (A). (C) ChIP-qPCR analysis of the EDS1 promoter sequence under normal and RL conditions. (D) ChIP-qPCR assays were performed to analyse the binding of HY5 protein to the G-box motifs of EDS1 promoter. Error bars indicate ±SD values. Statistical analysis was performed with Student's paired t-test: *P < 0.05, **P < 0.01 in compared with control.
Discussion
LSD1 and HY5 antagonistically regulate PCD associated with ROS and SA production triggered by RL
In this study, the lsd1-2 mutant displays an enhanced cell death, whereas the hy5-215 mutant shows a suppressed cell death in response to RL (Figure 1), as indicated by trypan blue staining and ion leakage measurement; this result suggested that LSD1 and HY5 serve as negative and positive regulators of RL-mediated PCD, respectively. RL also induces ROS release and SA accumulation. The lsd1-2 mutant also accumulates higher ROS and SA concentrations, whereas the hy5-215 mutant accumulates lower ROS and SA concentrations under RL; this result indicated that LSD1 and HY5 play different roles in regulating ROS and SA signaling in response to RL (Figure 2).
LSD1 prevents cell death and enhances light acclimation; LSD1 is also involved in modulating ROS and hormone signaling (Mateo et al., 2004; Muhlenbock et al., 2008). Light-dependent cell death is related to H2O2 and SA accumulation in the lsd1 mutant (Li et al., 2013). LSD1 positively modulates superoxide dismutase and CAT gene expression and CAT activities; therefore, LSD1 controls cellular ROS production (Kliebenstein et al., 1999; Mateo et al., 2004; Li et al., 2013). LSD1 negatively regulates RL-induced PCD by controlling ROS/SA signaling. Under EL stress, the onsets of PCD in the lsd1 mutant partly depends on the redox status of the PQ pool, which is required for EDS1 (Muhlenbock et al., 2008). The data showed that the lsd1-2 mutant and WT pre-treated with DCMU partly inhibit PCD and ROS/SA production exposed to RL (Figures 1, 2). The change in the redox status of the PQ pool may act upstream of LSD1 to regulate PCD in response to RL.
HY5, as a transcription factor that binds to the promoter ROS-response genes, promotes ROS production and cell death during seedling greening under long-term light conditions (Chen et al., 2013). HY5 is also involved in modulating the signaling of various hormones, including auxin, cytokinin, brassinosteroids, and gibberellins (Vandenbussche et al., 2007; Alabadi and Blazquez, 2008; Shi et al., 2011). HY5 integrates ROS with SA signaling to regulate RL response (Figure 2). HY5 is downstream of phyB in light signaling pathways (Osterlund et al., 2000). The phyAphyB mutant reduces cell death and blocks ROS and SA accumulation (Griebel and Zeier, 2008). The phyB plays a crucial roles in red light signaling (Neff et al., 2000). phyB also positively mediates PCD and ROS and SA accumulation under RL (Figure 4). HY5 may promote RL-triggered PCD by phyB signaling pathway and act independent of the PQ pool in the chloroplast.
LSD1 and HY5 had an antagonistic effect on SA accumulation by regulating SA-related gene expression in response to RL. It was observed that RL induced the expression of EDS1, non-expressor of pathogenesis-relatedgenes1 (NPR1) and isochorismate synthase1 (ICS1), but didn't influence the expression of phytoalexin deficient4 (PAD4) and phenylalanine ammonia-lyase (PAL). Meanwhile, EDS1 expression was only and antagonistically controlled by LSD1 and HY5 (Figure 6D and Supplementary Figure 5), which indicated that EDS1 played a key role in the regulation of RL-dependent SA signaling pathways.
RL prevents LSD1 activity by changing the LSD1 conformation
LSD1 that contains three zinc finger domains acts as a redox-sensor protein (Dietrich et al., 1997). LSD1 and Hsp33 also contain similar amino acid sequences of zinc finger domain, which is C-x-x-C zinc finger type. This sequence is sensitive to oxidized conditions. A previous study confirmed that Hsp33 releases zinc ion from the zinc finger domain, and the inactive monomer is converted into the active dimer under oxidizing conditions (Graf and Jakob, 2002). In our study, RL resulted in a more oxidized GSH pool in the cytoplasm by disrupting the PQ pool. Thus, RL caused a more oxidized cellular environment. Subsequently, the reduced monomeric LSD1 may transform into the oxidized dimeric LSD1 by removing zinc; the active LSD1 was converted into the inactive LSD1, as indicated by the CAT activity (Figure 3). The active LSD1 interacts with CATs (CAT1, CAT2, and CAT3) and positively controls CAT activity (Li et al., 2013). Therefore, LSD1 activity may be dependent on monomer conformation and complete zinc finger, which interact with other proteins.
Although LSD1 and HY5 play different roles in the PCD process in response to RL, these factors did not influence each other in terms of expression and protein levels (Supplementary Figure 2). The LSD1 protein contains three zinc finger domains and HY5 is a bZIP family protein (Oyama et al., 1997). AtbZIP10 interacts with LSD1 in the cytoplasm and suppresses AtbZIP10 activity by blocking the AtbZIP10 translocation to the nucleus (Kaminaka et al., 2006). To determine whether LSD1 interacted with HY5, Co-immunoprecipitation assays showed that LSD1 did not interact with HY5 (Supplementary Figure 2A). The localization of subcelluar LSD1 remained unchanged under RL treatment, and HY5 was only localized in the nucleus (Supplementary Figures 2E,F). Considering that LSD1 is not directly related to HY5, we investigated LSD1 conformation and activity in the hy5-215 mutant exposed to RL. The result indicated that HY5 partially changed LSD1 protein conformation and repressed activity (Figure 4). The PQ pool change combined with HY5 modulated ROS levels and disrupted the cellular redox status, resulting in changes in the LSD1 structure and activity in RL-induced PCD. However, the function of RL-induced PQ pool change was more important than that of HY5.
LSD1 suppresses EDS1 expression by upregulating SR1 expression
LSD1 negatively modulates EDS1-dependent cell death under long-term light, UVC, cold stress, and pathogen stress exposure (Muhlenbock et al., 2008; Huang et al., 2010; Wituszynska et al., 2013, 2015). The eds1 mutant displayed an inhibition of cell death and a decrease in ROS/SA production after RL treatment; this result indicated that EDS1 is required for PCD in response to RL (Figures 6A–C and Supplementary Figure 3). The EDS1 expression was significantly increased in response to RL (Figure 6D). Previous data showed that EDS1 expression levels are increased in the lsd1 mutant (Wituszynska et al., 2013). Our data implied that LSD1 could suppress EDS1 expression under RL condition. SR1 also directly combines with the EDS1 promoter and inhibits the EDS1 expression (Du et al., 2009). The sr1 mutant showed an enhanced EDS1 expression and exhibited a remarkable increase in cell death, as well as higher ROS/SA concentrations. This result was similar to that observed in the lsd1-2 mutant exposed RL (Figures 1, 6 and Supplementary Figure 3). LSD1 repressed the EDS1 expression by upregulating the SR1 expression. The LSD1 protein may act as a transcription regulator to modulate the SR1 gene expression; however, the underlying molecular mechanism requires further research. This study showed that the SR1 expression was dependent on the change in PQ pool exposed to RL. Thus, RL may repress LSD1 activity and inhibit SR1 expression by altering the redox status of the PQ pool.
HY5 interacts with phyB and promotes EDS1 expression during RL-induced PCD
phyB specifically functions in response to RL and enhances plant defense under RL (Zhao et al., 2014). Under RL, the inactive phyB turns into a photo-active phyB and is transferred from the cytoplasm to the nucleus (Nagatani, 2004). The interaction between phyB and COP1 rapidly inhibits the COP1 activity under RL (Luo et al., 2014). In darkness, HY5 interacts with COP1 and is degraded via the 26S proteasome pathway. However, RL promoted HY5 accumulation (Osterlund et al., 2000). The HY5 protein was decreased in the phyB mutant, but the HY5 protein was increased in the phyB-ox-YFP transgenic plant under RL treatment (Figure 4F). Meanwhile, it was found that phyB more significantly interacted with HY5 under RL condition, and the effect may be because of the increase of HY5 protein (Figure 5B). This process possibly involves the activation of phyB induced by RL, and the activated phyB may interact with HY5 and promoted HY5 accumulation by inhibiting COP1 activity.
The genes related to the SA-dependent defense pathway and the EDS1 expression were downregulated in the phyAphyBphyC mutant (Xie et al., 2011). The results indicated that phyB and HY5 positively controlled the EDS1 expression and induced PCD under RL (Figure 6D and Supplementary Figure 4). HY5 serves as a transcription factor, which combines with DNA sequence (G-box); thus, ROS-relative gene was regulated (Lee et al., 2007). The results showed that HY5 could more evidently bind to the G-box motif of the EDS1 gene promoter and positively regulate EDS1 expression under RL (Figures 6D, 8). Due to RL-induced the accumulation of HY5, the abundance of HY5 might increase the chance to bind EDS1. Hence, phyB-mediated RL signaling is dependent on ROS and SA signaling modulated by HY5.
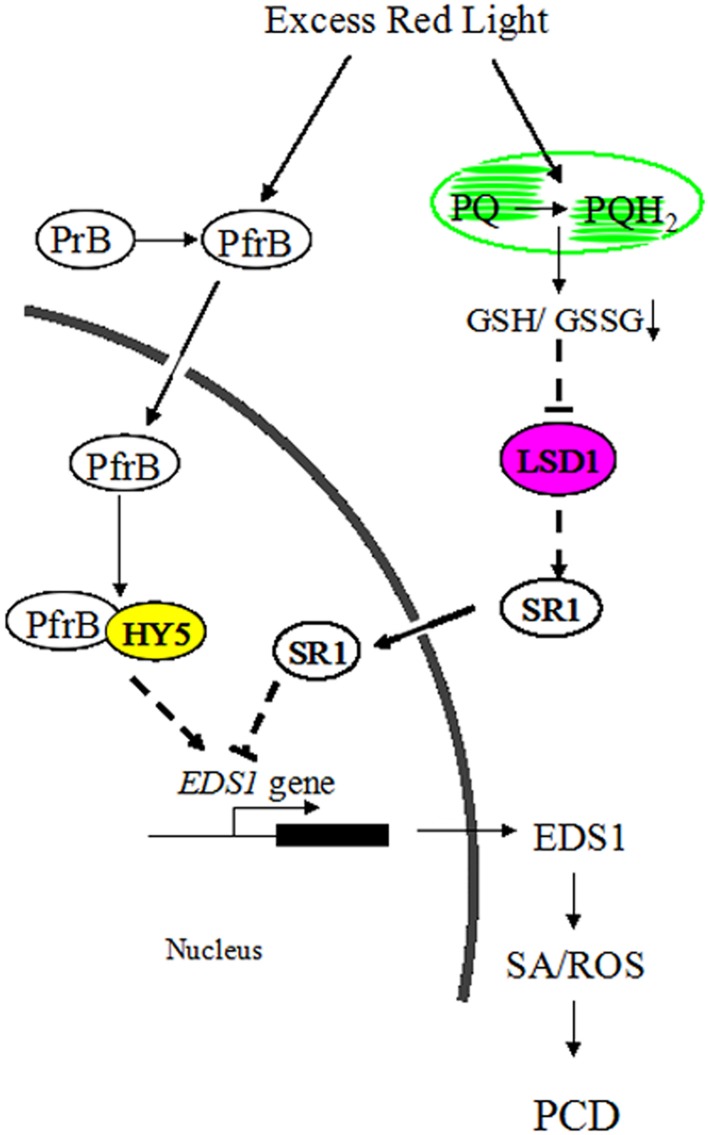
Figure 8.
A possible model for the roles of LSD1 and HY5 in antagonistically regulating PCD induced by RL inArabidopsis.
Although ROS signaling could be integrated into SA signaling to regulate adaptive responses in plants, components connecting ROS with SA pathways remain unknown (Mittler et al., 2011). In this study, light signaling could interact with ROS and SA signaling, and LSD1 and HY5 represent a key convergence point between these signaling pathways. RL resulted in a more oxidized environment by altering PQ pool, where the active monomer of LSD1 was converted into the inactive dimer of LSD1. However, the inactive LSD1 did not contribute to the SR1 expression and relieved inhibition of the EDS1 expression. RL also positively modulated HY5 by activating phyB. Subsequently, HY5 interacted with phyB and enhanced the EDS1 expression by binding to the G-box of the EDS1 promoter. As a result, plants induced ROS release, SA accumulation, and cell death in response to RL (Figure 8). Indeed, the molecular mechanism of RL signaling with other cellular pathways provides novel insights into these resulting processes.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Petra Epple for providing Arabidopsislsd1-2 mutant seeds and Prof. Xingwang Deng for kindly providing anti-HY5 antibody and seeds of Arabidopsishy5-215 mutant. We thank Prof. Roman Ulma for providing Arabidopsishy5-ox-YFP seeds, thank Prof. Hongquan Yangfor providing ArabidopsisphyB-ox-YFP seeds, and thank Prof. Mike Thomashow for providing Arabidopsissr1 mutant seeds. This research is supported by the Programs for Changjiang Scholars and Innovative Research Team in University (IRT0829), the Key Program of NSFC-Guangdong Joint Funds of China (U0931005), and the National High Technology Research and Development Program of China (863 Program) (2007AA10Z204).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00292/abstract
RL-induced change of cellular redox status.
LSD1 and HY5 don't influence on each other under RL.
EDS1 and SR1 influence on ROS and SA production in response to RL.
The phyB positively regulates EDS1 expression.
LSD1 and HY5 have different effects on SA-related genes expression under RL.
References
- Abbas N., Maurya J. P., Senapati D., Gangappa S. N., Chattopadhyay S. (2014). Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell 26, 1036–1052. 10.1105/tpc.113.122515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alabadi D., Blazquez M. A. (2008). Integration of light and hormone signals. Plant Signal. Behav. 3, 448–449. 10.4161/psb.3.7.5558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. (1999). THE WATER-WATER CYCLE IN CHLOROPLASTS: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 601–639. 10.1146/annurev.arplant.50.1.601 [DOI] [PubMed] [Google Scholar]
- Busch F. A. (2014). Opinion: the red-light response of stomatal movement is sensed by the redox state of the photosynthetic electron transport chain. Photosyn. Res. 119, 131–140. 10.1007/s11120-013-9805-6 [DOI] [PubMed] [Google Scholar]
- Chen D., Xu G., Tang W., Jing Y., Ji Q., Fei Z., et al. (2013). Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25, 1657–1673. 10.1105/tpc.112.104869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll N. S., Vercammen D., Smidler A., Clover C., Van Breusegem F., Dangl J. L., et al. (2010). Arabidopsis type I metacaspases control cell death. Science 330, 1393–1397. 10.1126/science.1194980 [DOI] [PubMed] [Google Scholar]
- Derong Y., Yuanyuan Z., Dongguo L. (2005). Fast computation of multiscale morphological operations for local contrast enhancement. Conf. Proc. IEEE Eng. Med. Biol. Soc. 3, 3090–3092. 10.1109/IEMBS.2005.1617128 [DOI] [PubMed] [Google Scholar]
- Dietrich R. A., Richberg M. H., Schmidt R., Dean C., Dangl J. L. (1997). A novel zinc finger protein is encoded by the Arabidopsis LSD1 gene and functions as a negative regulator of plant cell death. Cell 88, 685–694. 10.1016/S0092-8674(00)81911-X [DOI] [PubMed] [Google Scholar]
- Du L., Ali G. S., Simons K. A., Hou J., Yang T., Reddy A. S., et al. (2009). Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. 10.1038/nature07612 [DOI] [PubMed] [Google Scholar]
- Franklin K. A., Quail P. H. (2010). Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24. 10.1093/jxb/erp304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L., Spira A. P., Hernandez B., Mather C., Sheikh J., Ancoli-Israel S., et al. (2012). Brief morning light treatment for sleep/wake disturbances in older memory-impaired individuals and their caregivers. Sleep Med. 13, 546–549. 10.1016/j.sleep.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon Y., Nave R., Boyce J. M., Nachmias D., Knight M. R., Fromm H. (2008). Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 582, 943–948. 10.1016/j.febslet.2008.02.037 [DOI] [PubMed] [Google Scholar]
- Galon Y., Snir O., Fromm H. (2010). How calmodulin binding transcription activators (CAMTAs) mediate auxin responses. Plant Signal. Behav. 5, 1311–1314. 10.4161/psb.5.10.13158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li G. J., Wang S. B., Zhu H., Zhu T., Wang X., et al. (2007). AtNUDT7, anegative regulator of basal immunity in Arabidopsis, modulates twodistinct defense response pathways and is involved in maintaining redoxhomeostasis. Plant Physiol. 145, 204–215. 10.1104/pp.107.103374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev T. S., Van Breusegem F., Stone J. M., Denev I., Laloi C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28, 1091–1101. 10.1002/bies.20493 [DOI] [PubMed] [Google Scholar]
- Graf P. C., Jakob U. (2002). Redox-regulated molecular chaperones. Cell. Mol. Life Sci. 59, 1624–1631. 10.1007/PL00012489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griebel T., Zeier J. (2008). Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 147, 790–801. 10.1104/pp.108.119503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S., Tan G., Liu Q., Huang K., Ren J., Zhang X., et al. (2011). The LSD1-interacting protein GILP is a LITAF domain protein that negatively regulates hypersensitive cell death in Arabidopsis. PLoS ONE 6:e18750. 10.1371/journal.pone.0018750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Li Y., Zhang X., Zuo J., Yang S. (2010). The Arabidopsis LSD1 gene plays an important role in the regulation of low temperature-dependent cell death. New Phytol. 187, 301–312. 10.1111/j.1469-8137.2010.03275.x [DOI] [PubMed] [Google Scholar]
- Hwang S. G., Lee H. C., Lee D. W., Kim Y. S., Joo W. H., Cho Y. K., et al. (2001). Induction of apoptotic cell death by a p53-independent pathway in neuronal SK-N-MC cells after treatment with 2,2′,5,5′-tetrachlorobiphenyl. Toxicology 165, 179–188. 10.1016/S0300-483X(01)00432-2 [DOI] [PubMed] [Google Scholar]
- Junming Z., Yuanyuan Z., Fang F., Weiming F., Ryan H., Jianmin Z., et al. (2014). Anterior corpus callosotomy combined with anterior temporal resection with amygdalohippocampectomy: outcome in a patient with congenital bilateral perisylvian syndrome. Turk. Neurosurg. 24, 70–74. 10.5137/1019-5149.JTN.6362-12.1 [DOI] [PubMed] [Google Scholar]
- Kaminaka H., Nake C., Epple P., Dittgen J., Schutze K., Chaban C., et al. (2006). bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 25, 4400–4411. 10.1038/sj.emboj.7601312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. J., Dietrich R. A., Martin A. C., Last R. L., Dangl J. L. (1999). LSD1 regulates salicylic acid induction of copper zinc superoxide dismutase in Arabidopsis thaliana. Mol. Plant Microbe Interact. 12, 1022–1026. 10.1094/MPMI.1999.12.11.1022 [DOI] [PubMed] [Google Scholar]
- Lee J., He K., Stolc V., Lee H., Figueroa P., Gao Y., et al. (2007). Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19, 731–749. 10.1105/tpc.106.047688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Li G., Wang H., Wang Deng X. (2011). Phytochrome signaling mechanisms. Arabidopsis Book 9, e0148. 10.1199/tab.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen L., Mu J., Zuo J. (2013). LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 163, 1059–1070. 10.1104/pp.113.225805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Yue H., Xing D. (2012). MAP Kinase 6-mediated activation of vacuolar processing enzyme modulates heat shock-induced programmed cell death in Arabidopsis. New Phytol. 195, 85–96. 10.1111/j.1469-8137.2012.04131.x [DOI] [PubMed] [Google Scholar]
- Luo Q., Lian H. L., He S. B., Li L., Jia K. P., Yang H. Q. (2014). COP1 and phyB Physically Interact with PIL1 to Regulate Its Stability and Photomorphogenic Development in Arabidopsis. Plant Cell 26, 2441–2456. 10.1105/tpc.113.121657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackintosh F. C., Zhu J. X., Pine D. J., Weitz D. A. (1989). Polarization memory of multiply scattered light. Phys. Rev. B Condens. Matter 40, 9342–9345. 10.1103/PhysRevB.40.9342 [DOI] [PubMed] [Google Scholar]
- Maret W. (2006). Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid. Redox Signal. 8, 1419–1441. 10.1089/ars.2006.8.1419 [DOI] [PubMed] [Google Scholar]
- Mateo A., Muhlenbock P., Rusterucci C., Chang C. C., Miszalski Z., Karpinska B., et al. (2004). LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136, 2818–2830. 10.1104/pp.104.043646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel A., Levine A. (2001). Induction of cell death in arabidopsis by superoxide in combination with salicylic acid or with protein synthesis inhibitors. Free Radic. Biol. Med. 30, 98–106. 10.1016/S0891-5849(00)00452-4 [DOI] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Suzuki N., Miller G., Tognetti V. B., Vandepoele K., et al. (2011). ROS signaling: The new wave? Trends Plant Sci. 16, 300–309. 10.1016/j.tplants.2011.03.007 [DOI] [PubMed] [Google Scholar]
- Muhlenbock P., Szechynska-Hebda M., Plaszczyca M., Baudo M., Mateo A., Mullineaux P. M., et al. (2008). Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20, 2339–2356. 10.1105/tpc.108.059618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7, 708–711. 10.1016/j.pbi.2004.09.010 [DOI] [PubMed] [Google Scholar]
- Neff M. M., Fankhauser C., Chory J. (2000). Light: an indicator of time and place. Genes Dev. 14, 257–271. 10.1101/gad.14.3.257 [DOI] [PubMed] [Google Scholar]
- Oravecz A., Baumann A., Mate Z., Brzezinska A., Molinier J., Oakeley E. J., et al. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18, 1975–1990. 10.1105/tpc.105.040097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M. T., Hardtke C. S., Wei N., Deng X. W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. 10.1038/35013076 [DOI] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. 10.1101/gad.11.22.2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusterucci C., Aviv D. H., Holt B. F., III, Dangl J. L., Parker J. E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13, 2211–2224. 10.1105/tpc.13.10.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q. M., Yang X., Song L., Xue H. W. (2011). Arabidopsis MSBP1 is activated by HY5 and HYH and is involved in photomorphogenesis and brassinosteroid sensitivity regulation. Mol. Plant 4, 1092–1104. 10.1093/mp/ssr049 [DOI] [PubMed] [Google Scholar]
- Strasser B., Sanchez-Lamas M., Yanovsky M. J., Casal J. J., Cerdan P. D. (2010). Arabidopsis thaliana life without phytochromes. Proc. Natl. Acad. Sci. U.S.A. 107, 4776–4781. 10.1073/pnas.0910446107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szechynska-Hebda M., Kruk J., Gorecka M., Karpinska B., Karpinski S. (2010). Evidence for light wavelength-specific photoelectrophysiological signaling and memory of excess light episodes in Arabidopsis. Plant Cell 22, 2201–2218. 10.1105/tpc.109.069302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao L. I., Su T. P. (2001). Hibernation-induction peptide and cell death: [D-Ala2,D-Leu5]enkephalin blocks Bax-related apoptotic processes. Eur. J. Pharmacol. 428, 149–151. 10.1016/S0014-2999(01)01346-2 [DOI] [PubMed] [Google Scholar]
- Ulm R., Baumann A., Oravecz A., Mate Z., Adam E., Oakeley E. J., et al. (2004). Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 1397–1402. 10.1073/pnas.0308044100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungureanu M., Zazpe R., Golmar F., Stoliar P., Llopis R., Casanova F., et al. (2012). A light-controlled resistive switching memory. Adv. Mater. Weinheim. 24, 2496–2500. 10.1002/adma.201200382 [DOI] [PubMed] [Google Scholar]
- Vandenbussche F., Habricot Y., Condiff A. S., Maldiney R., Van Der Straeten D., Ahmad M. (2007). HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 49, 428–441. 10.1111/j.1365-313X.2006.02973.x [DOI] [PubMed] [Google Scholar]
- Vandewalle G., Gais S., Schabus M., Balteau E., Carrier J., Darsaud A., et al. (2007). Wavelength-dependent modulation of brain responses to a working memory task by daytime light exposure. Cereb. Cortex 17, 2788–2795. 10.1093/cercor/bhm007 [DOI] [PubMed] [Google Scholar]
- Vani J., Justin J., Nagasuma R. C., Nayak R., Shaila M. S. (2007). Peptidomimics of antigen are present in variable region of heavy and light chains of anti-idiotypic antibody and function as surrogate antigen for perpetuation of immunological memory. Mol. Immunol. 44, 3345–3354. 10.1016/j.molimm.2007.02.015 [DOI] [PubMed] [Google Scholar]
- Wang F. F., Lian H. L., Kang C. Y., Yang H. Q. (2010). Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana. Mol. Plant 3, 246–259. 10.1093/mp/ssp097 [DOI] [PubMed] [Google Scholar]
- Wang L., Tsuda K., Truman W., Sato M., Nguyen Le V., Katagiri F., et al. (2011). CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67, 1029–1041. 10.1111/j.1365-313X.2011.04655.x [DOI] [PubMed] [Google Scholar]
- Wituszynska W., Slesak I., Vanderauwera S., Szechynska-Hebda M., Kornas A., Van Der Kelen K., et al. (2013). Lesion simulating disease1, enhanced disease susceptibility1, and phytoalexin deficient4 conditionally regulate cellular signaling homeostasis, photosynthesis, water use efficiency, and seed yield in Arabidopsis. Plant Physiol. 161, 1795–1805. 10.1104/pp.112.208116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wituszynska W., Szechynska-Hebda M., Sobczak M., Rusaczonek A., Kozlowska-Makulska A., Witon D., et al. (2015). LESION SIMULATING DISEASE 1 and ENHANCED DISEASE SUSCEPTIBILITY 1 differentially regulate UV-C-induced photooxidative stress signalling and programmed cell death in Arabidopsis thaliana. Plant Cell Environ. 38, 315–330. 10.1111/pce.12288 [DOI] [PubMed] [Google Scholar]
- Xie X. Z., Xue Y. J., Zhou J. J., Zhang B., Chang H., Takano M. (2011). Phytochromes regulate SA and JA signaling pathways in rice and are required for developmentally controlled resistance to Magnaporthe grisea. Mol. Plant 4, 688–696. 10.1093/mp/ssr005 [DOI] [PubMed] [Google Scholar]
- Xing F., Li Z., Sun A., Xing D. (2013). Reactive oxygen species promote chloroplast dysfunction and salicylic acid accumulation in fumonisin B1-induced cell death. FEBS Lett. 587, 2164–2172. 10.1016/j.febslet.2013.05.034 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhou J., Xing D. (2014). Phytochrome B-mediated activation of lipoxygenase modulates an excess red light-induced defence response in Arabidopsis. J. Exp. Bot. 65, 4907–4918. 10.1093/jxb/eru247 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RL-induced change of cellular redox status.
LSD1 and HY5 don't influence on each other under RL.
EDS1 and SR1 influence on ROS and SA production in response to RL.
The phyB positively regulates EDS1 expression.
LSD1 and HY5 have different effects on SA-related genes expression under RL.