Abstract
Plant cells biosynthesize primary cell walls (PCW) in all cells and produce secondary cell walls (SCWs) in specific cell types that conduct water and/or provide mechanical support, such as xylem vessels and fibers. The characteristic mechanical stiffness, chemical recalcitrance, and hydrophobic nature of SCWs result from the organization of SCW-specific biopolymers, i.e., highly ordered cellulose, hemicellulose, and lignin. Synthesis of these SCW-specific biopolymers requires SCW-specific enzymes that are regulated by SCW-specific transcription factors. In this review, we summarize our current knowledge of the transcriptional regulation of SCW formation in plant cells. Advances in research on SCW biosynthesis during the past decade have expanded our understanding of the transcriptional regulation of SCW formation, particularly the functions of the NAC and MYB transcription factors. Focusing on the NAC-MYB-based transcriptional network, we discuss the regulatory systems that evolved in land plants to modify the cell wall to serve as a key component of structures that conduct water and provide mechanical support.
Keywords: land plant evolution, MYB transcription factor, NAC transcription factor, secondary cell wall, network
Introduction
The cell wall, a characteristic feature of plant cells, consists of biopolymers, such as polysaccharides, phenolic compounds, and various proteins, which impart mechanical strength and rigidity. The structure of the cell wall determines the characteristics of plant cells, thus directly affecting organ development and responses to environmental stimuli (Hamant and Traas, 2010; Wolf et al., 2012).
Plant cells have two types of cell wall, primary cell wall (PCW) and secondary cell wall (SCW), based on their biosynthetic composition and cellular location (Figure 1). Every plant cell has a PCW, a relatively thin and extensible wall that the cell synthesizes during cell division. The force generated by the PCW functions as a critical regulator of cell elongation and expansion; thus, PCW biosynthesis fundamentally conditions the shape and size of cells (Geitmann, 2010; Hamant and Traas, 2010). By contrast, the relatively thick and rigid SCW forms in specific types of cells, such as xylem cells, and cells of valve margin and anther endothecium. The cell produces the SCW between the PCW and the plasma membrane during cell differentiation, and the SCW imparts additional mechanical stiffness and/or hydrophobicity to the cell (Cosgrove and Jarvis, 2012). Cellulose, also called (1,4)-β-d-glucan, contains >500 β-d-glucose residues polymerized with glycosidic bonds into a chain; cellulose microfibrils contain ~40 cellulose chains formed into bundles. Cellulose constitutes the main component of the PCW and SCW, but the cellulose of the PCW and SCW shows key structural differences. In the PCW, cellulose has a relatively low degree of polymerization (e.g., 2000–6000 β-d-glucose residues in cotton, Gossypium hirsutum) and microfibril widths of 2–2.5 nm. By contrast, in the SCW, cellulose has a high degree of polymerization (e.g., 13,000 β-d-glucose residues in cotton and approximately 8000 in wood; Marx-Figini, 1969) and microfibril widths of 5–10 nm (Heyn, 1955, 1965, 1966). Other components of the cell wall also differ between the PCW and SCW. For example, the PCW typically contains xyloglucan as the major hemicellulose (i.e., the polysaccharide component that is soluble in alkali), but the SCW contains xylan. In addition, the PCW is rich in gel-like pectin, whereas lignin and specific phenolic polymers are more abundant in the SCW (Figure 1). These differences in cell wall composition impart different physical properties to the cell wall, rendering the PCW flexible and the SCW mechanically and biologically robust.
Figure 1.
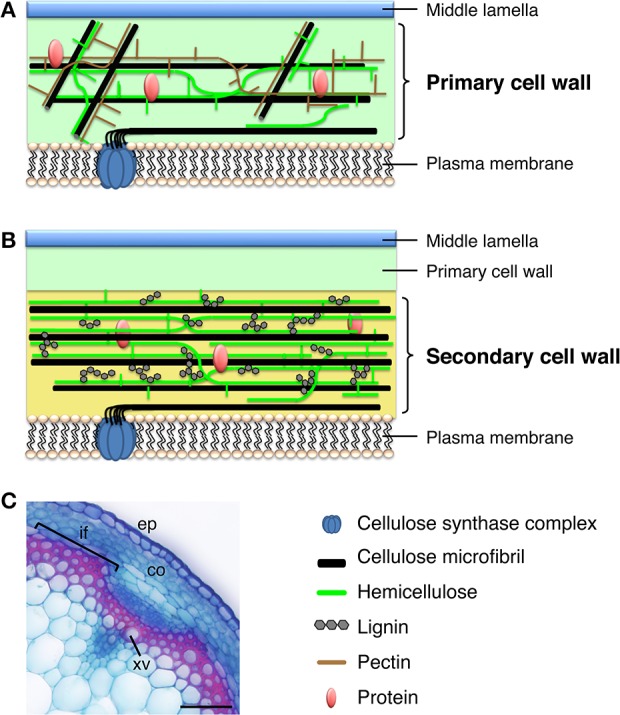
Plant cell walls. (A) Model of the primary cell wall. Cellulose microfibrils in the primary cell wall are relatively short and thin, compared with those in the secondary cell wall, and hemicellulose in the primary cell wall is composed of xyloglucan. The primary cell wall is rich in pectin. (B) Model of the secondary cell wall, which is deposited between the primary cell wall and the plasma membrane. The secondary cell wall mainly contains relatively long and thick cellulose microfibrils, hemicellulosic xylan, and lignin. (C) Cross section of an Arabidopsis inflorescence stem stained with Safranin, which stains lignin red, and Astra blue. co, cortex; ep, epidermis; if, interfascicular fiber; xv, xylem vessel. Bar = 50 μm.
The differences in cell wall components between the SCW and PCW suggest that plants have a set of SCW-specific biosynthetic genes. Indeed, transcriptome analysis of xylem tissues in tree species identified many genes thought to be involved in the biosynthesis of SCW-specific polymers during xylem development in loblolly pine (Pinus taeda, Allona et al., 1998; Lorenz and Dean, 2002), poplar (Populus, spp., Sterky et al., 2004), white spruce (Picea glauca, Pavy et al., 2005), and eucalyptus (Eucalyptus gunnii, Rengel et al., 2009). A series of molecular genetic studies on Arabidopsis thaliana irregular xylem (irx) mutants, in which xylem cells are disrupted due to stunted SCW formation (Turner and Somerville, 1997), expanded our knowledge of SCW-specific enzymes. For example, functional molecular and co-expression analysis of IRX genes identified SCW-specific cellulose synthase subunit A (CesA) genes (IRX1/CesA8, IRX3/CesA7, and IRX5/CesA4), SCW-specific hemicellulose biosynthetic genes (IRX7, IRX8, IRX9, IRX10, IRX14, and IRX15), and lignin biosynthetic genes (IRX4 and IRX12) (Turner and Somerville, 1997; Jones et al., 2001; Brown et al., 2005, 2009, 2011; Lee et al., 2007; Peña et al., 2007; Wu et al., 2009; Jensen et al., 2011). These findings also suggested that the upregulation of SCW-specific enzyme genes promotes SCW formation, and that SCW formation requires this SCW-specific transcriptional regulatory system.
In 2005, a milestone year for SCW biosynthesis research, multiple studies identified transcriptional regulators of SCW biosynthesis. For example, Kubo et al. used an in vitro cell culture system to identify the plant-specific NAM, ATAF1,2, and CUC2 (NAC) transcription factors VASCULAR-RELATED NAC-DOMAIN1-7 (VND1-7) as master regulators of xylem vessel cell differentiation (Kubo et al., 2005). Also, Mitsuda et al. reported that NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1) and NST2, members of a sister group to the VNDs, regulate SCW formation in anther cells (Mitsuda et al., 2005). Subsequent work showed that NST1 and NST3 (also called SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN1 [SND1]) function as master switches of fiber cell differentiation in Arabidopsis (Zhong et al., 2006; Mitsuda et al., 2007). These findings revealed the regulation of SCW biosynthesis at the molecular level, showing that plants have specific transcriptional switches that regulate SCW biosynthesis, and these factors belong to the NAC family, including VND and NST transcription factors (Yamaguchi and Demura, 2010; Zhong et al., 2010a; Wang and Dixon, 2011; Hussey et al., 2013). After these reports on NAC proteins, additional reports implicated several MYB-type transcription factors as secondary master regulators of SCW formation (McCarthy et al., 2009; Ko et al., 2012, 2014; Zhong and Ye, 2012; Hussey et al., 2013), and proposed an intricate network of transcription factors that regulate SCW formation in Arabidopsis (Figure 2).
Figure 2.
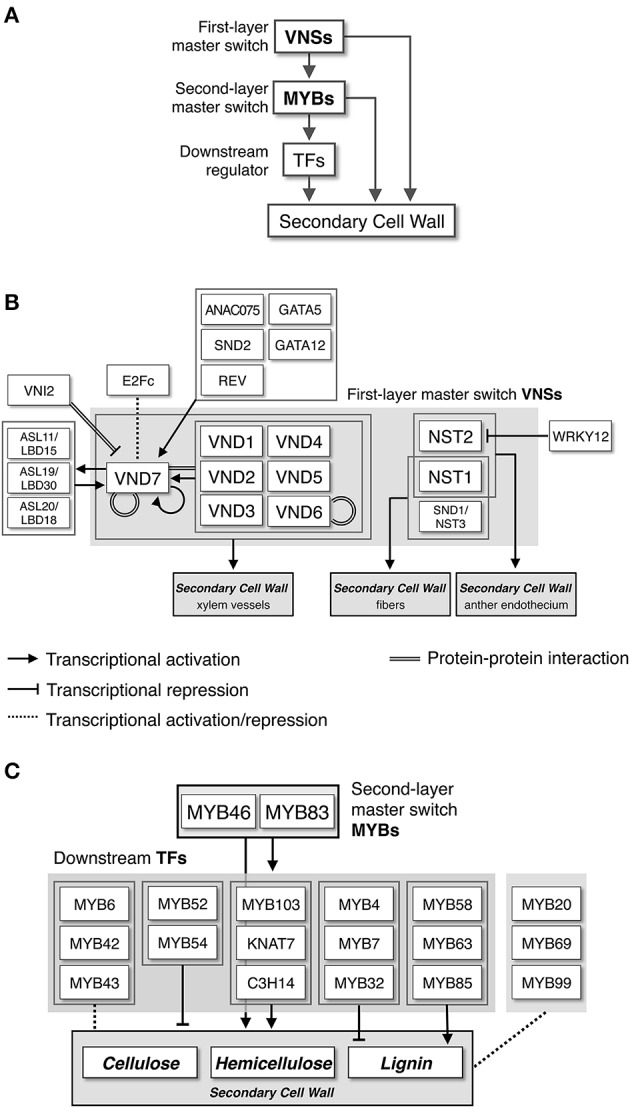
Transcriptional network regulating secondary cell wall formation. (A) NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis. Some metabolic genes for secondary cell wall biosynthesis are targeted by both NACs and MYBs, producing “feed-forward” regulation. (B) Transcriptional regulatory network around VNS proteins, first-layer master switches for secondary cell wall formation, based on work in Arabidopsis. (C) Transcriptional regulatory network around MYB proteins, second-layer master switches for secondary cell wall formation, based on work in Arabidopsis.
In this review, we describe the transcriptional regulation of SCW formation based on information accumulated in the decade since 2005, focusing on the well-studied NAC and MYB transcription factors. An analysis of the NAC-MYB-based transcriptional regulatory system of the SCW reveals clues to how plant cells modify cell wall biosynthesis to conduct water (xylem vessels) and/or provide support (fibers).
The function of NAC (NAM, ATAF1,2 and CUC2) proteins in SCW formation
VNS (VND, NST/SND, and SMB related) proteins function as master regulators of SCW formation
Members of the NAC domain transcription factor family have a highly conserved N-terminal NAC domain, which has been implicated in nuclear localization, DNA binding, and homo- and/or heterodimer formation with other NAC domain proteins (Olsen et al., 2005). NAC transcription factors consist of a large gene family with more than 100 members in A. thaliana (Ooka et al., 2003), Oryza sativa (Ooka et al., 2003; Nuruzzaman et al., 2010), Glycine max (Le et al., 2011), Populus trichocarpa (Hu et al., 2010), and Eucalyptus grandis (Hussey et al., in press). The NAC proteins have been reported to participate in many developmental processes (Krizek and Fletcher, 2005; Olsen et al., 2005; Petricka et al., 2012), including SCW formation (Yamaguchi and Demura, 2010) and biotic and abiotic stress responses (Fang et al., 2008; Nakashima et al., 2012; Puranik et al., 2012).
The first clear indication of NAC protein function in SCW formation came from studies of in vitro transdifferentiation of tracheary elements using Zinnia elegans mesophyll cells. Demura and co-workers found that expression of the NAC domain transcription factor Z567 increased during transdifferentiation (Demura et al., 2002). They further established an in vitro system for xylem vessel cell differentiation with Arabidopsis suspension culture cells, and showed that expression of seven NAC transcription factors with high sequence similarity to Z567 also increased, beginning in the early stages of cell differentiation (Kubo et al., 2005). They named these proteins VASCULAR-RELATED NAC-DOMAIN1 (VND1) to VND7 (Figure 3A, Table 1). All the VND genes are preferentially expressed in developing vascular tissues, although their expression patterns differ; promoter analysis suggested that VND7 regulates all types of xylem vessels in roots and shoots, whereas the other VND proteins might differentially regulate vessel formation (Kubo et al., 2005; Yamaguchi et al., 2008). Overexpression of VND genes induces ectopic deposition of patterned SCW, which is characteristic of xylem vessel cells (Kubo et al., 2005; Zhou et al., 2014; Endo et al., 2015; Figures 3B–G). Conversely, overexpression of a dominant chimeric repressor constructed by fusing VND6 or VND7 to the SRDX transcriptional repression domain, severely inhibited xylem vessel cell differentiation (Kubo et al., 2005; Reusche et al., 2012). Together, these findings indicate that the VND proteins act as master regulators of xylem vessel cell differentiation.
Figure 3.
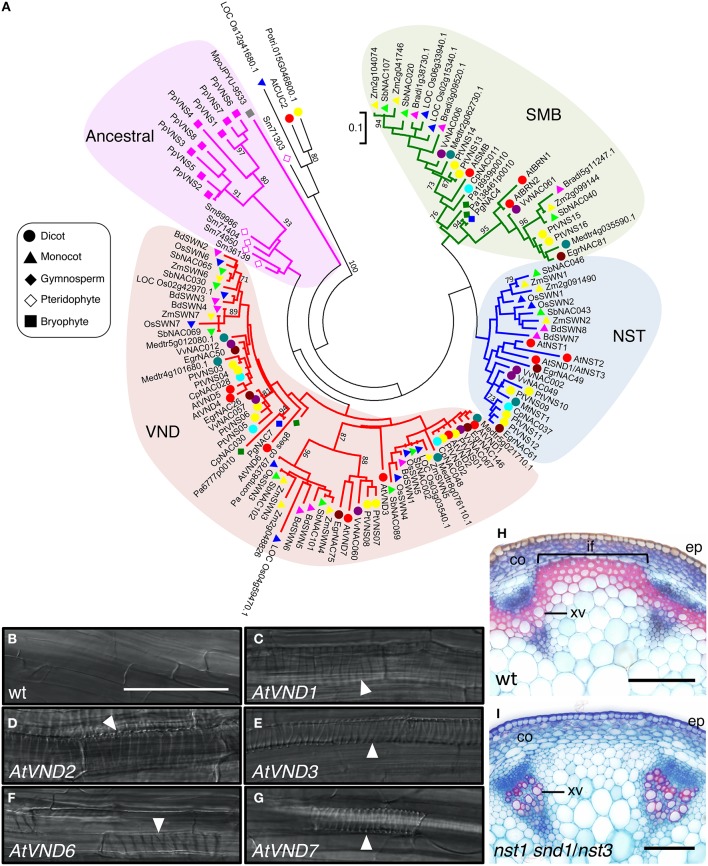
The VNS genes function as first-layer master switches for secondary cell wall formation. (A) Phylogenetic tree of VNS proteins. The unrooted phylogenetic tree was constructed with amino acid sequences of the NAC domain (sequences provided in Table S1) by the maximum-likelihood method. Numbers indicate bootstrap values for the clades that received support values of over 70% (1000 resamplings). Scale (0.1) represents a 10% change in sequences. Based on the tree, the VNS proteins are classified into four groups, VND, NST/SND, SMB, and Ancestral groups. (B–G) Seven-day-old Arabidopsis roots of wild type (wt, B) and transgenic plants, in which AtVND1 (C), AtVND2 (D), AtVND3 (E), AtVND6 (F), and AtVND7 (G) were overexpressed by an inducible system. Ectopic xylem elements formed in the transgenic roots (white arrowheads). Data were adapted from Endo et al. (2015). (H,I) Cross sections of Arabidopsis inflorescence stems stained with Safranin, which stains lignin red, and Astra blue. In the wild type (wt), both xylem vessel cells and interfascicular fiber cells have lignin-containing secondary cell wall, thus they stain red (H). By contrast, the mutant nst1 snd1/nst3 lacks secondary cell wall in interfascicular fiber cells, thus only xylem vessel cells were stained by Safranin (I), as described in Mitsuda et al. (2007). co, cortex; ep, epidermis; if, interfascicular fiber; xv, xylem vessel. Bars = 100 μm (B–I).
Table 1.
Known VNS genes.
| VNS GENES | References | |||
|---|---|---|---|---|
| VND | NST | SMB | Ancestral | |
| Arabidopsis thaliana | ||||
| AtVND1 | AtNST1 | AtBRN1 | – | Kubo et al., 2005 |
| AtVND2 | AtNST2 | AtBRN2 | Mitsuda et al., 2005, 2007 | |
| AtVND3 | AtNST3/AtSND1 | AtSMB | Zhong et al., 2006 | |
| AtVND4 | Bennett et al., 2010 | |||
| AtVND5 | ||||
| AtVND6 | ||||
| AtVND7 | ||||
| Carica papaya | ||||
| CpNAC028 | CpNAC037 | CpNAC011 | – | Zhu et al., 2012 |
| CpNAC030 | ||||
| CpNAC048 | ||||
| Populus trichocarpa | ||||
| PtVNS01/PtrWND5A/PtrVND6-C1 | PtVNS09/PtrWND2A/PtrSND1-B1 | PtVNS13/PtrSND1-L-1 | – | Zhong et al., 2010b |
| PtVNS02/PtrWND5B/PtrVND6-C2 | PtVNS10/PtrWND2BPtrSND1-B2 | PtVNS14/PtrSND1-L-2 | Ohtani et al., 2011 | |
| PtVNS03/PtrWND4A/PtrVND6-B2 | PtVNS11/PtrWND1B/PtrSND1-A2 | PtVNS15 | Li et al., 2012b | |
| PtVNS04/PtrWND4B/PtrVND6-B1 | PtVNS12/PtrWND1A/PtrSND1-A1 | PtVNS16 | ||
| PtVNS05/PtrWND3A/PtrVND6-A1 | ||||
| PtVNS06/PtrWND3B/PtrVND6-A2 | ||||
| PtVNS07/PtrWND6A/PtrVND7-2 | ||||
| PtVNS08/PtrWND6B/PtrVND7-1 | ||||
| Medicago truncatula | ||||
| Medtr4g101680.1 | MtNST1 | Medtr2g062730.1 | – | Zhao et al., 2010 |
| Medtr5g012080.1 | Medtr4g035590.1 | Phytozome v9.1 | ||
| Medtr5g021710.1 | (http://www.phytozome.net/) | |||
| Medtr8g076110.1 | ||||
| Eucalyptus grandis | ||||
| EgrNAC26 | EgrNAC49 | EgrNAC81 | - | Hussey et al., in press |
| EgrNAC50 | EgrNAC61 | |||
| EgrNAC75 | ||||
| EgrNAC146 | ||||
| Vitis vinifera | ||||
| VvNAC012 | VvNAC002 | VvNAC006 | – | Zhu et al., 2012 |
| VvNAC057 | VvNAC049 | VvNAC061 | ||
| VvNAC060 | ||||
| VvNAC067 | ||||
| Zea mays | ||||
| ZmSWN3 | ZmSWN1 | Zm2g041746 | – | Zhong et al., 2011 |
| ZmSWN4 | ZmSWN2 | Zm2g099144 | Zhu et al., 2012 | |
| ZmSWN5 | Zm2g091490 | Zm2g104074 | ||
| ZmSWN6 | ||||
| ZmSWN7 | ||||
| Zm2g048826 | ||||
| Sorghum bicolor | ||||
| SbNAC002 | SbNAC043 | SbNAC020 | – | Zhu et al., 2012 |
| SbNAC030 | SbNAC046 | SbNAC040 | ||
| SbNAC065 | SbNAC107 | |||
| SbNAC069 | ||||
| SbNAC089 | ||||
| SbNAC101 | ||||
| SbNAC102 | ||||
| Oryza sativa | ||||
| OsSWN3 | OsSWN1 | LOC_Os02g15340.1 | – | Zhong et al., 2011 |
| OsSWN4 | OsSWN2 | LOC_Os06g33940.1 | Zhu et al., 2012 | |
| OsSWN5 | ||||
| OsSWN6 | ||||
| OsSWN7 | ||||
| LOC_Os02g42970.1 | ||||
| LOC_Os03g03540.1 | ||||
| LOC_Os04g59470.1 | ||||
| Brachypodium distachyon | ||||
| BdSWN1 | BdSWN7 | Bradi1g38730.1 | – | Valdivia et al., 2013 |
| BdSWN2 | BdSWN8 | Bradi3g09520.1 | Phytozome v9.1 | |
| BdSWN3 | Bradi5g11247.1 | (htt://www.phytozome.net/) | ||
| BdSWN4 | ||||
| BdSWN5 | ||||
| BdSWN6 | ||||
| Picea abies | ||||
| Pa_comp83767_c0_seq8 | – | Pa138461p0010 | – | Nystedt et al., 2013 |
| Pa6777p0010 | Pa18939p0010 | |||
| Picea glauca | ||||
| PgNAC7 | – | PgNAC4 | – | Duval et al., 2014 |
| Selaginella moellendorffii | ||||
| – | – | – | Sm36139 | Xu et al., 2014 |
| Sm71404 | ||||
| Sm74950 | ||||
| Sm89986 | ||||
| Physcomitrella patens | ||||
| – | – | – | PpVNS1 | Xu et al., 2014 |
| PpVNS2 | ||||
| PpVNS3 | ||||
| PpVNS4 | ||||
| PpVNS5 | ||||
| PpVNS6 | ||||
| PpVNS7 | ||||
| PpVNS8 | ||||
| Marchantia polymorpha | ||||
| – | – | – | MpoJPYU-9533 | Xu et al., 2014 |
In A. thaliana, the VND-related proteins NAC SECONDARY WALL THICKENING PROMOTING FACTOR1 (NST1), NST2, and NST3/SECONDARY WALL-ASSOCIATED NAC DOMAIN PROTEIN 1 (SND1)/ARABIDOPSIS NAC DOMAIN CONTAINING PROTEIN012 (ANAC012) (Figure 3A, Table 1) regulate the differentiation of SCW-containing cells other than xylem vessel cells, such as anther endothecium (NST1 and NST2; Mitsuda et al., 2005), fiber cells (NST1 and NST3; Zhong et al., 2006, 2007b; Mitsuda et al., 2007; Figures 3H,I, Table 1), and silique cells (NST1 and NST3; Mitsuda and Ohme-Takagi, 2008). Moreover, other VND-related Arabidopsis proteins, namely SOMBRERO (SMB), BEARSKIN1 (BRN1), and BRN2, induce ectopic SCW deposition when overexpressed, although in wild type cells, they are expressed in root cap regions where SCW is not deposited (Willemsen et al., 2008; Bennett et al., 2010; Figure 3A, Table 1). These results indicate that the capacity to induce SCW biosynthesis is conserved among the VND, NST, SMB, and BRN proteins, and that these genes likely evolved from a common ancestral gene, acquiring the capacity to regulate wall modification during the differentiation of specific cell types.
The NAC protein subfamily, including VND, NST, SMB and BRN of Arabidopsis has been termed the VNS (VND, NST/SND, SMB related protein) family (Ohtani et al., 2011; Xu et al., 2014). The same subfamily was called “subfamily Ic” by Zhu et al. (2012), and is conserved among wide range of plant species, including non-vascular land plant species such as Bryophytes (Zhu et al., 2012; Xu et al., 2014; Figure 3A, Table 1).
VNS proteins are well-conserved among vascular plants
As shown in Table 1, many VNS proteins have been identified in land plant species. The VNS proteins in vascular plants can be classified into VND, NST, or SMB groups by phylogenetic analysis (Figure 3A), suggesting that the diversification of VNS genes occurred within the vascular plant lineage.
Black cottonwood (P. trichocarpa; Pt or Ptr in different studies), the first tree species with a high-quality, annotated genomic sequence (Tuskan et al., 2006), contains 16 PtVNS genes, including eight genes of the VND group, four genes of the NST group, and four genes of the SMB group (Zhong et al., 2010b; Ohtani et al., 2011; Li et al., 2012b; Figure 3A, Table 1). The PtVNS genes include PtrWND, PtrVND6, PtrVND7, and PtrSND1 and all PtVNS genes of the VND and NST groups are expressed in developing xylem and phloem fiber regions, although in primary vessels in the stem, only PtVNS genes of the VND group are expressed (Zhong et al., 2010b; Ohtani et al., 2011). The SMB-group PtVNS genes are not expressed in the xylem tissues, but rather are expressed in root tissues (Zhong et al., 2010b; Ohtani et al., 2011), indicating that the molecular functions of VNS genes in the SMB group in root tissue development might be conserved in Arabidopsis and poplar. Overexpression of PtVNS genes of the VND and NST groups in Arabidopsis and poplar caused ectopic deposition of SCW (Zhong et al., 2010b; Ohtani et al., 2011), and AtNST3 promoter-driven PtVNS genes can rescue fiber cell formation in nst1 snd1/nst3 double mutant stems (Zhong et al., 2010b; Figures 3H,I). Thus, the PtVNS proteins appear to possess the full potential to induce SCW biosynthesis.
Monocot VNS proteins have been also studied in terms of their expression patterns and molecular functions (Zhong et al., 2011; Valdivia et al., 2013; Yoshida et al., 2013). The members of the VND and NST groups in rice, maize (Zea mays), and Brachypodium distachyon are named SECONDARY WALL-ASSOCIATED NAC (SWN) proteins, and are expressed in SCW-forming cells such as xylem vessels, cortical fibers, and bundle sheath fibers (Zhong et al., 2011; Valdivia et al., 2013; Yoshida et al., 2013). Heterologous overexpression of the SWN genes can induce ectopic SCW deposition (Zhong et al., 2011; Valdivia et al., 2013), like the AtVNS and PtVNS genes; thus the SWN proteins are sufficient to promote the downstream events of SCW formation.
In addition to poplar and monocots, comparative genomics research has identified VNS genes in many plant species. The numbers of VNS gene vary by plant species without apparent correlation to genome size or the presence of woody tissues (Zhu et al., 2012; Figure 3A, Table 1). Recently, genome sequencing has been completed for other tree species, such as Picea abies (Nystedt et al., 2013) and E. grandis (Myburg et al., 2014). These tree species have only a few VNS genes: four in P. abies (Nystedt et al., 2013), six in E. grandis (Hussey et al., in press; Myburg et al., 2014), and two in P. glauca (Duval et al., 2014). It is noteworthy that no VNS genes of the NST group have been identified in gymnosperms at present (Figure 3A); thus, the NST group might have evolved within the angiosperm lineage, or lost in the gymnosperm lineage. The wood of gymnosperms is composed of single cell tracheids that function in water conduction and provide mechanical strength to the axis, whereas the wood of angiosperms is composed of xylem vessels and fibers, cells that are specialized for water conduction and providing mechanical strength, respectively (Pallady, 2008). Phylogenetic analysis suggests that gymnosperm tracheid cell differentiation could be regulated by the VND-type VNS genes (Figure 3A). Further studies on gymnosperm VNS genes will give insights into how woody cells developed during land plant evolution.
Target genes of VNS for SCW formation
As mentioned above, the overexpression of VNS genes induces ectopic SCW deposition (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2010a,b, 2011; Bennett et al., 2010; Yamaguchi et al., 2010a; Ohtani et al., 2011; Valdivia et al., 2013; Endo et al., 2015; Xu et al., 2014; Figures 3B–G). This observation demonstrates that VNS proteins likely share target genes to produce SCW. Indeed, the overexpression of AtVND and AtNST commonly upregulates the genes involved in the biosynthesis of SCW components, such as cellulose, hemicellulose and lignin (Kubo et al., 2005; Mitsuda et al., 2005, 2007; Zhong et al., 2006; Ko et al., 2007). However, xylem vessel cells and fiber cells differ in specific cell wall characteristics, such as the syringyl/guaiacyl (S/G) ratio of lignin subunits (Saito et al., 2012). In addition, expression of AtVND7 under the control of the AtSND1/NST3 promoter in the nst1 snd1/nst3 double mutant rescues the lack of SCW in mutant fiber cells (Mitsuda et al., 2007; Zhong et al., 2007b; Yamaguchi et al., 2011; Figures 3H,I). However, the SCW formed in the fibers differs from that produced by expression AtNST3; expression of AtVND7 causes formation of the patterned SCW characteristic of xylem vessel cells, even in the fiber cells (Yamaguchi et al., 2011). Accordingly, parts of the downstream SCW-related pathway appear to differ between AtVND and AtNST.
Around 2010, several groups independently identified the direct target genes of AtVND6, AtVND7, and AtSND1/NST3 (Ohashi-Ito et al., 2010; Zhong et al., 2010c; Yamaguchi et al., 2011). These studies did not identify identical sets of genes, reflecting the different genes and different experimental strategies, but the sets showed some overlap. First, common targets of AtVND and AtSND1/NST3 include transcription factors such as MYB and ASYMMETRIC LEAVES2-LIKE/LATERAL ORGAN BOUNDARIES DOMAIN (ASL/LBD) (for details, please see sections below; Figure 2), and the genes encoding enzymes involved in SCW formation, such as the IRX genes (Taylor et al., 2003). Second, the VND proteins preferentially target genes involved in programmed cell death (nucleases, proteases, and metacaspases), and signal transduction (receptor-like kinases). Additionally, even for common targets, the transcriptional activation activities of AtVND and AtSND1/NST3 sometimes differ (Ohashi-Ito et al., 2010; Zhong et al., 2010c; Yamaguchi et al., 2011). In silico and in vitro analyses of cis-elements targeted by VNS revealed the 11-bp tracheary element-regulating cis-elements (TEREs; Pyo et al., 2007) and the 19-bp secondary wall NAC-binding elements (SNBEs; Zhong et al., 2010c), which partly overlap. Both cis-elements are enriched in promoter regions of the genes directly regulated by AtVND and AtSND1/NST3 (Pyo et al., 2007; Ohashi-Ito et al., 2010; Zhong et al., 2010c), and are recognized by both AtVND and AtSND1/NST3 in transient expression experiments (Zhong et al., 2010c), suggesting that the determination of target preference between the VND group and the NST group must be regulated by cis-elements other than TERE or SNBE.
The overall characteristics of VNS targets identified in Arabidopsis, including the cis-elements TERE and SNBE, are basically conserved in poplar, rice, maize, B. distachyon, and Medicago truncatula (Zhao et al., 2010; Zhong et al., 2010b, 2011; Ohtani et al., 2011; Valdivia et al., 2013). However, notably, the expression specificity depending on gene group that occurs in Arabidopsis was not detected in the other plant species. For example, in Arabidopsis stems, AtVND genes are preferentially expressed in xylem vessel cells, while AtNST genes are expressed in interfascicular fibers (Mitsuda et al., 2005, 2007; Zhong et al., 2006; Yamaguchi et al., 2008). By contrast, in poplar, rice, and maize, the VNS genes of both VND and NST groups are expressed in vessels and fibers (Zhong et al., 2010b, 2011; Ohtani et al., 2011). Thus, SCW formation in xylem vessels and fibers in those species must involve distinct layers of regulation for downstream events, in addition to transcriptional control. Work on the spatial organization of SCW in xylem vessel cells could inform our understanding of this. In the current model, the patterns of SCW deposition in xylem vessel cells can be regulated by the balance of assembly and disassembly of the cortical microtubule array, determining the spatial orientation of the CesA complex (Oda and Fukuda, 2013b). The interactions of specific proteins with microtubules and/or ROP-GTPase activities control this balance (Oda and Fukuda, 2012, 2013a). Certain SCW-related enzymes likely function in the apoplastic regions of plant cells (Schuetz et al., 2014), so we might have to think about the regulation of spatial activities of enzymes in each plant species.
Transcriptional- and post-transcriptional regulation of VNS genes
Following the upregulation of VNS genes, the cells begin to differentiate as SCW-forming cells, such as xylem vessel cells and fibers. These cells are dead at maturity; therefore, VNS expression and/or activity must be well regulated in accordance with developmental programs and/or environmental signals.
During plant development, xylem vessel cells differentiate from vascular stem cells of the procambium and cambium (Fukuda, 1997; Demura and Fukuda, 2007). Phytohormones, especially auxin, provide one of first cues for vascular stem cell initiation. Several transcription factors function in the regulation of initiation, maintenance, and differentiation of vascular stem cells downstream of auxin signals (De Rybel et al., 2013; Ohashi-Ito et al., 2013a,b). Screening for factors upstream of AtVND7 in a transient expression system recently identified one such transcription factor, REV, a member of the Class III HD-Zip proteins (Carlsbecker et al., 2010; Miyashima et al., 2011; Endo et al., 2015). In addition, LBD18/ASL20 and LBD30/ASL19, which are expressed in xylem vessels, can induce ectopic SCW deposition in various types of cells through the upregulation of AtVND6 and AtVND7 (Soyano et al., 2008). LBD18/ASL20 and LBD30/ASL19 are upregulated by auxin and by AtVND6 and AtVND7 (Soyano et al., 2008), and LBD15/ASL11 and LBD30/ASL19 are also direct targets of AtVND6 and/or AtVND7 (Ohashi-Ito et al., 2010; Zhong et al., 2010c; Yamaguchi et al., 2011). This indicates the existence of auxin-mediated feedback regulation between VND and LBD/ASL (Figure 2). Moreover, all AtVND genes can induce AtVND7 expression by direct interaction with the VND7 promoter region through regions containing the SMBE/TERE motif (Zhou et al., 2014; Endo et al., 2015). Thus, once the precursor cells initiate xylem vessel cell differentiation, the positive feedback transcriptional regulation efficiently upregulates VND activity. Transient expression assays identified GATA family members (GATA5 and GATA12), and other NAC proteins (ANAC075 and SND2) as upstream factors of AtVND7 (Endo et al., 2015), suggesting that multiple signals contribute to the induction of VND activity through many types of transcription factors. Infection by the soil-born fungal pathogen Verticillium longisporum induces AtVND7 expression, resulting in the transdifferentiation of vascular bundle cells into tracheary elements (Reusche et al., 2012). Recent work also showed that AtVND6 and AtVND7 expression increased in response to high salinity and iron depletion (Taylor-Teeples et al., 2015). Such abiotic and/or biotic stress signals might be mediated by specific types of transcription factors functioning upstream of VNS genes.
In addition to many positive regulators of VNS genes, as described above, other studies have identified negative regulators of VNS, including the WRKY-type transcription factor, WRKY12 (Wang et al., 2010). In the loss-of-function wrky12 mutant, ectopic SCW formation occurred in pith parenchyma cells of inflorescence stems, and the expression of NST2 increased. Recombinant WRKY12 protein can bind to the NST2 promoter sequence in vitro; thus, WRKY12 negatively regulates SCW formation by directly inhibiting NST2 expression in pith parenchyma cells (Wang et al., 2010). Additionally, protein-DNA interaction analysis in xylem-expressed transcription factors of Arabidopsis showed that E2Fc, a member of a transcription factor family conserved in eukaryotes and a negative regulator of endoreduplication in plants, may function as a key regulator of SCW formation (Taylor-Teeples et al., 2015). Notably, E2Fc seems to function as both an activator and a repressor of AtVND6 and AtVND7 expression, depending on the situation. E2Fc also can bind to the promoters of many kinds of xylem-expressed transcription factors in addition to the AtVND promoters (Taylor-Teeples et al., 2015). These complex interactions among transcription factors suggest that vascular plants have developed a robust transcriptional regulation system to promote xylem vessel cell differentiation, which is vital for land plants.
Post-transcriptional regulation also likely plays an important role in modulating VNS activity. A VNS gene in the poplar P. trichocarpa, PtrWND1B/PtVNS11/PtrSND1-A2, has alternative splicing variants that vary in abundance in different tissues (Li et al., 2012b; Zhao et al., 2014). The predicted protein product of the short and minor variant lacks the C-terminal region, but can bind to full-length PtVNS proteins. As a result, the truncated PtrWND1B/PtVNS11/PtrSND1-A2 inhibits transcriptional activation by PtVNS proteins (Li et al., 2012b), and can suppress the SCW thickening of fiber cells in poplar (Zhao et al., 2014). This alternative splicing regulation is completely dependent on the intron sequence of PtrWND1B/PtVNS11/PtrSND1-A2, and such regulation could be specific to poplar. Understanding the contributions of regulation of splicing to the control of VNS activity will require further survey of VNS genes.
NAC domain proteins form homo- and/or hetero-dimers (Olsen et al., 2005; Weiner et al., 2012). Indeed, yeast two-hybrid screens showed that AtVND and AtNST can bind each other to form hetero-dimers as well as forming homo-dimers (Yamaguchi et al., 2008; Li et al., 2012b). Transient expression assays on poplar VNS genes indicate that transactivation activity varies by PtVNS, even between “twin” genes that possess more than 90% similarity in amino acid sequence (Ohtani et al., 2011). Thus, the VNS hetero-dimers and homo-dimers should have different transactivation activities and we should consider the effects of dimerization, particularly when multiple VNS are expressed. Yeast two-hybrid screening for proteins that interact with VND7 also identified the NAC domain protein VND-INTERACTING2 (VNI2) as a key regulator of VND7 activity (Yamaguchi et al., 2010b). VNI2 acts solely as a transcriptional repressor and inhibits transcriptional activation activities of AtVND7 and xylem vessel cell differentiation, probably through direct interaction via the NAC domain regions (Yamaguchi et al., 2010b). A Recent work also reported ANAC103 as a possible interactor with AtVND7 in vascular tissues (Yamaguchi et al., in press). Taken together with the fact that proteasome-mediated proteolysis actively regulates AtVND7 (Yamaguchi et al., 2008), these findings show that multiple layers of regulation also affect AtVND7 at the protein level.
SCW-related MYB proteins
MYB transcription factors as lignin biosynthesis regulators
MYB transcription factors occur widely in eukaryotes and have characteristic, highly conserved DNA-binding domains, called the R1, R2, and R3 domains, at their N-termini. In plants, the majority of MYB proteins have only two domains and thus are called R2R3-MYB proteins; R2R3-MYB proteins are encoded by 126 genes in Arabidopsis (Stracke et al., 2001; Dubos et al., 2010), 109 in rice (Yanhui et al., 2006), 141 in eucalyptus (Soler et al., in press), and 192 in poplar (Wilkins et al., 2009). These R2R3-MYB family genes function in a wide range of developmental processes, stress responses, and metabolism (Jin and Martin, 1999; Stracke et al., 2001; Larkin et al., 2003; Grotewold, 2006; Lepiniec et al., 2006; Valliyodan and Nguyen, 2006; Bergmann and Sack, 2007; Chinnusamy et al., 2007; Ishida et al., 2008; Dubos et al., 2010; De Geyter et al., 2012; Grima-Pettenati et al., 2012; Muñoz-Nortes et al., 2014).
Studies in the 1990s revealed the involvement of MYBs in biosynthesis of phenylpropanoids. Since then, promoter analysis of phenylpropanoid biosynthetic genes, including PAL (encoding phenylalanine ammonia-lyase) and 4CL (encoding 4-coumarate CoA ligase), revealed several important cis-elements in their promoters (Lois et al., 1989; Ohl et al., 1990; Becker-André et al., 1991; Leyva et al., 1992; Hauffe et al., 1993; Hatton et al., 1995; Wanner et al., 1995). One of them is the AC element, also known as the C1-motif, PAL-box, or H-box, which is rich in the sequence AC and is critical for xylem-specific expression of PAL and 4CL (Leyva et al., 1992; Hauffe et al., 1993; Bell-Lelong et al., 1997). Bioinformatic and biochemical analyses showed that the AC elements share sequence similarity to the motif recognized by MYB transcription factors, and that, indeed, some MYBs bind to the AC elements to regulate gene expression (Romero et al., 1998). These findings led to genome-wide in silico analysis of presumed lignin biosynthesis genes of Arabidopsis, which found that almost all lignin biosynthesis genes have AC elements in their promoter regions (Weisshaar and Jenkins, 1998; Rogers and Campbell, 2004). Taken together, these findings suggest that MYB genes coordinately regulate lignin biosynthesis genes through the AC elements. Moreover, functional analysis of MYB genes expressed in secondary xylem of P. taeda (PtMYB4) and E. gunnii (EgMYB2) demonstrated that they can enhance lignin biosynthesis and/or SCW thickening when overexpressed (Patzlaff et al., 2003a; Goicoechea et al., 2005; Table 2). MYB proteins have been reported to be involved in lignin biosynthesis in many plant species (Grima-Pettenati et al., 2012); in this review we limit our focus to Arabidopsis MYB genes.
Table 2.
MYB genes related to SCW formation.
| Gene Name | Locus or Transcript ID | Accession No. | Transcriptional activity | References |
|---|---|---|---|---|
| Arabidopsis thaliana | ||||
| AtMYB4 | At4g38620 | Repressor | Jin et al., 2000 | |
| Wang and Dixon, 2011 | ||||
| AtMYB6 | At4g09460 | Unknown | Zhong and Ye, 2012 | |
| AtMYB7 | At2g16720 | Repressor | Wang and Dixon, 2011 | |
| Zhong and Ye, 2012 | ||||
| AtMYB20 | At1g66230 | Unknown | Zhong et al., 2008 | |
| Nakano et al., 2010 | ||||
| AtMYB32 | At4g34990 | Repressor | Preston et al., 2004 | |
| Wang and Dixon, 2011 | ||||
| AtMYB42 | At4g12350 | Unknown | Zhong et al., 2008 | |
| AtMYB43 | At5g16600 | Unknown | Zhong et al., 2008 | |
| Nakano et al., 2010 | ||||
| AtMYB46 | At5g12870 | Activator | Ko et al., 2009 | |
| Zhong et al., 2007a | ||||
| Nakano et al., 2010 | ||||
| AtMYB52 | At1g17950 | Activator/ | Zhong et al., 2008 | |
| Repressor | Nakano et al., 2010 | |||
| Cassan-Wang et al., 2013 | ||||
| AtMYB54 | At1g73410 | Activator | Zhong et al., 2008 | |
| AtMYB58 | At1g16490 | Activator | Zhou et al., 2009 | |
| AtMYB63 | At1g79180 | Activator | Zhou et al., 2009 | |
| Nakano et al., 2010 | ||||
| AtMYB69 | At4g33450 | Activator | Zhong et al., 2008 | |
| AtMYB83 | At3g08500 | Activator | McCarthy et al., 2009 | |
| AtMYB85 | At4g22680 | Activator | Zhong et al., 2008 | |
| Nakano et al., 2010 | ||||
| AtMYB99 | At5g62320 | Unknown | Nakano et al., 2010 | |
| AtMYB103 | At1g63910 | Activator | Zhong et al., 2008 | |
| Nakano et al., 2010 | ||||
| Öhman et al., 2013 | ||||
| Eucalyptus gunnii | ||||
| EgMYB2 | AJ576023 | Activator | Goicoechea et al., 2005 | |
| Oryza sativa | ||||
| OsMYB46 | Os12g0515300/ LOC_Os12g33070 | JN634084 | Activator | Zhong et al., 2011 |
| Pinus taeda | ||||
| PtMYB1 | AY356372 | Activator | Bomal et al., 2008 | |
| Patzlaff et al., 2003b | ||||
| PtMYB4 | AY356371 | Activator | Patzlaff et al., 2003a | |
| PtMYB8 | DQ399057 | Activator | Bomal et al., 2008 | |
| Populus trichocarpa | ||||
| PtrMYB002 | Potri.001G258700 | KF148677 | Activator | McCarthy et al., 2010 |
| PtrMYB003 | Potri.001G267300 | KF148675 | Activator | Wilkins et al., 2009 |
| PtrMYB020 | Potri.009G061500 | KF148676 | Activator | Zhong et al., 2013 |
| PtrMYB021 | Potri.009G053900 | KF148678 | Activator | |
| Zea mays | ||||
| ZmMYB46 | JN634085 | Activator | Zhong et al., 2011 | |
AtMYB46 and AtMYB83: second-layer master switches of SCW biosynthesis
After the emergence of the first sets of evidence showing the involvement of pine and Eucalyptus MYBs in SCW formation in woody species (Patzlaff et al., 2003a,b; Goicoechea et al., 2005), their Arabidopsis closest functional orthologs MYB46 and MYB83 (AtMYB46 and AtMYB83) were reported to function as key regulators of SCW formation (Zhong et al., 2007a; Ko et al., 2009; McCarthy et al., 2009; Table 2). These genes are preferentially expressed in xylem tissues, and their overexpression induced ectopic deposition of SCW. Conversely, expression of the chimeric repressors for AtMYB46 or AtMYB83 inhibited SCW deposition in xylem (Zhong et al., 2007a; McCarthy et al., 2009). Promoter activity of AtMYB46 was found in both protoxylem-type and metaxylem-type vessel cells, suggesting the involvement of these MYBs in xylem vessel cell formation (Nakano et al., 2010). Consistent with this observation, in the double mutant myb46 myb83, SCW deposition in vessel cells is severely affected, leading to seedling growth arrest in the mutant (McCarthy et al., 2009; Figure 4). These findings indicate that AtMYB46 and AtMYB83 redundantly regulate SCW formation in Arabidopsis (McCarthy et al., 2009). Importantly, AtVND and/or AtNST/SND, the master regulators of the differentiation of SCW-containing cells, directly target these MYB genes (Zhong et al., 2007a, 2010c; McCarthy et al., 2009; Ohashi-Ito et al., 2010; Yamaguchi et al., 2011). Thus, AtMYB46 and AtMYB83 act as second layer-master switches of SCW biosynthesis (Figure 2).
Figure 4.
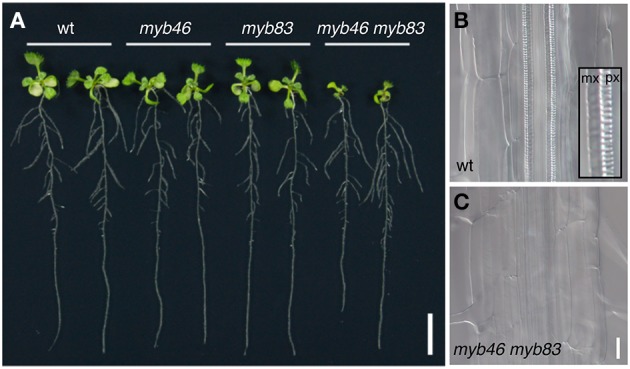
The MYB46/83 genes function as second-layer master switches for secondary cell wall formation. (A) Ten-day-old Arabidopsis seedlings of wild type (wt), myb46, myb83, and myb46 myb83 mutants. The myb46 myb83 mutant shows growth inhibition in aerial parts. (B,C) Xylem vessels in the roots of the wild type (wt, B) and myb46 myb83 mutants (C). In the wild type, thick secondary cell wall is deposited in protoxylem-type (px) and metaxylem-type (mx) vessel cells (inset in B). In the myb46 myb83 mutant, secondary cell wall deposition in xylem vessel cells is strongly inhibited, as described in McCarthy et al. (2009). Bars = 1 cm (A) and 25 μm (B,C).
Downstream genes of second-layer master switch MYBs
Biosynthetic genes of SCW components
The regulatory target genes of AtMYB46 include many downstream factors involved in SCW formation (Zhong et al., 2007a; Zhong and Ye, 2012; Ko et al., 2009, 2012; Kim et al., 2012, 2013a,b, 2014a,b). Overexpression analysis of AtMYB46 and time-course transcriptome analysis using an AtMYB46-dependent SCW formation induction system revealed that AtMYB46 can upregulate a number of SCW-biosynthesis genes (Zhong et al., 2007a; Zhong and Ye, 2012; Kim et al., 2013a,b, 2014a,b). Recently, independent groups defined the cis-element sequence recognized by MYB46 and/or MYB83 as the secondary wall MYB-responsive element [SMRE; ACC(A/T)A(A/C)(T/C), Zhong and Ye, 2012] or MYB46-responsive cis-regulatory element [M46RE; (T/C)ACC(A/T)A(A/C)(T/C), Kim et al., 2012], both of which contain the AC element sequence originally identified in the PAL gene promoter (Lois et al., 1989; Ohl et al., 1990; Leyva et al., 1992; Hatton et al., 1995; Wanner et al., 1995). A genomic survey of these cis-elements and DNA-protein binding assays suggest that the direct targets of AtMYB46/83 include genes encoding transcription factors (KNAT7, MYBs, and AtC3H14), the suite of SCW biosynthetic genes, including SCW-specific cellulose synthase genes (CesA4, CesA7, and CesA8), xylan biosynthetic genes (IRX7/FRA8, IRX8, IRX9, and IRX14), a mannan synthesis gene (CSLA9), and lignin biosynthetic genes (PAL1, C4H, 4CL1, C3H1, HCT, CCoAOMT, CCR1, F5H1, CAD6, and laccases), and genes related to cytoskeleton regulation and signal transduction (Zhong and Ye, 2012; Kim et al., 2013a,b, 2014a,b,c). Of note, some AtMYB46/83 direct targets overlap with the direct targets of VNS proteins, such as CesA4 and CesA8 (Ohashi-Ito et al., 2010; Zhong et al., 2010c; Yamaguchi et al., 2011). In vitro DNA-protein binding assays revealed that such “feed-forward” regulation often occurs in the transcriptional regulatory network in xylem cells (Taylor-Teeples et al., 2015). However, in the case of Arabidopsis, AtMYB46/83 appears to have a greater contribution than VNS, at least for cellulose synthase gene activation, as demonstrated by the severe defects in SCW deposition in xylem vessels of the myb46 myb83 double mutant (McCarthy et al., 2009; Figure 4) and the failed complementation of the cesa mutant phenotype by M46RE-mutated promoter-driven CesA genes (Kim et al., 2013a). Further study on the mechanisms by which VNS and MYB46/83 generate SCW will fill the gap between the in vitro DNA-protein interactions and in vivo mutant phenotypes.
MYB transcription factors downstream of AtMYB46/83; intermediate regulators of SCW formation
In addition to the genes for biosynthesis of SCW components, transcriptome analysis identified more than 40 transcription factors downstream of AtMYB46/83 (Ko et al., 2009; Kim et al., 2012; Zhong and Ye, 2012). This list includes KNAT7, NACs, MYBs, and AtC3H14, some of which function in SCW formation (Zhong et al., 2008; Ko et al., 2009; Zhou et al., 2009; Li et al., 2011, 2012a; Wang and Dixon, 2011; Kim et al., 2012; Zhong and Ye, 2012; Cassan-Wang et al., 2013; Öhman et al., 2013). Here we focus on AtMYB transcription factors downstream of AtMYB46/83 and describe the other factors in subsequent sections.
Before their identification as direct targets of AtMYB46/83, several AtMYBs had been reported to function in lignin biosynthesis. AtMYB4, an active repressor, regulates the expression of C4H, which encodes a cinnamic acid 4-hydroxylase required for biosynthesis of all types of lignin monomers, and AtMYB32 and AtMYB7, close homologs of AtMYB4, also negatively regulate several lignin biosynthesis genes (Jin et al., 2000; Preston et al., 2004). These repressor MYBs downregulate the expression of AtNST3/SND1 in vitro, and AtNST3/SND1 directly regulates AtMYB32 (Wang and Dixon, 2011). Based on this observation, negative feedback regulation of the VNS-MYB network for fine-tuning of SCW formation has been suggested (Wang and Dixon, 2011; Figure 2). Moreover, the lignin-specific MYBs, AtMYB58, AtMYB63, and AtMYB85, regulate lignin biosynthesis, but not cellulose and hemicellulose deposition, because their overexpression induced ectopic deposition of only lignin (Zhong et al., 2008; Zhou et al., 2009). These genes are expressed in lignifying cells such as xylem vessels and fibers, and are probably regulated by both AtVNS and AtMYB46/83 (Zhong et al., 2006, 2007a,b; Ko et al., 2009; Kim et al., 2012; Zhong and Ye, 2012). AtMYB58 and AtMYB63 can bind the AC element, and AtMYB85 can activate the promoter activity of 4CL; thus they can activate the monolignol biosynthesis pathway (Zhong et al., 2008; Zhou et al., 2009). Recent work on AtMYB103 revealed that the myb103 mutant shows a strong reduction of syringyl lignin, possibly due to a decrease in F5H expression (Öhman et al., 2013). These results suggest that the primary function of AtMYB103 is in regulation of lignin biosynthesis and that AtMYB103 functions as one of the lignin-specific MYBs, although in vitro transient expression assays showed the AtMYB103 can upregulate CesA8 promoter activity (Zhong et al., 2008).
The last group of MYB46/83-downstream MYBs contains AtMYB42, AtMYB43, AtMYB52, and AtMYB54, which are preferentially expressed in xylem tissues (Zhong et al., 2008; Nakano et al., 2010). However, their functions in SCW formation remain controversial. Overexpression of dominant-repressor forms of MYB52 and MYB54 inhibit SCW deposition in interfascicular fibers and vessels, but overexpression of MYB52 and MYB54 produced no significant changes (Zhong et al., 2008). Recently, AtMYB52 was suggested to negatively regulate SCW formation, because the myb52 mutant showed ectopic lignin deposition and the expression of AtMYB52 and SCW-related genes showed a high degree of correlation (Cassan-Wang et al., 2013). AtMYB46 and AtMYB83 can upregulate AtMYB43 (Nakano et al., 2010), but a detailed functional analysis remains to be performed.
Other MYBs that function in SCW formation
Several additional MYB transcription factors also participate in the regulation of SCW biosynthesis, probably in an MYB46/83-independent manner. Zhong et al. demonstrated that AtMYB20 and AtMYB69 also function downstream of AtNST1 and/or AtNST3/SND1, and are preferentially expressed in xylem cells (Zhong et al., 2006, 2007a,b, 2008). Dominant-repression analysis showed AtMYB69 is involved in the regulation of SCW formation (Zhong et al., 2008), and AtMYB69 is among the top 60 genes co-expressed with AtMYB52 (Cassan-Wang et al., 2013). In addition, AtMYB75 has been shown to be involved in SCW formation of xylem tissues (Bhargava et al., 2010). AtMYB75 is also called PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), because this gene was earlier identified as a positive regulator of anthocyanin biosynthesis (Borevitz et al., 2000). Phenotypes of loss-of-function mutant and overexpressor of AtMYB75 suggested that AtMYB75 negatively regulates SCW biosynthesis, especially for the branch of phenylpropanoid pathway connected to lignin biosynthesis (Bhargava et al., 2010). Nakano et al. (2010) identified AtMYB99 in a survey of factors upregulated during in vitro differentiation of xylem vessel cells. AtMYB99 is expressed in xylem vessel cells from early stages, suggesting its involvement in SCW formation in vessels (Nakano et al., 2010). However, the contribution of these genes to SCW formation in xylem tissues remains unknown. Further experiments will give us clues to elucidate the function of these MYBs.
In addition to the MYBs described above, AtMYB26 has been known to regulate SCW formation in anther endothecium (Wilson et al., 2011). Arabidopsis myb26 mutant, also known as male sterile35 mutant, lacks SCW in anther endothecium, resulting in a failure of anther dehiscence and male sterility (Dawson et al., 1999; Steiner-Lange et al., 2003). Similar phenotypes are found in nst1 nst2 double mutant (Mitsuda et al., 2005), and overexpression of AtMYB26 induced ectopic deposition of SCW (Yang et al., 2007) as the cases of AtNST1 and AtNST2 overexpression (Mitsuda et al., 2005). Interestingly, AtNST1 overexpression can induce AtMYB26 expression (Mitsuda et al., 2005), and adversely AtMYB26 overexpression can upregulate AtNST1 and AtNST2 expressions (Yang et al., 2007). AtMYB26 shares relatively high sequence homology with AtMYB46/83, second-layer master switches of SCW biosynthesis in xylem (Zhao and Bartley, 2014). Therefore, these findings seem to suggest that in the case of anther endothecium, the relationship between NAC and MYB had been changed to make a positive transcriptional feedback loop rather than the transcriptional regulation cascade, probably to make it possible to complete SCW biosynthesis in a short time of anther stage 11, which can be estimated up to 48 h (Smyth et al., 1990; Sanders et al., 1999), during another development.
Evolutionary conservation of MYB46/83 function as second-layer master switches for SCW formation
The data described above indicate a complex network of MYB-mediated transcriptional regulation of SCW formation (Figure 2). The VNS proteins act as the primary master switches of woody cell differentiation, and the MYB46/83 proteins act as secondary master switches of SCW formation. Downstream of the MYB46/83, several groups of MYBs mediate the transcriptional signals that regulate SCW biosynthetic processes; some MYBs specifically control lignin biosynthesis, and some MYBs repress or enhance SCW biosynthesis, at least partially (Figure 2). Signals can pass to the master switches from these MYBs (Wang and Dixon, 2011; Figure 2). SCW features, such as composition of cellulose, hemicellulose, and lignin, and S/G ratio of lignin, vary with tissue type, plant age, plant species, and environmental stress (Campbell and Sederoff, 1996; Knox, 2008; Vogel, 2008; Pauly and Keegstra, 2010). Complex networks of MYBs likely operate as a fine-tuning system for the formation of SCW with the appropriate composition for the specific situation.
Work to date has identified several orthologs and putative functional homologs of AtMYB46 from vascular plants, including poplar, pine, spruce, rice, maize, and switchgrass (Panicum virgatum), in addition to PtrMYB4 and EgMYB2 (Patzlaff et al., 2003a,b; Goicoechea et al., 2005; Bedon et al., 2007; Bomal et al., 2008; Zhong et al., 2011, 2013; Zhao and Bartley, 2014). Commonly, these MYBs can bind to the AC-rich elements, and overexpression of these MYBs upregulates lignin deposition (Patzlaff et al., 2003a,b; Goicoechea et al., 2005; Bomal et al., 2008; Zhong et al., 2011, 2013). These facts strongly suggest evolutionary conservation of second-layer master switch MYBs for SCW formation among vascular plant species. However, given the different characteristics of the SCW in different plant species and the fact that MYBs represent one of the most-expanded gene families in plants, the numbers and functions of intermediate MYBs downstream of MYB46/83 may have diversified among vascular plants. Engineering of lignin modification is an important target for industrial uses of plant materials (Pauly and Keegstra, 2010; Simmons et al., 2010; Zeng et al., 2014); thus we anticipate further studies of MYBs in crop species, particularly in woody species and biofuels feedstocks.
Other transcription factors involved in SCW formation
Finally, we would like to mention additional important transcription factors involved in SCW formation. Two NAC transcription factors, AtSND2 and AtSND3, which are expressed in SCW-associated tissues, function downstream of VNS proteins for SCW formation (Zhong et al., 2008). Hussey and his co-workers showed that AtSND2 can influence almost all the regulatory programs involved in SCW formation, i.e., biosynthesis of cellulose and hemicellulose, lignin polymerization, and signal transduction, in addition to the expression of AtNST3/SND1 (Hussey et al., 2011). Overexpression of AtSND2 in Eucalyptus increased the thickness of the SCW in fiber cells; thus the molecular function of SND2 is basically conserved between herbaceous and woody plants (Hussey et al., 2011). However, overexpression phenotypes differ in the woody species and transgenic lines; the effects of AtSND2 overexpression in Arabidopsis differed between Zhong et al. (2008) and Hussey et al. (2011), and the AtSND2 overexpression in Eucalyptus showed the increase in SCW thickness, while overexpression of PopNAC154, one of poplar genes homologous to AtSND2, did not change SCW thickness in xylem tissues of poplar (Grant et al., 2010; Hussey et al., 2011). These observations suggest that AtSND2 and its orthologs function as key modulators of SCW formation, and the effects of overexpression may change depending on the situation.
Transcriptome analysis of an AtMYB46-overexpressing line identified the plant-specific tandem CCCH zinc-finger gene AtC3H14 as a direct target of AtNST3/SND1 and AtMYB46 (Ko et al., 2009). AtC3H14 can activate transcription of the genes for cellulose, hemicellulose, and lignin biosynthesis (Ko et al., 2009; Kim et al., 2014b). Also, a recent study proposed an additional role of AtC3H14 in post-transcriptional regulation (Kim et al., 2014b). AtC3H14 can directly bind to mRNA in a target sequence-specific manner, similar to animal tandem CCCH zinc-finger proteins (Blackshear, 2002), and some cell wall-related genes seem to be binding targets of AtC3H14. Thus, AtC3H14 might participate in post-transcriptional and transcriptional regulation of cell wall biosynthetic genes (Kim et al., 2014b). Further functional analysis of AtC3H14 might reveal a new regulatory layer in the current model of SCW formation by transcription factors.
Moreover, other types of transcription factors, namely the homeodomain protein KNOTTED ARABIDOPSIS THALIANA7 (KNAT7) and OVATE FAMILY PROTEIN 4 (OFP4), have been described as negative regulators of SCW biosynthesis. Arabidopsis KNAT7 was first identified by co-expression analysis with SCW-related enzyme genes (Brown et al., 2005; Ehlting et al., 2005; Persson et al., 2005). The knat7 mutant showed a xylem phenotype similar to irx mutants; thus KNAT7 was also named IRX11 (Brown et al., 2005). AtNST3/SND1 and AtMYB46 directly target KNAT7 (Zhong et al., 2008; Ko et al., 2009). Li et al. revealed that KNAT7 functions as a transcriptional repressor, and that OFP4 can enhance KNAT7 activity via physical interaction. Both KANT7 and OFP4 are expressed in SCW-forming xylem cells (Li et al., 2011, 2012a), but, interestingly, the effects of knat7 loss-of-function mutation differ in xylem vessels and fiber cells. In xylem cells the SCW thickness decreased, leading to the irx phenotype (Brown et al., 2005; Li et al., 2012a). By contrast, the SCW thickness increased in fiber cells (Li et al., 2012a). Based on the identification of several interaction partners of KNAT7, including OFP4, MYB75/PAP1, and BELL1-LIKE HOMEODOMAIN6 proteins (Hackbusch et al., 2005; Bhargava et al., 2010; Li et al., 2011; Liu et al., 2014), KNAT7 may regulate specific aspects of SCW formation, depending on cell type, through interaction with different partners (Li et al., 2012a; Liu et al., 2014). The molecular function of KNAT7 is conserved with its poplar ortholog (Li et al., 2012a), and KNAT7 appears to have developed as a negative regulator to fine-tune SCW biosynthesis to fit the situation.
Conclusion and perspectives
During the last decade, extensive research has produced remarkable progress in our understanding of transcriptional regulation of SCW formation. Although some key modulators may remain unknown, the current model (Figure 2) covers the essential players of SCW formation regulation. SCW in wood tissues provides a major source of land biomass, and SCW formation thus is an important target for biomass engineering. Several trials using the transcription factors described in this review to target SCW properties have been already reported (Carpita, 2012; Yang et al., 2013; Sakamoto and Mitsuda, 2015). Such applied research will assume growing importance in studies of the regulation of SCW formation.
In 2015, Taylor-Teeples et al. reported a protein–DNA interaction-based network between transcription factors and SCW metabolic genes of Arabidopsis (Taylor-Teeples et al., 2015). This map showed complex interactions among transcription factors and SCW metabolic genes, with many instances of feed-forward regulation (Figure 2A). In the protein–DNA interaction network, many kinds of transcription factors other than those previously reported could recognize the promoter sequence of transcription factors and SCW metabolic genes (Taylor-Teeples et al., 2015). This observation implies that we are now at a turning point in research on the transcriptional regulation of SCW biosynthesis: in addition to continuing our efforts to identify genes involved in SCW formation and reveal the interactions between them on a one-on-one basis, we must move to the next steps. Thus, future research must aim to reveal the dynamism of the network itself based on observations of what happens in vivo, because modeling based on in vitro data only tells us the many possibilities of the network. The transcriptional regulatory network of SCW formation could become a good model for such advanced analysis in plants.
Comparative transcriptomic work revealed that the xylem transcriptomes of vascular plants are more highly conserved than the overall transcriptomes (Li et al., 2010), indicating the ancient origin of the xylem transcriptome. In accordance with this idea, recent work showed that the scheme of VNS-based transcriptional regulation of cell differentiation for wall thickening is conserved in the moss Physcomitrella patens (Figure 5), although P. patens does not have vascular plant-type SCW (Xu et al., 2014). The P. patens genome has eight VNS loci (Figure 3A), and the triple mutant ppvns1 ppvns6 ppvns7 of P. patens showed reduced wall thickness in stereid cells, which serve as supporting cells in mosses (Figure 5). Transcriptome analysis of P. patens overexpressing PpVNS7 indicated that PpVNS regulates many putative orthologs of the direct targets of AtVNS, including putative orthologs of AtMYB46/83/103 and AtMYB85 (Xu et al., 2014). These findings suggest that the genes downstream of VNS are evolutionarily conserved, and that the VNS-MYB-based transcriptional regulatory system of wall modification has an ancient root, at least at the common ancestors of mosses and vascular plants. Land plants would have developed this core regulatory scheme of cell wall modification during evolution to adapt to new environments. As reviewed here, we now have extensive knowledge on the factors governing SCW formation in a wide range of plant species, which probably includes some species-specific elements. What is the particular regulatory scheme for each species? Which parts of the common regulatory module are conserved among land plants? Future work will provide clues to answer these questions.
Figure 5.
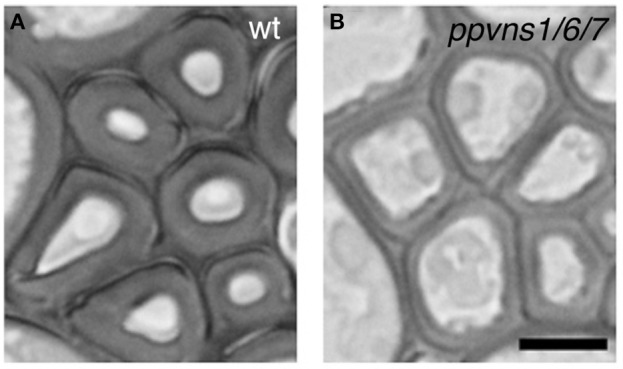
The PpVNS genes function in cell wall thickening in the moss P. patens. Stereid cells in leaf vein of the wild type (A) and ppvns1 ppvns6 pvns7 triple mutant (B). In the triple mutants, the stereid cell walls were significantly less thick, suggesting the importance of PpVNS proteins in cell wall thickening in the moss. Data were adapted from Xu et al. (2014). Bar = 5 μm.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Taku Demura (Nara Institute of Science and Technology) for critical discussions. We also thank Dr. Bo Xu and Mr. Nobuhiro Akiyoshi (Nara Institute of Science and Technology) for kindly providing the data of plant sections. This work was supported in part by Grants-in-Aid from the MEXT NC-CARP project to Y.N., and by a Start-up Grant for Women Researchers from the Nara Institute of Science and Technology to M.O., The Naito Foundation Subsidy for Female Researchers after Maternity Leave, and The Sumitomo Foundation for Grant for Basic Science Research Projects to M.O., and by Grants-in-Aid from the Japan Society for the Promotion of Science (grant numbers 24770052 and 25114520 to M.O, and 25840098 to M.Y.).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00288/abstract
References
- Allona I., Quinn M., Shoop E., Swope K., St Cyr S., Carlis J., et al. (1998). Analysis of xylem formation in pine by cDNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 95, 9693–9698. 10.1073/pnas.95.16.9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-André M., Schulze-Lefert P., Hahlbrock K. (1991). Structural comparison, modes of expression, and putative cis-acting elements of the two 4-coumarate: CoA ligase genes in potato. J. Biol. Chem. 266, 8551–8559. [PubMed] [Google Scholar]
- Bedon F., Grima-Pettenati J., Mackay J. (2007). Conifer R2R3-MYB transcriotion factors: sequence analyses and gene expression in wood-forming tissues of white spruce (Picea glauca). BMC Plant Biol. 7:17. 10.1186/1471-2229-7-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Lelong D. A., Cusumano J. C., Meyer K., Chapple C. (1997). Cinnamate-4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol. 113, 729–738. 10.1104/pp.113.3.729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett T., van den Toorn A., Sanchez-Perez G. F., Campiho A., Willemsen V., Snel B., et al. (2010). SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis. Plant Cell 22, 640–654. 10.1105/tpc.109.072272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann D., Sack F. D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58, 163–181. 10.1146/annurev.arplant.58.032806.104023 [DOI] [PubMed] [Google Scholar]
- Bhargava A., Mansfield S. D., Hall H. C., Douglas C. J., Ellis B. E. (2010). MYB75 functions in regulation of secondary cell wall formation in the Arabidopsis inflorescence stem. Plant Physiol. 154, 1428–1438. 10.1104/pp.110.162735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear P. J. (2002). Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans. 30, 945–952. 10.1042/BST0300945 [DOI] [PubMed] [Google Scholar]
- Bomal C., Bedon F., Caron S., Mansfield S. D., Levasseur C., Cooke J. E. K., et al. (2008). Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wallbiogenesis: a comparative in planta analysis. J. Exp. Bot. 59, 3925–3939. 10.1093/jxb/ern234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J. O., Xia Y., Blount J., Dixon R. A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. 10.1105/tpc.12.12.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. M., Zeef L. A. H., Ellis J., Goodacre R., Turner S. R. (2005). Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17, 2281–2295. 10.1105/tpc.105.031542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. M., Zhang Z., Stephens E., Dupree P., Turner S. R. (2009). Characterization of IRX10 and IRX10-like reveals an essential role in glucuronoxylan biosynthesis in Arabidopsis. Plant J. 57, 732–746. 10.1111/j.1365-313X.2008.03729.x [DOI] [PubMed] [Google Scholar]
- Brown D. M., Wightman R., Zhang Z., Gomez L. D., Atanassov I., Bukowski J.-P., et al. (2011). Arabidopsis genes IRREGULAR XYLEM (IRX15) and IRX15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J. 66, 401–413. 10.1111/j.1365-313X.2011.04501.x [DOI] [PubMed] [Google Scholar]
- Campbell M. M., Sederoff R. R. (1996). Variation in lignin content and composition. Plant Physiol. 110, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsbecker A., Lee J. Y., Roberts C. J., Dettmer J., Lehesranta S., Zhou J., et al. (2010). Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 465, 316–321. 10.1038/nature08977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita N. C. (2012). Progress in the biological synthesis of the plant cell wall: new ideas for improving biomass for bioenergy. Curr. Opin. Biotechnol. 23, 330–337. 10.1016/j.copbio.2011.12.003 [DOI] [PubMed] [Google Scholar]
- Cassan-Wang H., Goué N., Saidi M. N., Legay S., Sivadon P., Goffner D., et al. (2013). Identification of novel transcription factors regulating secondary cell wall formation in Arabidopsis. Front. Plant Sci. 4:189. 10.3389/fpls.2013.00189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. 10.1016/j.tplants.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Cosgrove D. J., Jarvis M. C. (2012). Comparative structure and biomechanics of primary and secondary cell walls. Front. Plant Sci. 3:204. 10.3389/fpls.2012.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson J., Sözen E., Vizir I., van Waeyenberge S., Wilson Z. A., Mulligan B. J. (1999). Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol. 144, 213–222 10.1046/j.1469-8137.1999.00507.x [DOI] [Google Scholar]
- Demura T., Fukuda H. (2007). Transcriptional regulation in wood formation. Trends Plant Sci. 12, 64–70. 10.1016/j.tplants.2006.12.006 [DOI] [PubMed] [Google Scholar]
- Demura T., Tashiro G., Horiguchi G., Kishimoto N., Kubo M., Matsuoka N., et al. (2002). Visualization by comprehensive microarray analysis of gene expression programs during transdifferentiation of mesophyll cells into xylem cells. Proc. Natl. Acad. Sci. U.S.A. 99, 15794–15799. 10.1073/pnas.232590499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Geyter N., Gholami A., Goormachtig S., Goossens A. (2012). Transcriptional machineries in jasmonate-elicited plant secondary metabolism. Trends Plant Sci. 17, 349–359. 10.1016/j.tplants.2012.03.001 [DOI] [PubMed] [Google Scholar]
- De Rybel B., Möller B., Yoshida S., Grabowicz I., Barbier de Reuille P., Boeren S., et al. (2013). A bHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev. Cell 24, 426–437. 10.1016/j.devcel.2012.12.013 [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Duval I., Lachance D., Giguère I., Bomal C., Morency M. J., Pelletier G., et al. (2014). Large-scale screening of transcription factor–promoter interactions in spruce reveals a transcriptional network involved in vascular development. J. Exp. Bot. 65, 2319–2333. 10.1093/jxb/eru116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J., Mattheus N., Aeschliman D. S., Li E., Hamberger B., Cullis I. F., et al. (2005). Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J. 42, 618–640. 10.1111/j.1365-313X.2005.02403.x [DOI] [PubMed] [Google Scholar]
- Endo H., Yamaguchi M., Tamura T., Nakano Y., Nishikubo N., Yoneda A., et al. (2015). Multiple classes of transcription factors regulate the expression of VASCULAR-RELATED NAC-DOMAIN7, a master switch of xylem vessel differentiation. Plant Cell Physiol. 56, 242–254. 10.1093/pcp/pcu134 [DOI] [PubMed] [Google Scholar]
- Fang Y., You J., Xie K., Xie W., Xiong L. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563. 10.1007/s00438-008-0386-6 [DOI] [PubMed] [Google Scholar]
- Fukuda H. (1997). Tracheary element differentiation. Plant Cell 9, 1147–1156. 10.1105/tpc.9.7.1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann A. (2010). Mechanical modeling and structural analysis of the primary plant cell wall. Curr. Opin. Plant Biol. 13, 693–699. 10.1016/j.pbi.2010.09.017 [DOI] [PubMed] [Google Scholar]
- Goicoechea M., Lacombe E., Legay S., Mihaljevic S., Rech P., Jauneau A., et al. (2005). EgMYB2, a new transcriptional activator from Eucalyptus xylem, regulates secondary cell wall formation and lignin biosynthesis. Plant J. 43, 553–567. 10.1111/j.1365-313X.2005.02480.x [DOI] [PubMed] [Google Scholar]
- Grant E. H., Fujino T., Beers E. P., Brunner A. M. (2010). Characterization of NAC domain transcription factors implicated in control of vascular cell differentiation in Arabidopsis and Populus. Planta 232, 337–352. 10.1007/s00425-010-1181-2 [DOI] [PubMed] [Google Scholar]
- Grima-Pettenati J., Soler M., Camargom E., Wang H. (2012). Transcriptional regulation of the lignin biosynthetic pathway revisited: new players and insights, in Advances in Botanical Research, Vol. 61, eds Jouanin L., Lapierre C. (Burlington, VT: Academic Press; ), 173–218. [Google Scholar]
- Grotewold E. (2006). The genetics and biochemistry of floral pigments. Ann. Rev. Plant Biol. 57, 761–780. 10.1146/annurev.arplant.57.032905.105248 [DOI] [PubMed] [Google Scholar]
- Hackbusch J., Richter K., Muller J., Salamini F., Uhrig J. F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 4908–4912. 10.1073/pnas.0501181102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O., Traas J. (2010). The mechanics behind plant development. New Phytol. 185, 369–385. 10.1111/j.1469-8137.2009.03100.x [DOI] [PubMed] [Google Scholar]
- Hatton D., Sablowski R., Yung M. H., Smith C., Schuch W., Bevan M. (1995). Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. Plant J. 7, 859–876. 10.1046/j.1365-313X.1995.07060859.x [DOI] [PubMed] [Google Scholar]
- Hauffe K. D., Lee S. P., Subramaniam R., Douglas C. J. (1993). Combinatorial interactions between positive and negative cis-acting elements control spatial patterns of 4CL-1 expression in transgenic tobacco. Plant J. 4, 235–253. 10.1046/j.1365-313X.1993.04020235.x [DOI] [PubMed] [Google Scholar]
- Heyn A. N. J. (1955). Small particle X-ray scattering by fibers, size and shape of microcrystallites. J. Appl. Phys. 26, 519–526 10.1063/1.1722032 [DOI] [Google Scholar]
- Heyn A. N. J. (1965). Observations on the size and shape of the cellulose microcrystallite in cotton fiber by electron staining. J. Appl. Phys. 36, 2088 10.1063/1.1714417 [DOI] [Google Scholar]
- Heyn A. N. J. (1966). The microcrystalline structure of cellulose in cell walls of cotton, ramie, and jute fibers as revealed by negative staining of sections. J. Cell Biol. 29, 181–197. 10.1083/jcb.29.2.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R., Qi G., Kong Y., Kong D., Gao Q., Zhou G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10:145. 10.1186/1471-2229-10-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey S. G., Mizrachi E., Creux N. M., Myburg A. A. (2013). Navigating the transcriptional roadmap regulating plant secondary cell wall deposition. Front. Plant Sci. 4:325. 10.3389/fpls.2013.00325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey S. G., Mizrachi E., Spokevicius A. V., Bossinger G., Berger D. K., Myburg A. A. (2011). SND2, a NAC transcription factor gene, regulates genes involved in secondary cell wall development in Arabidopsis fibres and increases fibre cell area in Eucalyptus. BMC Plant Biol. 11:173. 10.1186/1471-2229-11-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey S. G., Saïdi M. N., Hefer C. A., Myburg A. A., Grima-Pettenati J. (in press). Structural, evolutionary and functional analysis of the NAC domain protein family in Eucalyptus. New Phytol. 10.1111/nph.13139 [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59, 365–386. 10.1146/annurev.arplant.59.032607.092949 [DOI] [PubMed] [Google Scholar]
- Jensen J. K., Kim H., Cocuron J. C., Orler R., Ralph J., Wilkerson C. G. (2011). The DUF579 domain containing proteins IRX15 and IRX15-L affect xylan synthesis in Arabidopsis. Plant J. 66, 387–400. 10.1111/j.1365-313X.2010.04475.x [DOI] [PubMed] [Google Scholar]
- Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., et al. (2000). Trancriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. 10.1093/emboj/19.22.6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Martin C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. 10.1023/A:1006319732410 [DOI] [PubMed] [Google Scholar]
- Jones L., Ennos A. R., Turner S. R. (2001). Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis. Plant J. 2, 205–216 10.1046/j.1365-313x.2001.01021.x [DOI] [PubMed] [Google Scholar]
- Kim W. C., Kim J. Y., Ko J. H., Kim J., Han K. H. (2013a). Transcription factor MYB46 is an obligate component of the transcriptional regulatory complex for functional expression of secondary wall-associated cellulose synthases in Arabidopsis thaliana. J. Plant Physiol. 170, 1374–1378. 10.1016/j.jplph.2013.04.012 [DOI] [PubMed] [Google Scholar]
- Kim W. C., Ko J. H., Han K. H. (2012). Identification of a cis-acting regulatory motif recognized by MYB46, a master regulator of secondary wall biosynthesis. Plant Mol. Biol. 78, 489–501. 10.1007/s11103-012-9880-7 [DOI] [PubMed] [Google Scholar]
- Kim W. C., Ko J. H., Kim J. Y., Kim J., Bae H. J., Han K. H. (2013b). MYB46 directly regulated the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J. 73, 26–36 10.1111/j.1365-313x.2012.05124.x [DOI] [PubMed] [Google Scholar]
- Kim W. C., Kim J. Y., Ko J. H., Kang H., Han K. H. (2014a). Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis. Plant Mol. Biol. 85, 589–599. 10.1007/s11103-014-0205-x [DOI] [PubMed] [Google Scholar]
- Kim W. C., Kim J. Y., Ko J. H., Kang H., Kim J., Han K. H. (2014b). AtC3H14, a plant-specific tandem CCCH zinc-finger protein, binds to its target mRNAs in a sequence-specific manner and affects cell elongation in Arabidopsis thaliana. Plant J. 80, 772–784. 10.1111/tpj.12667 [DOI] [PubMed] [Google Scholar]
- Kim W. C., Reca I. B., Kim Y. S., Prk S., Thomashow M. F., Keegstra K., et al. (2014c). Transcription factors that directly regulate the expression of CSLA9 encoding mannan synthase in Arabidopsis thaliana. Plant Mol. Biol. 84, 577–587. 10.1007/s11103-013-0154-9 [DOI] [PubMed] [Google Scholar]
- Knox J. P. (2008). Revealing the structural and functional diversity of plant cell walls. Curr. Opin. Plant Biol. 11, 308–313. 10.1016/j.pbi.2008.03.001 [DOI] [PubMed] [Google Scholar]
- Ko J. H., Jeon H. W., Kim W. C., Han K. H. (2014). The MYB46/MYB83-mediated transcriptional regulatory programme is a gatekeeper of secondary wall biosynthesis. Ann. Bot. 114, 1099–1107. 10.1093/aob/mcu126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. H., Kim W. C., Han K. H. (2009). Ectopic expression of MYB46 identifies transcriptional regulatory genes involved in secondary wall biosynthesis in Arabidopsis. Plant J. 60, 649–665. 10.1111/j.1365-313X.2009.03989.x [DOI] [PubMed] [Google Scholar]
- Ko J. H., Kim W. C., Kim J. Y., Ahn S. J., Han K. H. (2012). MYB46-mediated transcriptional regulation of secondary cell wall biosynthesis. Mol. Plant 5, 961–963. 10.1093/mp/sss076 [DOI] [PubMed] [Google Scholar]
- Ko J. H., Yang S. H., Park A. H., Lerouxel O., Han K. H. (2007). ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J. 50, 1035–1048. 10.1111/j.1365-313X.2007.03109.x [DOI] [PubMed] [Google Scholar]
- Krizek B. A., Fletcher J. C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6, 688–698. 10.1038/nrg1675 [DOI] [PubMed] [Google Scholar]
- Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., et al. (2005). Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860. 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J. C., Brown M. L., Schiefelbein J. (2003). How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 54, 403–430. 10.1146/annurev.arplant.54.031902.134823 [DOI] [PubMed] [Google Scholar]
- Le D. T., Nishiyama R., Watanabe Y., Mochida K., Yamaguchi-Shinozaki K., Shinozaki K., et al. (2011). Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18, 263–276. 10.1093/dnares/dsr015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., O'Neill M. A., Tsumuraya Y., Darvill A. G., Ye Z. H. (2007). The irregular xylem9 mutant is deficient in xylan xylosyltransferase activity. Plant Cell Physiol. 48, 1624–1634. 10.1093/pcp/pcm135 [DOI] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J.-M., Baudry A., Pourcel L., Nesi N., et al. (2006). Genetics and biochemistry of seed flavonoids. Ann. Rev. Plant Biol. 57, 405–430. 10.1146/annurev.arplant.57.032905.105252 [DOI] [PubMed] [Google Scholar]
- Leyva A., Liang X., Pintor-Toro J. A., Dixon R. A., Lamb C. J. (1992). cis-Element combinations determine phenylalanine ammonia-lyase gene tissue-specific expression patterns. Plant Cell 4, 263–271. 10.1105/tpc.4.3.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E., Bhargava A., Qiang W., Friedmann M. C., Forneris N., Savidge R. A., et al. (2012a). The Class II KNOX gene KNAT7 negatively regulates secondary wall formation in Arabidopsis and is functionally conserved in Populus. New Phytol. 194, 102–115. 10.1111/j.1469-8137.2011.04016.x [DOI] [PubMed] [Google Scholar]
- Li E., Wang S., Liu Y., Chen J. G., Douglas C. J. (2011). OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 67, 328–341. 10.1111/j.1365-313X.2011.04595.x [DOI] [PubMed] [Google Scholar]
- Li Q., Lin Y. C., Sun Y. H., Song J., Chen H., Zhang X. H., et al. (2012b). Splice variant of the SND1 transcription factor is a dominant negative of SND1 members and their regulation in Populus trichocarpa. Proc. Natl. Acad. Sci. U.S.A. 109, 14699–14704. 10.1073/pnas.1212977109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu H. X., Southerton S. G. (2010). Comparative genomics reveals conservative evolution of the xylem transcriptome in vascular plants. BMC Evol. Biol. 10:190. 10.1186/1471-2148-10-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., You S., Taylor-Teeples M., Li W. L., Schuetz M., Brady S. M., et al. (2014). BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 26, 4843–4861. 10.1105/tpc.114.128322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois R., Dietrich A., Hahlbrock K., Schulz W. (1989). A phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 8, 1641–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz W. W., Dean J. F. D. (2002). SAGE Profiling and demonstration of differential gene expression along the axial developmental gradient of lignifying xylem in loblolly pine (Pinus taeda). Tree Physiol. 22, 301–310. 10.1093/treephys/22.5.301 [DOI] [PubMed] [Google Scholar]
- Marx-Figini M. (1969). On the biosynthesis of cellulose in higher and lower plants. J. Polym. Sci. Part C 28, 57–67 10.1002/polc.5070280108 [DOI] [Google Scholar]
- McCarthy R. L., Zhong R., Fowler S., Lyskowski D., Piyasena H., Carleton K., et al. (2010). The poplar MYB transcription factors, PtrMYB3 and PtrMYB20, are involved in the regulation of secondary wall biosynthesis. Plant Cell Physiol. 51, 1084–1090. 10.1093/pcp/pcq064 [DOI] [PubMed] [Google Scholar]
- McCarthy R. L., Zhong R., Ye Z. H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. 10.1093/pcp/pcp139 [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Iwase A., Yamamoto H., Yoshida M., Seki M., Shinozaki K., et al. (2007). NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19, 270–280. 10.1105/tpc.106.047043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N., Ohme-Takagi M. (2008). NAC transcription factors NST1 and NST3 regulate pod shattering in a partially redundant manner by promoting secondary wall formation after the establishment of tissue identity. Plant J. 56, 768–778. 10.1111/j.1365-313X.2008.03633.x [DOI] [PubMed] [Google Scholar]
- Mitsuda N., Seki M., Shinozaki K., Ohme-Takagi M. (2005). The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17, 2993–3006. 10.1105/tpc.105.036004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S., Koi S., Hashimoto T., Nakajima K. (2011). Non-cell-aoutonomous microRNA165 acts in a dose-dependent manner to regulate multiple differentiation status in the Arabidopsis root. Development 138, 2303–2313. 10.1242/dev.060491 [DOI] [PubMed] [Google Scholar]
- Muñoz-Nortes T., Wilson-Sánchez D., Candela H., Micol L. (2014). Symmetry, asymmetry, and the cell cycle in plants: known knowns and some known unknowns. J. Exp. Bot. 65, 2645–2655. 10.1093/jxb/ert476 [DOI] [PubMed] [Google Scholar]
- Myburg A. A., Grattapaglia D., Tuskan G. A., Hellsten U., Hayes R. D., Grimwood J., et al. (2014). The genome of Eucalyptus grandis. Nature 510, 356–362. 10.1038/nature13308 [DOI] [PubMed] [Google Scholar]
- Nakano Y., Nishikubo N., Goué N., Ohtani M., Yamaguchi M., Katayama Y., et al. (2010). MYB transcription factors orchestrating the developmental program of xylem vessels in Arabidopsis roots. Plant Biotechnol. 27, 267–272 10.5511/plantbiotechnology.27.267 [DOI] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta. 1819, 97–103. 10.1016/j.bbagrm.2011.10.005 [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M., Manimekalai R., Sharoni A. M., Satoh K., Kondoh H., Ooka H., et al. (2010). Genome-wide analysis of NAC transcription factor family in rice. Gene 465, 30–44. 10.1016/j.gene.2010.06.008 [DOI] [PubMed] [Google Scholar]
- Nystedt B., Street N. R., Wetterbom A., Zuccolo A., Lin Y. C., Scofield D. G., et al. (2013). The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584. 10.1038/nature12211 [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2012). Initiation of cell wall pattern by a rho- and microtubule-driven symmetry breaking. Science 337, 1333–1336. 10.1126/science.1222597 [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2013a). Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell call patterns. Plant Cell 25, 4439–4450. 10.1105/tpc.113.117853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2013b). The dynamic interplay of plasma membrane domains and cortical microtubules in secondary cell wall patterning. Front. Plant Sci. 4:511. 10.3389/fpls.2013.00511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Matsukawa M., Fukuda H. (2013a). An atypical bHLH transcription factor regulates early xylem development downstream of auxin. Plant Cell Physiol. 54, 398–405. 10.1093/pcp/pct013 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oda Y., Fukuda H. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 22, 3461–3473. 10.1105/tpc.110.075036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K., Oguchi M., Kojima M., Sakakibara H., Fukuda H. (2013b). Auxin-associated initiation of vascular cell differentiation by LONESOME HIGHWAY. Development 140, 765–769. 10.1242/dev.087924 [DOI] [PubMed] [Google Scholar]
- Ohl S., Hedrick S. A., Chory J., Lamb C. J. (1990). Functional properties of a phenylalanine ammonia-lyase promoter from Arabidopsis. Plant Cell 2, 837–848. 10.1105/tpc.2.9.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhman D., Demedts B., Kumar M., Gerber L., Gorzsás A., Goeminne G., et al. (2013). MYB103 is required for FERULATE-5-HYDROXYLASE expression and syringyl lignin biosynthesis in Arabidopsis stems. Plant J. 73, 63–76. 10.1111/tpj.12018 [DOI] [PubMed] [Google Scholar]
- Ohtani M., Nishikubo N., Xu B., Yamaguchi M., Mitsuda N., Goue N., et al. (2011). A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J. 67, 499–512. 10.1111/j.1365-313X.2011.04614.x [DOI] [PubMed] [Google Scholar]
- Olsen A. N., Ernst H. A., Leggio L. L., Skriver K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. 10.1016/j.tplants.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Ooka H., Satho K., Doi K., Nagata T., Otomo Y., Murakami K., et al. (2003). Comprehensive analyisis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10, 239–247. 10.1093/dnares/10.6.239 [DOI] [PubMed] [Google Scholar]
- Pallady S. G. (2008). Chapter 2 – The woody plant body, in Physiology of woody plants, 3rd Edn (San Diego, CA: Academic Press; ), 9–38 10.1016/B978-012088765-1.50003-8 [DOI] [Google Scholar]
- Patzlaff A., McInnis S., Courtenay A., Surman C., Newman L. J., Smith C., et al. (2003a). Characterization of a pine MYB that regulates lignification. Plant J. 36, 743–754. 10.1046/j.1365-313X.2003.01916.x [DOI] [PubMed] [Google Scholar]
- Patzlaff A., Mewman L. J., Dubos C., Whetten R. W., Smith C., McInnis S., et al. (2003b). Characterization of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 53, 597–608. 10.1023/B:PLAN.0000019066.07933.d6 [DOI] [PubMed] [Google Scholar]
- Pauly M., Keegstra K. (2010). Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 13, 304–311. 10.1016/j.pbi.2009.12.009 [DOI] [PubMed] [Google Scholar]
- Pavy N., Paule C., Parsons L., Crow J. A., Morencey M. J., Cooke J., et al. (2005). Generation, annotation, analysis and database integration of 16,500 white spruce EST clusters. BMC Genomics 6:144. 10.1186/1471-2164-6-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña M. J., Zhong R., Zhou G. K., Richardson E. A., O'Neil M. A., Darvill A. G., et al. (2007). Arabidopsis irregular xylem8 and irregular xylem9: implications for the complexity of glucuronoxylan biosynthesis. Plant Cell 19, 549–563. 10.1105/tpc.106.049320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Wei H., Milne J., Page G. P., Somerville C. R. (2005). Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc. Natl. Acad. Sci. U.S.A. 102, 8633–8638. 10.1073/pnas.0503392102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka J. J., Winter C. M., Benfey P. N. (2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590. 10.1146/annurev-arplant-042811-105501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J., Wheeler J., Heazlewood J., Li S. F., Parish R. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40, 979–995. 10.1111/j.1365-313X.2004.02280.x [DOI] [PubMed] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Pyo H., Demura T., Fukuda H. (2007). TERE; a novel cis-element responsible for a coordinated expression of genes related to programmed cell death and secondary wall formation during differentiation of tracheary elements. Plant J. 51, 955–965. 10.1111/j.1365-313X.2007.03180.x [DOI] [PubMed] [Google Scholar]
- Rengel D., Clemente H. S., Servant F., Ladouce N., Paux E., Wincker P., et al. (2009). A new genomic resource dedicated to wood formation in Eucalyptus. BMC Plant Biol. 9:36. 10.1186/1471-2229-9-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusche M., Thole K., Janz D., Truskina J., Rindfleisch S., Drubert C., et al. (2012). Verticillium infection triggers VASCULAR-RELATED NAC DOMAIN7-dependent de novo xylem formation and enhances drought tolerance in Arabidopsis. Plant Cell 24, 3823–3837. 10.1105/tpc.112.103374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers L. A., Campbell M. M. (2004). The genetic control of lignin deposition during plant growth and development. New Phytol. 1, 17–30 10.1111/j.1469-8137.2004.01143.x [DOI] [PubMed] [Google Scholar]
- Romero I., Fuertes A., Benito M. J., Malpica J. M., Leyva A., Paz-Ares J. (1998). More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14, 273–284. 10.1046/j.1365-313X.1998.00113.x [DOI] [PubMed] [Google Scholar]
- Sakamoto S., Mitsuda N. (2015). Reconstitution of a secondary cell wall in a secondary cell wall-deficient Arabidopsis mutant. Plant Cell Physiol. 56, 299–310. 10.1093/pcp/pcu208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K., Watanabe Y., Shirakawa M., Matsushita Y., Imai T., Koike T., et al. (2012). Direct mapping of morphological distribution of syringyl and guaiacyl lignin in the xylem of maple by time-of-flight secondary ion mass spectrometry. Plant J. 69, 542–552. 10.1111/j.1365-313X.2011.04811.x [DOI] [PubMed] [Google Scholar]
- Sanders P. M., Bui A. Q., Weterings K., McIntire K. N., Hsu Y.-C., Lee P. Y., et al. (1999). Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 11, 297–322. [Google Scholar]
- Schuetz M., Benske A., Smith R., Watanabe Y., Tobimatsu Y., Ralph J., et al. (2014). Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 166, 798–807. 10.1104/pp.114.245597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons B. A., Loqué D., Ralph J. (2010). Advances in modifying lignin for enhanced biofuel production. Curr. Opin. Plant Biol. 13, 312–319. 10.1016/j.pbi.2010.03.001 [DOI] [PubMed] [Google Scholar]
- Smyth D. R., Bowman J. L., Meyerowitz E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. 10.1105/tpc.2.8.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler M., Camargo E. L. O., Carocha V., Cassan-Wang H., Clemente H. S., Savelli B., et al. (in press). The Eucalyptus grandis R2R3-MYB transcription factor family: evidence for woody growth-related evolution and function. New Phytol. 10.1111/nph.13039 [DOI] [PubMed] [Google Scholar]
- Soyano T., Thitamadee S., Machida Y., Chua N. H. (2008). ASYMMETRIC LEAVES2-LIKE19/LATERAL ORGAN BOUNDARIES DOMAIN30 and ASL20/LBD18 regulate tracheary element differentiation in Arabidopsis. Plant Cell. 20, 3359–3373. 10.1105/tpc.108.061796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner-Lange S., Unte U. S., Eckstein L., Yang C., Wilson Z. A., Schmelzer E., et al. (2003). Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 34, 519–528. 10.1046/j.1365-313X.2003.01745.x [DOI] [PubMed] [Google Scholar]
- Sterky F., Bhalerao R. R., Unneberg P., Segerman B., Nilsson P., Brunner A. M., et al. (2004). A populus EST resources for plant functional genomics. Proc. Natl. Acad. Sci. U.S.A. 101, 13951–13956. 10.1073/pnas.0401641101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Taylor N. G., Howells R. M., Huttly A. K., Vickers K., Turner S. R. (2003). Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. U.S.A. 100, 1450–1455. 10.1073/pnas.0337628100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Teeples M., Lin L., de Lucas M., Turco G., Toal T. W., Gaudinier A., et al. (2015). An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517, 571–575. 10.1038/nature14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. R., Somerville C. R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. 10.1105/tpc.9.5.689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuskan G. A., DiFazio S., Jansson S., Bohlmann J., Grigoriev I., Hellsten U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 15, 1596–1604. 10.1126/science.1128691 [DOI] [PubMed] [Google Scholar]
- Valdivia E. R., Herrera M. T., Gianzo C., Fidalgo J., Revilla G., Zarra I., et al. (2013). Regulation of secondary wall synthesis and cell death by NAC transcription factors in the monocot Brachypodium distachyon. J. Exp. Bot. 64, 1333–1343. 10.1093/jxb/ers394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valliyodan B., Nguyen H. (2006). Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 9, 189–195. 10.1016/j.pbi.2006.01.019 [DOI] [PubMed] [Google Scholar]
- Vogel J. (2008). Unique aspects of the grass cell wall. Curr. Opin. Plant Biol. 11, 301–307. 10.1016/j.pbi.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Wang H., Avci U., Nakashima J., Hahn M. G., Chen F., Dixon R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proc. Natl. Acad. Sci. U.S.A. 107, 22338–22343. 10.1073/pnas.1016436107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Z., Dixon R. A. (2011). On-off switches for secondary cell wall biosynthesis. Mol. Plant. 5, 297–303. 10.1093/mp/ssr098 [DOI] [PubMed] [Google Scholar]
- Wanner L. A., Li G., Ware D., Somssich I. E., Davis K. R. (1995). The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 27, 327–338. 10.1007/BF00020187 [DOI] [PubMed] [Google Scholar]
- Weiner D. H., Lindemose S., Grossmann J. G., Møllegaard N. E., Olsen A. N., Helgstrand C., et al. (2012). DNA binding by the plant-specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors. Biochem. J. 444, 395–404. 10.1042/BJ20111742 [DOI] [PubMed] [Google Scholar]
- Weisshaar B., Jenkins G. I. (1998). Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1, 251–257. 10.1016/S1369-5266(98)80113-1 [DOI] [PubMed] [Google Scholar]
- Wilkins O., Nahal H., Foong J., Provart N. J., Campbell M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149, 981–993. 10.1104/pp.108.132795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V., Bauch M., Bennett T., Campiho A., Wolkenfelt H., Xu J., et al. (2008). The NAC domain transcription factors FEZ and SOMBRERO control the orientation of cell division plane in Arabidopsis root stem cells. Dev. Cell 15, 923–922. 10.1016/j.devcel.2008.09.019 [DOI] [PubMed] [Google Scholar]
- Wilson Z. A., Song J., Taylor B., Yang C. (2011). The final split: the regulation of anther dehiscence. J. Exp. Bot. 62, 1633–1649. 10.1093/jxb/err014 [DOI] [PubMed] [Google Scholar]
- Wolf S., Hématy K., Höfte H. (2012). Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 63, 381–407. 10.1146/annurev-arplant-042811-105449 [DOI] [PubMed] [Google Scholar]
- Wu A. M., Rihouey C., Seveno M., Hörnblad E., Singh S. K., Matsunaga T., et al. (2009). The Arabidopsis IRX10 and IRX10-LIKE glycosyltransferases are critical for glucuronoxylan biosynthesis during secondary cell wall formation. Plant J. 57, 718–731. 10.1111/j.1365-313X.2008.03724.x [DOI] [PubMed] [Google Scholar]
- Xu B., Ohtani M., Yamaguchi M., Toyooka K., Wakazaki M., Sato M., et al. (2014). Contribution of NAC transcription factors of plant adaptation to land. Science 343, 1505–1508. 10.1126/science.1248417 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Demura T. (2010). Transcriptional regulation of secondary wall formation controlled by NAC domain proteins. Plant Biotechnol. 27, 237–242 10.5511/plantbiotechnology.27.237 [DOI] [Google Scholar]
- Yamaguchi M., Goué N., Igarashi H., Ohtani M., Nakano Y., Mortimer J. C., et al. (2010a). VASCULAR-RELATED NAC-DOMAIN6 and VASCULAR-RELATED NAC-DOMAIN7 effectively induce transdifferentiation into xylem vessel elements under control of an induction system. Plant Physiol. 153, 906–914. 10.1104/pp.110.154013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Kubo M., Fukuda H., Demura T. (2008). VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55, 652–664. 10.1111/j.1365-313X.2008.03533.x [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Mitsuda N., Ohtani M., Ohme-Takagi M., Demura T. (2011). VASCULAR-RELATED NAC-DOMAIN 7 directly regulates the expression of broad range of genes for xylem vessel formation. Plant J. 66, 579–590. 10.1111/j.1365-313X.2011.04514.x [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Nagahage I. S. P., Ohtani M., Ishikawa T., Uchimiya H., Kawai-Yamada M., et al. (in press). Arabidopsis NAC domain proteins VND-INTERACTING1 and ANAC103 interact with multiple NAC domain proteins. Plant Biotechnol. 10.5511/plantbiotechnology.15.0208a [DOI] [Google Scholar]
- Yamaguchi M., Ohtani M., Mitsuda N., Kubo M., Ohme-Takagi M., Fukuda H., et al. (2010b). VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22, 1249–1263. 10.1105/tpc.108.064048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Xu Z., Song J., Conner K., Vizcay Barrena G., Wilson Z. A. (2007). Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534–548. 10.1105/tpc.106.046391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Mitra P., Zhang L., Prak L., Verhertbruggen Y., Kim J. S., et al. (2013). Engineering secondary cell wall deposition in plants. Plant Biotechnol. J. 11, 325–335. 10.1111/pbi.12016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanhui C., Xiaoyuan Y., Kun H., Meihua L., Jigang L., Zhaofeng G., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. 10.1007/s11103-005-2910-y [DOI] [PubMed] [Google Scholar]
- Yoshida K., Sakamoto S., Kawai T., Kobayashi Y., Sato K., Ichinose Y., et al. (2013). Engineering the Oryza sativa cell wall with rice NAC transcription factors regulating secondary wall formation. Front. Plant Sci. 4:383. 10.3389/fpls.2013.00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Zhao S., Yang S., Ding S. Y. (2014). Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr. Opin. Biotechnol. 27, 38–45. 10.1016/j.copbio.2013.09.008 [DOI] [PubMed] [Google Scholar]
- Zhao K., Bartley L. E. (2014). Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis, poplar, rice, maize, and switchgrass. BMC Plant Biol. 14:135. 10.1186/1471-2229-14-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Wang H., Yin Y., Xu Y., Chen F., Dixon R. A. (2010). Syringyl lignin biosynthesis is directly regulated by a secondary cell wall master switch. Proc. Natl. Acad. Sci. U.S.A. 107, 14496–14501. 10.1073/pnas.1009170107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Sun J., Xu P., Zhang R., Li L. (2014). Intron-mediated alternative splicing of WOOD-ASSOCIATED NAC TRANSCRIPTION FACTOR1B regulates cell wall thickening during fiber development in Populus species. Plant Physiol. 164, 765–776. 10.1104/pp.113.231134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Demura T., Ye Z. H. (2006). SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18, 3158–3170. 10.1105/tpc.106.047399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., McCarthy R. L., Reeves C. K., Jones E. G., Ye Z. H. (2011). Transcriptional activation of secondary wall biosynthesis by rice and maize NAC and MYB transcription factors. Plant Cell Physiol. 52, 1856–1871. 10.1093/pcp/pcr123 [DOI] [PubMed] [Google Scholar]
- Zhong R., Lee C., Ye Z. H. (2010a). Evolutionary conservation of the transcriptional network regulating secondary cell wall biosynthesis. Trends Plant Sci. 15, 625–632. 10.1016/j.tplants.2010.08.007 [DOI] [PubMed] [Google Scholar]
- Zhong R., Lee C., Ye Z. H. (2010b). Functional characterization of poplar wood associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055. 10.1104/pp.109.148270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Lee C., Ye Z. H. (2010c). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol. Plant. 3, 1087–1103. 10.1093/mp/ssq062 [DOI] [PubMed] [Google Scholar]
- Zhong R., Lee C., Zhou J., McCarthy R. L., Ye Z. H. (2008). A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 20, 2763–2782. 10.1105/tpc.108.061325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., McCarthy R. L., Haghighat M., Ye Z. H. (2013). The poplar MYB master switches bind to the SMRE site and activate the secondary wall biosynthetic program during wood formation. PLoS ONE 8:e69219 10.1371/journal.pone.0069219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E. A., Ye Z. H. (2007a). The MYB46 transcription factor is a direct target of SND1 and regulates secondary cell wall biosynthesis in Arabidopsis. Plant Cell 19, 2776–2791. 10.1105/tpc.107.053678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R., Richardson E. A., Ye Z. H. (2007b). Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225, 1603–1611. 10.1007/s00425-007-0498-y [DOI] [PubMed] [Google Scholar]
- Zhong R., Ye Z. H. (2012). MYB46 and MYB83 bind to the SMRE sites and directly activate a suit of transcription factors and secondary wall biosynthetic genes. Plant Cell Physiol. 53, 368–380. 10.1093/pcp/pcr185 [DOI] [PubMed] [Google Scholar]
- Zhou J., Lee C., Zhong R., Ye Z. H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266. 10.1105/tpc.108.063321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Zhong R., Ye Z. H. (2014). Arabidopsis NAC domain proteins, VND1 to VND5, are transcriptional regulators of secondary wall biosynthesis in vessels. PLoS ONE 9:e105726. 10.1371/journal.pone.0105726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Nevo E., Sun D., Peng J. (2012). Phylogenic analyses unravel the evolutionary of NAC proteins in plants. Evolution 66, 1833–1848. 10.1111/j.1558-5646.2011.01553.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.