Abstract
Staphylococcus aureus is an opportunistic pathogen that colonizes human hosts and causes a wide variety of diseases. Two interacting regulatory systems called agr (accessory gene regulator) and sar (staphylococcal accessory regulator) are involved in the regulation of virulence factors. The aim of this study was to evaluate the effect of vancomycin on hld and spa gene expression during the exponential and post-exponential growth phases in multidrug-resistant (MDR) S. aureus.
Methods: Antibiotic susceptibility was evaluated by the standard microdilution method. The phylogenetic profile was obtained by pulsed-field gel electrophoresis (PFGE). Polymorphisms of agr and SCCmec (staphylococcal cassette chromosome mec) were analyzed by multiplex polymerase chain reaction (PCR). The expression levels of hld and spa were analyzed by reverse transcription-PCR. An enzyme-linked immunosorbent assay (ELISA) was performed to detect protein A, and biofilm formation was analyzed via crystal violet staining.
Results: In total, 60.60% (20/33) of S. aureus clinical isolates were MDR. Half (10/20) of the MDR S. aureus isolates were distributed in subcluster 10, with >90% similarity among them. In the isolates of this subcluster, a high prevalence (100%) for the agrII and the cassette SCCmec II polymorphisms was found. Our data showed significant increases in hld expression during the post-exponential phase in the presence and absence of vancomycin. Significant increases in spa expression, protein A production and biofilm formation were observed during the post-exponential phase when the MDR S. aureus isolates were challenged with vancomycin.
Conclusion: The polymorphism agrII, which is associated with nosocomial isolates, was the most prevalent polymorphism in MDR S. aureus. Additionally, under our study conditions, vancomycin modified hld and spa expression in these clinical isolates. Therefore, vancomycin may regulate alternative systems that jointly participate in the regulation of these virulence factors.
Keywords: Staphylococcus aureus, multidrug-resistant, vancomycin, agr system, antibiotics
Introduction
Staphylococcus aureus is an opportunistic pathogen capable of causing a wide variety of diseases in humans, ranging from localized infections of the skin and soft tissues to life-threatening systemic infections (Archer, 1998; Shopsin and Kreiswirth, 2001; David and Daum, 2010; Sowash and Uhlemann, 2014). An infection is initiated when S. aureus has access to subcutaneous tissues and is disseminated by the circulatory system, and it infects nearly every organ, leading to severe osteomyelitis, sepsis, abscesses, endocarditis, pneumonia, and toxic shock syndrome (Archer, 1998; Shopsin and Kreiswirth, 2001; David and Daum, 2010; Sowash and Uhlemann, 2014). S. aureus can produce a remarkable array of wall surface and secreted virulence factors that contribute to the establishment and maintenance of infection (Novick et al., 1993). These cell surface virulence factors include microbial surface components that recognize extracellular matrix proteins, such as fibrinogen, laminin, plasminogen, vitronectin, fibronectin, thrombospondin, and bone sialoprotein (Falord et al., 2011; Yamamoto et al., 2013). The secreted virulence factors are generally produced during the post-exponential or stationary phase, and they include several extracellular toxins (i.e., alpha-, beta-, gamma-, and delta-hemolysin, enterotoxins, exfoliative toxins A and B, and toxic shock syndrome toxin-1) and exoenzymes (i.e., lipase, nucleases, proteases, hyaluronate lyase, and metalloproteases; Chan and Foster, 1998; Kolar et al., 2013).
The expression of virulence genes in S. aureus is regulated under the partial control of the two-component quorum-sensing system encoded by genes at the agr locus (Bronner et al., 2004). The P2 transcript (RNAII) encodes the quorum-sensing system, which consists of the following four proteins: AgrB (the secreted protein responsible for the export and processing of AgrD to its active form), AgrD (a signaling peptide), and AgrC-ArgA (a two-component system in which AgrC is the transmembrane receptor histidine kinase and AgrA is the DNA-binding response regulator; Novick et al., 1995; Gilot et al., 2002; Novick, 2003; Gilot and van Leeuwen, 2004). A high cell population density causes the activation of AgrA, which induces the transcription of the P3 promoter. Next, P3 drives the transcription of RNAIII, a regulatory RNA that is both a positive and negative regulator of virulence factor production. The activation of RNAIII transcription in response to an increase in cell population density induces a transition in gene expression correlated with metabolic changes and stress adaptations. Toxin- and extracellular enzyme-encoding genes are positively regulated by the agr-hld (δ-lysin gene) system, and the genes coding for protein A (spa) and coagulase are negatively regulated (Morfeldt et al., 1988). Protein A has a molecular weight of 42 kDa and is covalently anchored to the peptidoglycan of S. aureus (Palmqvist et al., 2002). Ninety percent of the molecule is localized in the cell wall, and 10% is in the bacterial cytoplasm. Protein A is an important virulence factor of S. aureus based on its ability to bind to a variety of ligands, including the Fc region of IgG, the von Willebrand factor, tumor necrosis factor receptor-1 (TNFR-1), the Fab-heavy chains of the Vh3 subclass, and the epidermal growth factor receptor (EGFR; Cedergren et al., 1993; Viau and Zouali, 2005; Gómez et al., 2006; O’Seaghdha et al., 2006). An increase in protein A during the post-exponential phase has been observed in agr-deleted S. aureus strains (Novick, 2003).
In addition, AgrA activation leads to increased transcription of the δ-lysin gene (hld), which is located immediately upstream of the agr operon (Janzon and Arvidson, 1990). δ-lysin is a small polypeptide of only 26 amino acids. It is secreted without a signal peptide, and it makes cation-selective channels in the phospholipid bilayers (Lee and Birkbeck, 1984). δ-lysin is a virulence factor with lytic activity in a wide range of cells, such as neutrophils, macrophages, mammalian erythrocytes, and bacterial protoplasts, as well as in cellular organelles (Julander et al., 1983).
The aim of this study was to assess agr system expression by quantifying hld and spa expression in multidrug-resistant (MDR) S. aureus clinical isolates cultured from the exponential to post-exponential growth phases in the presence of vancomycin. In addition, the agr group I–IV polymorphisms were evaluated as a factor that predisposes the permanence and survival of MDR S. aureus clinical isolates during nosocomial or hospital-acquired infections in the Hospital Infantil de México Federico Gómez (HIMFG).
Materials and Methods
Bacterial Isolates
Thirty-three S. aureus from different infections were obtained from a clinical isolates collection at the Central Laboratory of the HIMFG. The S. aureus isolates were collected from January 2006 to June 2007. They were cultured on 5% sheep blood agar plates (Becton Dickinson, East Rutherford, NJ, USA) at 37°C under 5% CO2 for 24 h and kept at -70°C in skim milk (Becton Dickinson, East Rutherford, NJ, USA).
Diagnostic Tests for Identifying S. aureus
The bacteria were grown on blood agar, and identification was performed using conventional bacteriological techniques, such as colony morphology examination, catalase assays, coagulase assays, Gram staining (Sigma-Aldrich, St. Louis, MO, USA), mannitol fermentation, and Brain Heart Infusion (BHI) broth growth assays (Becton Dickinson, East Rutherford, NJ, USA) with 15% NaCl (MacFadin, 1996).
Antimicrobial Susceptibility
The antibiotic susceptibility profiles of the S. aureus isolates were determined by the Minimum Inhibitory Concentration (MIC) technique with the microdilution method in Mueller-Hinton broth (MH; Becton Dickinson, East Rutherford, NJ, USA), as recommended by the Clinical and Laboratory Standards Institute (2014). The MIC tests were conducted with vancomycin, ciprofloxacin, erythromycin (MP Biomedicals, Solon, OH, USA), clarithromycin (Grünenthal Gmbh, Aachen, Germany), oxacillin, clindamycin, linezolid (Sigma-Aldrich, St. Louis, MO, USA), meropenem (AstraZeneca Pharmaceuticals LP, Wilmington, DE, USA), trimethoprim, sulfamethoxazole (Roche, Basel, Switzerland), and gentamicin (Schering-Plough Pharmaceuticals, Kenilworth, NJ, USA). To identify methicillin-resistant S. aureus clinical isolates, the bacteria were tested for oxacillin resistance by the oxacillin-salt screening method. Oxacillin is a more stable antibiotic than methicillin, although they are chemically identical. S. aureus strain ATCC 29213 (American Type Culture Collection, Manassas, VA, USA) was used as a positive control.
Molecular Genotyping Assays
Pulsed-field gel electrophoresis (PFGE) was performed using a previously described protocol (Pereira et al., 2014). The chromosomal DNA from MDR and sensitive S. aureus isolates was digested with the Sma I restriction enzyme (Thermo Fisher Scientific Inc., Life Technologies, Grand Island, NY, USA) and subjected to electrophoresis on 1% agarose gels (Promega; Madison, WI, USA) using the following parameters: 200 V (6 v/cm) at 14°C for 21.5 h, with an initial change of 5 s and a final change of 40 s. The gels were stained with 0.5 μg/mL ethidium bromide solution (Sigma-Aldrich, St. Louis, MO, USA) and visualized using a gel imaging system (ChemiDocTM MP System, Biorad, Hercules, CA, USA). The DNA fragment patterns generated by PFGE were analyzed with NTSY-pc software (version 2.0, Applied Biostatistics, Inc., Port Jefferson, NY, USA; Ramazanzadeh et al., 2013) using the Sørensen–Dice similarity coefficient and the unweighted pair group method with arithmetic mean (UPGMA) clustering approach (Dice, 1945).
Multiplex Polymerase Chain Reaction (PCR) Conditions
The S. aureus clinical isolates were recovered from frozen stock onto BHI agar plates and incubated at 37°C for 18–24 h. Genomic DNA extraction was performed with a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) from a bacterial culture grown in BHI broth. Briefly, the bacterial culture pellet was mixed with TE buffer (10 mM Tris HCl and 1 mM EDTA, pH 8.0), lysozyme (0.25 mg/mL; Sigma-Aldrich, St. Louis, MO, USA), proteinase K (0.0125 mg/mL; Sigma-Aldrich, St. Louis, MO, USA), and lysostaphin (0.062 mg/mL; Sigma-Aldrich, St. Louis, MO, USA). Multiplex polymerase chain reaction (PCR) assays for detecting agr polymorphisms (agrI, agrII, agrIII, and agrIV; Table 1) were prepared according to the protocol for Go Taq Green Master Mix (Promega, Madison, WI, USA). Multiplex PCR reactions were prepared in a final volume of 25 μL as follows: 12.5 μL of Go Taq Green Master Mix 2x (Promega, Madison, WI, USA), 2 μL of bacterial DNA (100 ng/μL), 5 μL of agr primers (agrI, agrII, agrIII, and agrIV) at 10 pg/μL, and 5.5 μL of nuclease-free water. The DNA amplification was performed in a Veriti 96-Well Thermal Cycler-Life Technologies (Applied Biosystems, Foster City, CA, USA) with the following parameters: an initial denaturation at 94°C for 5 min followed by 26 amplification cycles (denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 60 s), ending with a final extension at 72°C for 7 min. An external positive control [DNA extracted from S. aureus strains USA300 (agrI), 1749 (agrII), and ATCC 25923 (agrIII)] and an external negative control (DNase/RNase-free distilled water) were included with each run. PCR amplicons (10 μL) were loaded into a 1.5% (wt/v) agarose gel (Promega, Madison, WI, USA) using a 100 bp DNA ladder (Promega, Madison, WI, USA), and electrophoresis was performed in 1x TAE buffer at 100 V for 1 h. The bands were visualized using a gel imaging system (ChemiDocTM MP System, Biorad, Hercules, CA, USA).
Table 1.
Primers used in agr and SCCmec typing by multiplex PCR.
| Gene | Primer | Sequence (5′–3′) | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| agrI |
agr I-F agr I-R |
ATGCACATGGTGCACATGC GTCACAAGTACTATAAGCTGCGAT |
441 | Gilot et al. (2002) |
| agrII |
agr II-F agr II-R |
ATGCACATGGTGCACATGC TATTACTAATTGAAAAGTGGCCATAGC |
575 | Gilot et al. (2002) |
| agrIII |
agr III-F agr III-R |
ATGCACATGGTGCACATGC GTAATGTAATAGCTTGTAAAAAGTGGCCATAGC |
323 | Gilot et al. (2002) |
| agrIV |
agr IV-F agr IV-R |
ATGCACATGGTGCACATGC CGATAATGCCGTAATACCCG |
659 | Gilot et al. (2002) |
| SCCmec I |
mec I-F mec I-R |
GCTTTAAAGAGTGTCGTTACAGG GTTCTCTCATAGTATGACGTCC |
613 | Zhang et al. (2005) |
| SCCmec II |
mec II-F mec II-R |
CGTTGAAGATGATGAAGCG CGAAATCAATGGTTAATGGACC |
398 | Zhang et al. (2005) |
| SCCmec III |
mec III-F mec III-R |
CCATATTGTGTACGATGCG CCTTAGTTGTCGTAACAGATCG |
280 | Zhang et al. (2005) |
| SCCmec IVa |
mec IVa-F mec IVa-R |
GCCTTATTCGAAGAAACCG CTACTCTTCTGAAAAGCGTCG |
776 | Zhang et al. (2005) |
| SCCmec V |
mec V-F mec V-R |
GAACATTGTTACTTAAATGAGCG TGAAAGTTGTACCCTTGACACC |
325 | Zhang et al. (2005) |
| mecA |
mec 147-F mec 147-R |
GTGAAGATATACCAAGTGATT ATGCGCTATAGATTGAAAGGAT |
147 | Zhang et al. (2005) |
F, forward; R, reverse.
The SCCmec genes (I, II, III, and IVa) were characterized by multiplex PCR according to Cázares-Domínguez et al. (2015).
RNA Extraction from S. aureus Clinical Isolates
Staphylococcus aureus isolates that were grown overnight were adjusted to an optical density of 0.05 at 600 nm and incubated in BHI with 1 μg/mL vancomycin. The bacterial cultures were grown for 4 h until reaching the exponential phase (OD600 of 0.6–0.8) and for 11 h just in the post-exponential phase (OD600 of 1.2–1.4). Aliquots of bacterial cultures were harvested by centrifugation at 10,000 × g for 3 min at 4°C. Each pellet was washed in an equal volume of TE buffer (10 mM Tris HCl and 1 mM EDTA, pH 8.0) three times and lysed with the same buffer supplemented with 0.25 mg/mL lysozyme, 0.0125 mg/mL proteinase K, and 0.062 mg/mL lysostaphin. Total bacterial RNA was isolated (TRIzol, Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s directions. After purification, contaminating DNA was removed with RNase-free DNase I (2 U/10 μg of total bacterial RNA at 37°C for 30 min). The RNA was then re-purified with RNeasy Minicolumns (Qiagen Incorporated, Ln Valencia, CA, USA). The amount of recovered RNA was determined spectrophotometrically, and the samples were then stored at -80°C.
Transcriptional Expression Analyses of the spa and hld Genes
The relative expression levels of the spa and hld genes were determined by cDNA-PCR. The purified RNA of all S. aureus clinical isolates was employed for reverse transcription (RT)-PCR assays with the GeneAmp RNA PCR Kit (Applied Biosystems, Foster City, CA, USA), using specific primers for the spa (encoding protein A) and hld (encoding a delta toxin) genes (Table 2). A GeneAmp RNA PCR Kit was used with 0.2 μg of total RNA per reaction as a template for PCR amplification. Reactions containing S. aureus cells alone, only RNA, or no reverse transcriptase were used as negative controls. Specific primers were used for the amplification of 16S RNA, which was used as an internal control (Table 2).
Table 2.
Primers used in hld and spa expression analysis by RT-PCR.
| Gene | Primer sequence (5′–3′) | Product size (bp) | Reference |
|---|---|---|---|
| spa | TATCTGGTGGCGTAACACCTG GATGAAGCCGTTACGTTGTTC |
322 | Goerke et al. (2000) |
| hld | GAAGGAGTGTTTCAATGG TAAGAAAATACATAGCACTGAG |
260 |
Goerke et al. (2000) |
| 16S | TCCGGAATTATTGGGCGTAA CCACTTTCCTCTTCTGCACTCA |
121 | Goerke et al. (2000) |
The expression levels of spa and hld transcripts from S. aureus clinical isolates were quantified by densitometric analysis with Bio-Rad image software (Bio-Rad chemi-doc, Quantity one 4.4.1). The data are expressed as the mean ± standard error of the means. A p-value of less than 0.05 was considered significant. All experiments were repeated at least three times, and a representative result is shown for each experiment.
Quantitative Measurements of Protein A
A qualitative screening test for the production of protein A was conducted by an enzyme linked immunosorbent assay (ELISA). Briefly, 96-well plates containing 200 μL of BHI were inoculated with 10 μL (1.5 × 108 bacteria/mL) of bacterial suspensions and incubated at 37°C in the presence or absence of vancomycin to the exponential phase (4 h) and post-exponential phase (11 h). Cell wall-associated protein A was identified using anti-protein A, followed by mouse anti-IgG antibodies, o-phenylenediamine (OPD) compounds, and ELISA as previously described (Ohkaru et al., 1995).
Quantitative Determination by Biofilm Assays
Biofilm formation was quantitatively analyzed according to the protocol described by Erdem et al. (2008). MDR and sensitive S. aureus clinical isolates were grown in BHI broth overnight at 37°C. Then, 96-well plates containing 200 μL of BHI were inoculated with 10 μL (1.5 × 108 bacteria/mL) of bacterial suspensions and grown at 37°C in the presence or absence of vancomycin in the exponential phase (4 h) and post-exponential phase (11 h). The biofilms that developed on the surfaces of the wells were gently washed three times with 1x phosphate-buffered saline (PBS; pH 7.4) and fixed with 2% formaldehyde at 4°C overnight. Wells with fixed biofilms were decanted, washed three times with PBS and stained with 200 μL of 1% crystal violet for 30 min. The excess crystal violet was removed, and the plates were washed twice with water. Crystal violet was subsequently solubilized in 70% methanol, and the absorbance was determined at 620 nm. Assays were performed in triplicate and repeated three consecutive times.
Results
The Antimicrobial Susceptibility Testing of S. aureus Clinical Isolates
Thirty-three S. aureus clinical isolates were tested for antimicrobial susceptibility. In total, 60.60% (20/33) of S. aureus clinical isolates were MDR, 100% (33/33) were sensitive to four antibiotics (vancomycin, trimethoprim/sulfamethoxazole, gentamicin, and linezolid), and 39.39% (13/33) were sensitive to all antimicrobials (Table 3).
Table 3.
MIC analysis for the MDR and sensitive S. aureus clinical isolates.
| Clinical isolates | Antibiotics (μg/mL) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CEC | CEP | VAN | CLA | CIP | STX | OXA | GEN | ERI | MEM | LZN | |
| 488H, 428H, 10H, 714H | ≥128 | ≥128 | 1 | ≥128 | 64 | 0.5 | 16 | 1 | ≥128 | 64 | 2 |
| 242H, 143H | ≥128 | ≥128 | 1 | ≥128 | ≥128 | 2 | ≥128 | 0.5 | ≥128 | 32 | 4 |
| 246H, 175H | ≥128 | ≥128 | 1 | ≥128 | ≥128 | 1 | ≥128 | 0.5 | ≥128 | 16 | 4 |
| 882HR | ≥128 | ≥128 | 1 | ≥128 | 64 | 1 | ≥128 | 1 | ≥128 | 32 | 2 |
| 425LCR | ≥128 | ≥128 | 1 | ≥128 | 64 | 0.5 | 16 | 0.5 | ≥128 | 64 | 2 |
| 330H | ≥128 | ≥128 | 1 | ≥128 | ≥128 | 2 | ≥128 | 1 | ≥128 | 32 | 4 |
| 783H | ≥128 | ≥128 | 1 | ≥128 | ≥128 | 2 | ≥128 | 1 | ≥128 | 32 | 4 |
| 902H | ≥128 | ≥128 | 1 | ≥128 | 64 | 1 | ≥128 | 2 | ≥128 | 32 | 4 |
| 54H | ≥128 | ≥128 | 0.5 | ≥128 | 64 | 0.5 | 16 | 0.5 | ≥128 | 32 | 2 |
| A-32 | ≥128 | ≥128 | 0.5 | ≥128 | ≥128 | 0.5 | ≥128 | 0.5 | ≥128 | 16 | 2 |
| 828H | 8 | ≥128 | 1 | ≥128 | 64 | 0.5 | 16 | 0.5 | ≥128 | 64 | 2 |
| 931H | 8 | ≥128 | 1 | ≥128 | 64 | 0.5 | 16 | 1 | ≥128 | 64 | 2 |
| 260H | 0.125 | 64 | 0.5 | 32 | 0.25 | 0.5 | 64 | 0.5 | ≥128 | 8 | 0.5 |
| 622H | 0.125 | ≥128 | 1 | 0.06 | 0.25 | 1 | 32 | 0.5 | 0.25 | 4 | 4 |
| 299H | 0.060 | ≥128 | 1 | ≥128 | 32 | 0,5 | 8 | 0.5 | ≥128 | 0.12 | 0.5 |
| 679H, 318LCR, 573H, 770H, 633H, 779H, 291H, 336H, 18H, 108H, 780H, 493H, 198H | ≤0.25 | ≤8 | ≤1 | ≤0.25 | ≤2 | ≤1 | ≤1 | ≤1 | ≤1 | ≤0.125 | ≤4 |
| CVR∗ | ≥32 | ≥32 | ≥16 | ≥8 | ≥4 | ≥4 | ≥4 | ≥16 | ≥8 | ≥16 | ≥8 |
CEC, cefaclor; CEP, cephalothin; VAN, vancomycin; CLA, clarithromycin; CIP, ciprofloxacin; STX, trimethoprim/sulfametoxazole; OXA, oxacillin; GEN, gentamicin; ERI, erythromycin; MEM, meropenem; LZN, linezolid. The MDR S. aureus clinical isolates are marked in gray. ∗Cut-off values for resistance to MIC (μg/mL; CVR).
Molecular Typing Analysis of the MDR and Sensitive S. aureus Isolates by PFGE
A total of 19 DNA pulsotypes grouped in four clusters (I–IV) were identified, revealing patterns that consisted of 11–18 DNA fragments ranging in size from 48.5 to 339.5 Kb (Figure 1). In total, 3.03% (1/33) of the S. aureus clinical isolates were identified as pulsotype F, and cluster I showed 38% genetic similarity when compared with other pulsotypes. Four pulsotypes (E, I, K, and M) belonging to cluster II included 21.21% (7/33) of the S. aureus clinical isolates with 64% genetic similarity. In addition, the nine pulsotypes (H, J, L, N, O, P, Q, R, and S) organized in cluster III included 54.55% (18/33) of the S. aureus clinical isolates with 56% genetic similarity. In this cluster, the S. aureus clinical isolates with >90% genetic similarity were grouped in subcluster 10 and distributed in the following order: two isolates in pulsotype Q, one isolate in pulsotype R, and eight isolates in pulsotype J (Figure 1). In addition, 21.21% (7/33) of the S. aureus clinical isolates were classified as cluster IV with 62% genetic similarity and distributed as pulsotypes A, B, C, D, and G (Figure 1).
FIGURE 1.
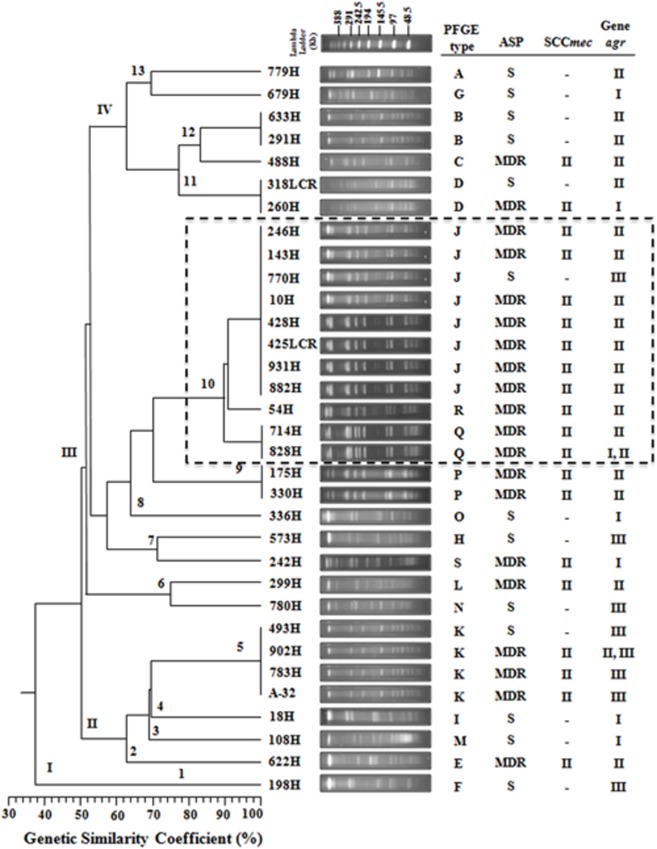
Dendrogram analysis of PFGE results showing the genetic relationships among the PFGE profiles and the presence of agr and SCCmec type among the 33 S. aureus clinical isolates. A phylogenetic analysis was conducted using the Sørensen–Dice similarity coefficient in association with the UPGMA algorithm as the grouping method. The dendrogram was evaluated by obtaining the cophenetic correlation coefficient with the Mantel test, which yielded an r-value of 0.9077. Antimicrobial susceptibility profile (ASP), multidrug-resistant (MDR), sensitive (S). The dotted line indicates that the clinical isolates that belong to subcluster 10 with >90% genetic similarity.
Only one S. aureus clinical isolate sensitive to all antibiotics was identified (pulsotype F of cluster I; Figure 1). Four MDR and three sensitive S. aureus clinical isolates were distributed in the four pulsotypes (E, I, K, and M) of cluster II (Figure 1). Fourteen MDR and four sensitive S. aureus clinical isolates were distributed into the nine pulsotypes (H, J, L, N, O, P, Q, R, and S) of cluster III. Interestingly, seven MDR and one sensitive S. aureus clinical isolates that were distributed in pulsotype J were organized in subcluster 10. In addition, two MDR and five sensitive S. aureus clinical isolates were distributed over the five pulsotypes (A, B, C, D, and G) of cluster IV (Figure 1).
Distributing agr, SCCmec, hld, and spa Genes by Multiplex PCR Endpoint Analysis
Multiplex PCR amplification assays were performed for agr-specific group identification in the selected MDR and sensitive S. aureus clinical isolates. Our results showed three different agr specificity groups (Figures 2A,B), which were identified according to the expected product sizes (Table 1). Briefly, the sensitive S. aureus clinical isolates were 30.76% (4/13) agrI, 30.76% (4/13) agrII, and 38.46% (5/13) agrIII (Figures 2B,C). The MDR S. aureus clinical isolates were 70% (14/20) agrII, 10% (2/20) agrI, and 10% (2/20) agrIII (Figures 2A,C). Furthermore, both the agrI/II and agrII/III polymorphism groups were identified in a single (1/20; 5%) clinical isolate of MDR S. aureus (Figures 2A,C). The expression of the agrIV was not identified in either the MDR or sensitive S. aureus clinical isolates.
FIGURE 2.
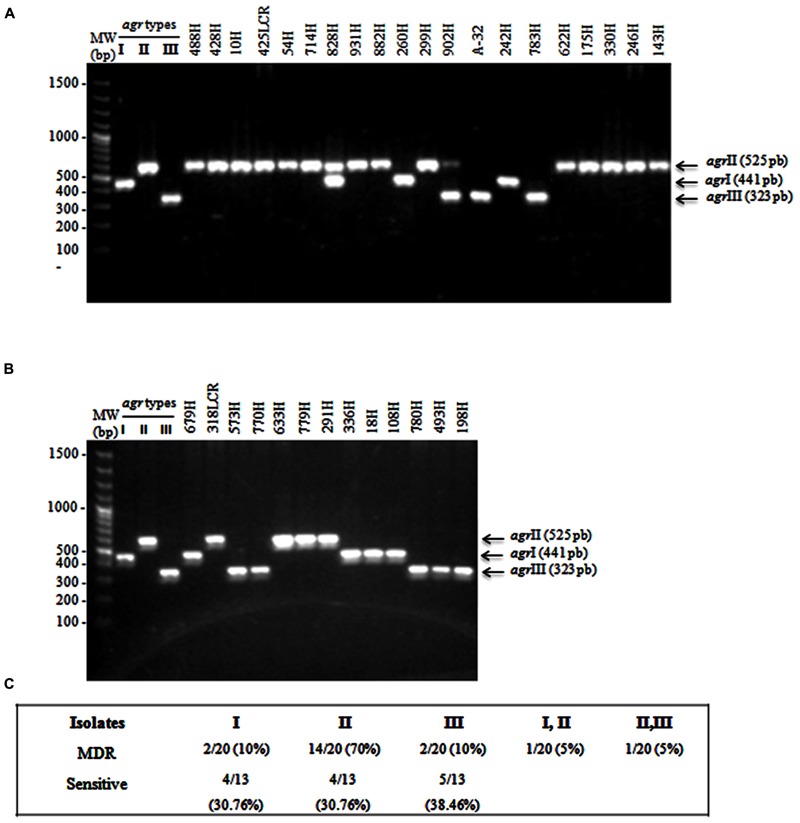
Multiplex PCR assays for identifying agr types. (A) MDR S. aureus clinical isolates showing the agrI (441 bp), agrII (575 bp), and agrIII (323 bp) types. (B) Sensitive S. aureus clinical isolates. (C) The percentages of agr types that were identified in MDR and sensible S. aureus clinical isolates. S. aureus strain USA300 (agrI), 1749 (agrII), and ATCC 25923 (agrIII) were used as reference strains. MW, molecular weight (bp).
A 398 bp product corresponding to the SCCmec group II polymorphism was present in 60.60% (20/33) of the MDR S. aureus clinical isolates that were distributed in the fourth cluster as determined by PFGE. A SCCmec II polymorphism was observed in 12.12% (4/33) of the isolates in cluster II, 42.42% (14/33) of the isolates in cluster III, and 6.06% (2/33) of the isolates in cluster IV. However, the SCCmec II polymorphism was not identified in cluster I (Figure 1). Moreover, SCCmec polymorphism types I, III, and IV were not identified in the MDR S. aureus clinical isolates (Figure 1). It is important to emphasize that the presence of the SCCmec gene explains methicillin (oxacillin) resistance; therefore, it was not identified in sensitive S. aureus clinical isolates (Figure 1).
The frequencies of the hld and spa genes were determined using specific primers to be 100% (33/33) in MDR and sensitive S. aureus clinical isolates. Our results showed two bands, which corresponded to hld at 260 bp and spa at 322 bp (data not shown).
hld and spa Gene Expression as Determined by RT-PCR
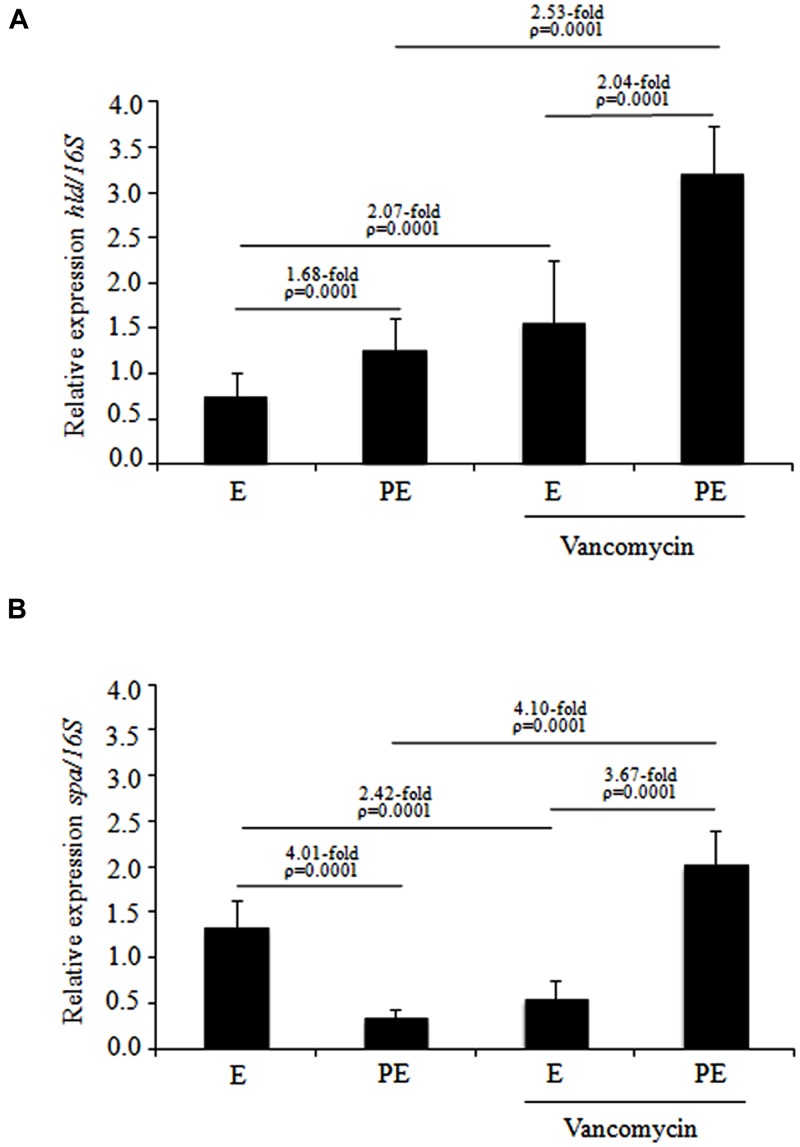
The specific transcripts of the spa and hld genes produced by the ten MDR S. aureus isolates that were distributed in subcluster 10 of cluster III were quantified by RT-PCR-densitometry. These MDR S. aureus isolates were treated with and without vancomycin during two different stages of the growth phase (exponential and post-exponential). The hld expression in MDR S. aureus clinical isolates showed a significant increase (1.68-fold; p = 0.0001) when cultured from the exponential to the post-exponential growth phase in the absence of vancomycin as well as a significant increase (2.04-fold; p = 0.0001) when grown in the presence of vancomycin (Figure 3A). Interestingly, under the same test conditions, a significant increase (2.07-fold; p = 0.0001) in hld expression was observed in MDR S. aureus clinical isolates in the exponential growth phase treated with vancomycin compared to those treated without vancomycin (Figure 3A). In addition, a significant increase (2.53-fold; p = 0.0001) in hld expression was observed in the MDR S. aureus clinical isolates cultured to the post-exponential growth phase when challenged with 1 μg/mL vancomycin compared to without vancomycin challenge (Figure 3A). The hld expression in sensitive S. aureus isolates showed increases from the exponential to the post-exponential growth phase in both the presence and absence of vancomycin (data not shown).
FIGURE 3.
Expression levels of the hld and spa genes determined by RT-PCR in MDR S. aureus clinical isolates cultured from the exponential to post-exponential growth phases in the presence of vancomycin. (A) hld expression; (B) spa expression. The densitometric results of the transcripts were quantified using the Quantity One program and analyzed by paired Student’s t-tests. E, exponential; PE, post-exponential.
The spa expression level showed a significant reduction (4.01-fold; p = 0.0001) when the MDR S. aureus clinical isolates were cultured from the exponential to post-exponential growth phases in the absence of vancomycin (Figure 3B). Interestingly, a remarkable increase (3.67-fold; p = 0.0001) in spa expression was observed from the exponential to the post-exponential phases in MDR S. aureus clinical isolates cultured in the presence of vancomycin (Figure 3B). During the exponential phase, a significant reduction in spa expression (2.42-fold; p = 0.0001) was observed when the MDR S. aureus clinical isolates were challenged with vancomycin compared to those grown in the absence of vancomycin; the MDR S. aureus clinical isolates in the post-exponential growth phase showed a significant increase (4.10-fold; p = 0.0001) in spa expression when challenged with vancomycin compared to those grown in the absence of vancomycin (Figure 3B). The spa expression level did not show a significant changes (reduction and/or increase) when the sensitive S. aureus clinical isolates were cultured in the exponential and post-exponential growth phases in the presence or absence of vancomycin (data not shown).
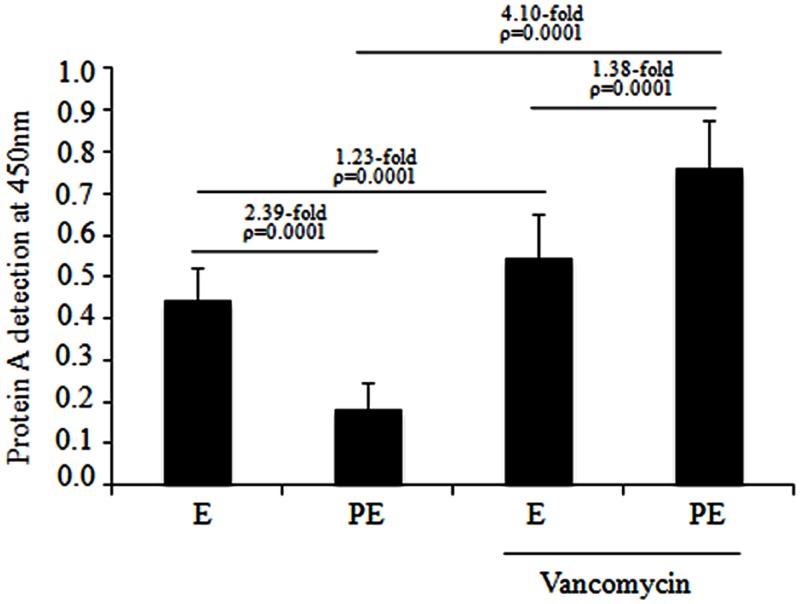
Protein A Immunodetection by ELISA
Protein A immunodetection assays were performed for the MDR S. aureus clinical isolates distributed in subcluster 10 of cluster III. Quantitative analysis showed a significant reduction (2.39-fold; p = 0.0001) in protein A expression in the post-exponential phase compared to the exponential phase in the absence of vancomycin (Figure 4). Furthermore, a significant increase (1.38-fold; p = 0.0001) in protein A expression was observed from the exponential to the post-exponential growth phase in the MDR S. aureus clinical isolates cultured the presence of 1 μg/mL vancomycin. Interestingly, a significant increase (4.10-fold; p = 0.0001) in protein A expression was observed in these clinical isolates cultured to the post-exponential growth phase in the presence of vancomycin compared to in the absence of vancomycin (Figure 4). Likewise, a significant increase (1.23-fold; p = 0.0001) in protein A expression was observed in the MDR S. aureus clinical isolates that were cultured to the exponential growth phase in the presence of vancomycin compared to in the absence of vancomycin.
FIGURE 4.
Protein A production in the MDR S. aureus clinical isolates cultured from the exponential to post-exponential growth phases in the presence of vancomycin. A significant reduction of 2.39-fold (p = 0.0001) was observed in the MDR S. aureus clinical isolates when they were cultured from the exponential to post-exponential growth phases in the absence of vancomycin and there was a significant increase of 1.38-fold (p = 0.0001) when cultured from the exponential to post-exponential growth phase when challenged with 1 μg/mL vancomycin.
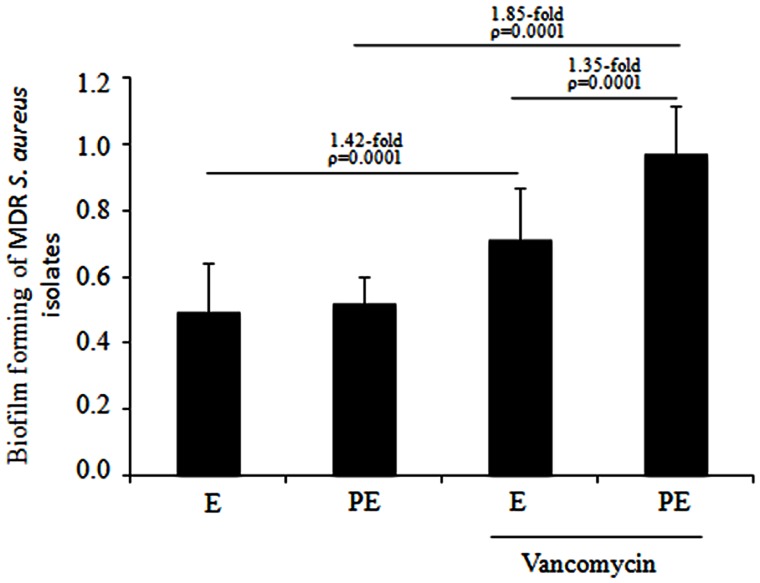
Clinical Strains of MDR and Sensitive S. aureus Isolates Produce Biofilms
A crystal-violet biofilm assay was performed for the MDR S. aureus clinical isolates that were distributed in subcluster 10 of cluster III. Quantitative analysis showed no difference in biofilm formation when the MDR S. aureus clinical isolates were cultured to the exponential and post-exponential growth phases in the absence of vancomycin. In the presence of vancomycin, a significant increase (1.42-fold; p = 0.0001) was observed in the biofilm formation of the MDR S. aureus clinical isolates cultured to the exponential growth phase and a significant increase (1.85-fold; p = 0.0001) was observed in the isolates cultured to the post-exponential growth phase. Furthermore, a significant increase (1.35-fold; p = 0.0001) in the biofilm formation of these isolates was observed from the exponential to post-exponential growth phases in the presence of vancomycin (Figure 5).
FIGURE 5.
Biofilm assays of the MDR S. aureus clinical isolates when cultured from the exponential to post-exponential growth phases in the presence of vancomycin. Interestingly, a significant increase (1.35-fold; p = 0.0001) in biofilm formation was observed in the MDR S. aureus clinical isolates cultured from the exponential to post-exponential growth phases in the presence of the vancomycin.
Discussion
In this study, 100% of S. aureus clinical isolates showed sensitive to vancomycin, trimethoprim/sulfamethoxazole, gentamicin, and linezolid. Low rates of resistance to tetracycline, gentamicin, and trimethoprim/sulfamethoxazole in clinical methicillin-resistant staphylococcus aureus (MRSA) isolates have been described in other studies (Bordon et al., 2010; Davis et al., 2013). However, the impressive ability of S. aureus to develop resistance against various antibiotics through point mutations, transposons, plasmids, and resistance cassettes is the most likely reason for the rapid increase in antimicrobial resistance worldwide in recent years (Sakoulas and Moellering, 2008). High levels of erythromycin resistance and increasing ciprofloxacin resistance in MRSA clinical isolates have been observed (Davis et al., 2013). The clinical condition, the administration route, and the resistance pattern of the organism are also risk factors that are considered in treating MRSA infections when drugs such as vancomycin, linezolid, daptomycin, clindamycin, and mupirocin are administered (Liu et al., 2011). The increase of vancomycin-resistant S. aureus (VRSA) isolates in the United States could be explained by the selective pressure originating from the excessive use of vancomycin to treat MRSA infections (David and Daum, 2010). The high frequency of resistance to the antibiotics tested in our study may reflect the fact that the public health service in Mexico prescribes all of these antibiotics for treating pediatric patients; these drugs are considered to be essential in this health sector.
According to the PFGE analysis, 33.33% (11/33) of the S. aureus clinical isolates distributed in subcluster 10 shared 90% similarity. In particular, 50% of the MDR S. aureus clinical isolates were distributed in subcluster 10 and showed closely related pulsotypes with three clones that were assigned as J, Q, and R according to the criteria of Tenover et al. (1995). These results suggest that MDR S. aureus isolates are associated with patients with infections acquired during their current hospital stay. MRSA isolates with a highly related PFGE type have been associated with an MDR profile to β-lactams, gentamicin, ciprofloxacin, clindamycin, and erythromycin (Velazquez-Meza et al., 2004). In total, 90.90% (10/11) of the S. aureus clinical isolates belonging to subcluster 10 were resistant to six antibiotics, namely cephalothin, clarithromycin, ciprofloxacin, oxacillin, erythromycin, and meropenem. Furthermore, 72.72% (8/11) of these clinical isolates were resistant to cefaclor.
Our data showed a high prevalence of the agr group II polymorphism, with a PCR-amplified product of 575 kb, in the MDR S. aureus clinical isolates belonging to subcluster 10 by PFGE analysis. Several studies performed in Japan and the USA described the agr group II polymorphism as the agr type that was most frequent in MDR S. aureus clinical isolates, and it has been associated with nosocomial infections from pediatric patients (Sakoulas et al., 2003). Similar findings indicated that all S. aureus clinical isolates from diverse geographic origins and those recovered from patients undergoing intubation showed the agr group II polymorphism (Sakoulas et al., 2002; Goerke et al., 2003; Moise et al., 2004). Furthermore, the agr group II polymorphism in MRSA predicts the failure of vancomycin therapy (Moise et al., 2007).
We further evaluated agr expression through indirect mechanisms by quantification of hld and spa expression in MDR S. aureus clinical isolates during the exponential and post-exponential growth phases and upon vancomycin challenge. Our RT-PCR analysis showed that hld activation occurs during the post-exponential growth phase, resulting in an increase of 1.68-fold without vancomycin and 2.04-fold with vancomycin compared with the exponential phase. The increased hld expression in the MDR S. aureus clinical isolates of subcluster 10 suggested that the activation of this gene is influenced by the growth phase and vancomycin challenge. Other studies have shown significant increases in hld expression at the end of the exponential growth phase by an agr-dependent mechanism that is involved in the regulation of virulence genes. agrA codes for a protein that can activate hld transcription as a response to the growth phase (Janzon and Arvidson, 1990). Our data indicated that the MDR S. aureus isolates were stimulated by vancomycin at subinhibitory concentrations, as indicated by increases in hld expression. These results indicate that the antibiotic stimulates the agr system. In a previous study, the over-expression of the hld transcript in vancomycin-resistant S. aureus strains associated with activation of the sigma factor was observed when these isolates were exposed to subinhibitory antibiotic concentrations (Chen et al., 2011). Recently, community-associated (CA) MRSA was challenged with subinhibitory concentrations of tetracycline and clindamycin, which had a strong stimulatory effect on the activity of the agr operon (Joo et al., 2010). These results, together with the data obtained in our studies, suggest that vancomycin also exerts a strong stimulatory effect on the activity of the agr operon. It is important to note that agr controls many virulence factors of S. aureus and that vancomycin is frequently used in Mexico for treating MRSA infections in pediatric patients.
Interestingly, a reduction in spa expression was observed during the exponential to post-exponential growth phases when MDR S. aureus clinical isolates were cultured without vancomycin. In contrast, a significant increase (3.67-fold) in spa expression was observed when MDR S. aureus clinical isolates were only cultured with vancomycin during the post-exponential growth phase. These data suggest that spa activation or repression is regulated by the growth phase and that expression of spa mRNA is modified by drugs such as vancomycin. In previous studies, CA-MRSA isolates treated with daptomycin and vancomycin showed no change in spa mRNA and SpA protein levels (Subrt et al., 2011; Otto et al., 2013). However, in a different study, the expression levels of different virulence factors in the CA-MRSA isolates were suppressed in the presence of clindamycin and linezolid (Otto et al., 2013).
The expression levels of the spa gene are directly correlated with protein A production because most of the gene transcript is translated as protein A. The localization of protein A in the cell wall could contribute to biofilm formation in MRSA clinical isolates during the post-exponential phase and when challenged with vancomycin. The XdrA regulator has almost as strong an activating effect on spa as SarS, and it acts on the same spa operator regions (identified potential cis-acting regulatory regions) as SarS or on closely overlapping regions (McCallum et al., 2010). The current evidence suggests that XdrA directly regulates spa transcripts independently of other well-characterized regulators. The over-expression of spa transcripts and protein A in the MDR S. aureus clinical isolates challenged with vancomycin could also be related to the action of the regulator XdrA, which should be examined in future studies. In addition, the variation in the levels of spa transcripts during different growth phases is likely due to the influence of other regulators and/or to the characteristics of clinical isolates.
Conclusion
Staphylococcus aureus clinical strains are influenced by intercellular signaling through the agr system, which modulates the activation and/or repression of many outer membrane proteins associated with biofilms (Cafiso et al., 2012). Vancomycin is the antibiotic of choice for treating nosocomial infections of MDR S. aureus clinical isolates in pediatric patients at the HIMFG. These data provide a direct evidence for evaluating the role of virulence genes (hld and spa) associated with agr regulation and suggest the participation of other regulatory elements; which are include an operon that activates many virulence factors that could be important during infection by this nosocomial pathogen. In our study, the polymorphism agrII was associated with nosocomial isolates and was the most prevalent polymorphism in MDR S. aureus. Our finding showed that vancomycin modified the hld and spa expression in the MDR S. aureus clinical isolates; suggesting that, vancomycin may regulate alternative systems that jointly participate in the regulation of virulence factors involved in bacteria pathogenesis, which allows spread and adaptation into a hospital environmental.
Author Contributions
Designed and conceived the experiments: VCD, SAO, ACC, NVG, and JXC. Performed the experiments: VCD, GE, and SAO. Analyzed the data: VCD, SAO, ALO, and GER. Contributed reagents/materials/analysis tools: VCD, SAO, ACC, JJOT, and JXC. Wrote and reviewed the manuscript: VCD, SAO, NVG, and JXC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grant number CONACYT 133451 and public Federal Funds HIM/2007/004. The study was reviewed and approved by the Research Committee (Dr. Onofre Muñoz Hernández), Ethics Committee (Dr. Amparo Faure Fontenla), and Biosecurity Committee (Dr. Herlinda Vera Hermosillo) of HIMFG, under permit number HIM/2007/004. We thank Karina Espinosa-Mazariego and Juana Rodriguez for their technical assistance with this work. The Central Laboratory of HIMFG provided the S. aureus clinical isolates for this study. The anonymous clinical information presented in this manuscript prior to analysis was obtained from patient medical records, and the records were chosen by considering the diagnosis and sample type. According to the institutional ethical, biosecurity and investigation evaluation, informed consent was not required.
References
- Archer G. L. (1998). Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26 1179–1181 10.1086/520289 [DOI] [PubMed] [Google Scholar]
- Bordon J., Master R. N., Clark R. B., Duvvuri P., Karlowsky J. A., Ayesu K., et al. (2010). Methicillin-resistant Staphylococcus aureus resistance to non-beta-lactam antimicrobials in the United States from 1996 to 2008. Diagn. Microbiol. Infect. Dis. 67 395–398 10.1016/j.diagmicrobio.2010.03.006 [DOI] [PubMed] [Google Scholar]
- Bronner S., Monteil H., Prévost G. (2004). Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28 183–200 10.1016/j.femsre.2003.09.003 [DOI] [PubMed] [Google Scholar]
- Cafiso V., Bertuccio T., Spina D., Purrello S., Campanile F., Di Pietro C., et al. (2012). Modulating activity of vancomycin and daptomycin on the expression of autolysis cell-wall turnover and membrane charge genes in hVISA and VISA strains. PLoS ONE 7:e29573 10.1371/journal.pone.0029573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cázares-Domínguez V., Cruz-Córdova A., Ochoa S. A., Escalona G., Espinosa M. S., Jiménez R., et al. (2015). Thickening of cell wall as an effect to vancomycin tolerantce of methicillin-resistant Staphylococcus aureus strains from pediatric patients. PLoS ONE 10:e0118791 10.1371/journal.pone.0118791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergren L., Andersson R., Jansson B., Uhlén M., Nilsson B. (1993). Mutational analysis of the interaction between staphylococcal protein A and human IgG1. Protein Eng. 6 441–448 10.1093/protein/6.4.441 [DOI] [PubMed] [Google Scholar]
- Chan P. F., Foster S. J. (1998). The role of environmental factors in the regulation of virulence-determinant expression in Staphylococcus aureus 8325-4. Microbiol. Read. Engl. 144 2469–2479 10.1099/00221287-144-9-2469 [DOI] [PubMed] [Google Scholar]
- Chen H.-Y., Chen C.-C., Fang C.-S., Hsieh Y.-T., Lin M.-H., Shu J.-C. (2011). Vancomycin activates σ(B) in vancomycin-resistant Staphylococcus aureus resulting in the enhancement of cytotoxicity. PLoS ONE 6:e24472 10.1371/journal.pone.0024472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty Fourth Informational Supplement M100-S24 Vol. 34 Wayne, PA: Clinical and Laboratory Standards Institute [Google Scholar]
- David M. Z., Daum R. S. (2010). Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23 616–687 10.1128/CMR.00081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. F., Peterson A. E., Julian K. G., Greene W. H., Price L. B., Nelson K., et al. (2013). Household risk factors for colonization with multidrug-resistant Staphylococcus aureus isolates. PLoS ONE 8:e54733 10.1371/journal.pone.0054733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice L. R. (1945). Measures of the amount of ecologic association between species. Ecology 26 297–302 10.2307/1932409 [DOI] [Google Scholar]
- Erdem A. K., Sanli-Yürüdü N. O., Arslan-Aydoǧdu E. Ö., Dogruoz N., Zeybek Z., Türetgen I., et al. (2008). Quantitative microbiological analysis of biofilm communities from the surfaces of different cooling tower materials. IUFS J. Biol. 67 9–16. [Google Scholar]
- Falord M., Mäder U., Hiron A., Débarbouillé M., Msadek T. (2011). Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS ONE 6:e21323 10.1371/journal.pone.0021323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilot P., Lina G., Cochard T., Poutrel B. (2002). Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40 4060–4067 10.1128/JCM.40.11.4060-4067.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilot P., van Leeuwen W. (2004). Comparative analysis of agr locus diversification and overall genetic variability among bovine and human Staphylococcus aureus isolates. J. Clin. Microbiol. 42 1265–1269 10.1128/JCM.42.3.1265-1269.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C., Campana S., Bayer M. G., Döring G., Botzenhart K., Wolz C. (2000). Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68 1304–1311 10.1128/IAI.68.3.1304-1311.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goerke C., Kümmel M., Dietz K., Wolz C. (2003). Evaluation of intraspecies interference due to agr polymorphism in Staphylococcus aureus during infection and colonization. J. Infect. Dis. 188 250–256 10.1086/376450 [DOI] [PubMed] [Google Scholar]
- Gómez M. I., O’Seaghdha M., Magargee M., Foster T. J., Prince A. S. (2006). Staphylococcus aureus protein A activates TNFR1 signaling through conserved IgG binding domains. J. Biol. Chem. 281 20190–20196 10.1074/jbc.M601956200 [DOI] [PubMed] [Google Scholar]
- Janzon L., Arvidson S. (1990). The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 9 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H.-S., Chan J. L., Cheung G. Y. C., Otto M. (2010). Subinhibitory concentrations of protein synthesis-inhibiting antibiotics promote increased expression of the agr virulence regulator and production of phenol-soluble modulin cytolysins in community-associated methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 54 4942–4944 10.1128/AAC.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander I. G., Granström M., Hedström S. A., Möllby R. (1983). The role of antibodies against alpha-toxin and teichoic acid in the diagnosis of staphylococcal infections. Infection 11 77–83 10.1007/BF01641071 [DOI] [PubMed] [Google Scholar]
- Kolar S. L., Ibarra J. A., Rivera F. E., Mootz J. M., Davenport J. E., Stevens S. M., et al. (2013). Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen 2 18–34 10.1002/mbo3.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y., Birkbeck T. H. (1984). In vitro synthesis of the delta-lysin of Staphylococcus aureus. Infect. Immun. 44 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Bayer A., Cosgrove S. E., Daum R. S., Fridkin S. K., Gorwitz R. J., et al. (2011). Clinical practice guidelines by the infectious diseases society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin. Infect. Dis. 52 285–292 10.1093/cid/cir034 [DOI] [PubMed] [Google Scholar]
- MacFadin J. F. (1996). Biochemical Tests for Identification of Medical Bacteria 3rd Edn. Baltimore: Williams and Wilkins. [Google Scholar]
- McCallum N., Hinds J., Ender M., Berger-Bächi B., Meier P. S. (2010). Transcriptional profiling of XdrA, a new regulator of spa transcription in Staphylococcus aureus. J. Bacteriol. 192 5151–5164 10.1128/JB.00491-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moise P. A., Sakoulas G., Eliopoulos G. M., Schentag J. J., Forrest A., Moellering R. C. (2004). Accessory gene regulator group II polymorphism in methicillin-resistant Staphylococcus aureus is predictive of failure of vancomycin therapy. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 38 1700–1705 10.1086/421092 [DOI] [PubMed] [Google Scholar]
- Moise P. A., Sakoulas G., Forrest A., Schentag J. J. (2007). Vancomycin in vitro bactericidal activity and its relationship to efficacy in clearance of methicillin-resistant Staphylococcus aureus bacteremia. Antimicrob. Agents Chemother. 51 2582–2586 10.1128/AAC.00939-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morfeldt E., Janzon L., Arvidson S., Löfdahl S. (1988). Cloning of a chromosomal locus (exp) which regulates the expression of several exoprotein genes in Staphylococcus aureus. Mol. Gen. Genet. 211 435–440 10.1007/BF00425697 [DOI] [PubMed] [Google Scholar]
- Novick R. P. (2003). Autoinduction and signal transduction in the regulation of Staphylococcal virulence. Mol. Microbiol. 48 1429–1449 10.1046/j.1365-2958.2003.03526.x [DOI] [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Kornblum J., Ross H. F., Ji G., Kreiswirth B., et al. (1995). The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248 446–458 10.1007/BF02191645 [DOI] [PubMed] [Google Scholar]
- Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkaru Y., Asayama K., Ishii H., Nishimura S., Sunahara N., Tanaka T., et al. (1995). Development of a sandwich enzyme-linked immunosorbent assay for the determination of human heart type fatty acid-binding protein in plasma and urine by using two different monoclonal antibodies specific for human heart fatty acid-binding protein. J. Immunol. Methods 178 99–111 10.1016/0022-1759(94)00248-U [DOI] [PubMed] [Google Scholar]
- O’Seaghdha M., van Schooten C. J., Kerrigan S. W., Emsley J., Silverman G. J., Cox D., et al. (2006). Staphylococcus aureus protein A binding to von Willebrand factor A1 domain is mediated by conserved IgG binding regions. FEBS J. 273 4831–4841 10.1111/j.1742-4658.2006.05482.x [DOI] [PubMed] [Google Scholar]
- Otto M. P., Martin E., Badiou C., Lebrun S., Bes M., Vandenesch F., et al. (2013). Effects of subinhibitory concentrations of antibiotics on virulence factor expression by community-acquired methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 68 1524–1532 10.1093/jac/dkt073 [DOI] [PubMed] [Google Scholar]
- Palmqvist N., Foster T., Tarkowski A., Josefsson E. (2002). Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb. Pathog. 33 239–249 10.1006/mpat.2002.0533 [DOI] [PubMed] [Google Scholar]
- Pereira V. C., Riboli D., da Cunha M. (2014). Characterization of the clonal profile of MRSA isolated in neonatal and pediatric intensive care units of a University Hospital. Ann. Clin. Microbiol. Antimicrob. 13:50 10.1186/s12941-014-0050-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazanzadeh R., Zamani S., Zamani S. (2013). Genetic diversity in clinical isolates of Escherichia coli by enterobacterial repetitive intergenic consensus (ERIC)-PCR technique in Sanandaj hospitals. Iran J. Microbiol. 5 126–131. [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G., Eliopoulos G. M., Moellering R. C., Novick R. P., Venkataraman L., Wennersten C., et al. (2003). Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187 929–938 10.1086/368128 [DOI] [PubMed] [Google Scholar]
- Sakoulas G., Eliopoulos G. M., Moellering R. C., Wennersten C., Venkataraman L., Novick R. P., et al. (2002). Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46 1492–1502 10.1128/AAC.46.5.1492-1502.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G., Moellering R. C. (2008). Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 46(Suppl. 5) S360–S367 10.1086/533592 [DOI] [PubMed] [Google Scholar]
- Shopsin B., Kreiswirth B. N. (2001). Molecular epidemiology of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 7 323–326 10.3201/eid0702.700323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowash M. G., Uhlemann A.-C. (2014). Community-associated methicillin-resistant Staphylococcus aureus case studies. Methods Mol. Biol. 1085 25–69 10.1007/978-1-62703-664-1_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrt N., Mesak L. R., Davies J. (2011). Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J. Antimicrob. Chemother. 66 979–984 10.1093/jac/dkr043 [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez-Meza M. E., Aires de Sousa M., Echaniz-Aviles G., Solorzano-Santos F., Miranda-Novales G., Silva-Sanchez J., et al. (2004). Surveillance of methicillin-resistant Staphylococcus aureus in a pediatric hospital in Mexico city during a 7-year period (1997 to 2003): clonal evolution and impact of infection control. J. Clin. Microbiol. 42 3877–3880 10.1128/JCM.42.8.3877-3880.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viau M., Zouali M. (2005). Effect of the B cell superantigen protein A from S. aureus on the early lupus disease of (NZBxNZW) F1 mice. Mol. Immunol. 42 849–855 10.1016/j.molimm.2004.07.047 [DOI] [PubMed] [Google Scholar]
- Yamamoto W, C. H. Takano T. A. N., Takano W. (2013). Genetic nature and virulence of community associated methicillin-resistant Staphylococcus aureus. BioMedicine 3 2–18 10.1016/j.biomed.2012.12.001 [DOI] [Google Scholar]
- Zhang K., McClure J.-A., Elsayed S., Louie T., Conly J. M. (2005). Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43 5026–5033 10.1128/JCM.43.10.5026-5033.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]