Abstract
We present a novel tensile testing system optimized for the mechanical loading of microliter volume protein hydrogels. Our apparatus incorporates a voice coil servoactuator capable of carrying out fixed velocity extension-relaxation cycles as well as extension step protocols. The setup is equipped with an acrylic cuvette permitting day-long incubations in solution. To demonstrate the functionality of the device, we photochemically crosslinked polyproteins of the I91 immunoglobulin domain from the muscle protein titin to create solid hydrogels that recapitulate elastic properties of muscle. We present data from tensile tests of these low volume biomaterials that support protein unfolding as a main determinant of the elasticity of protein hydrogels. Our results demonstrate the potential use of protein hydrogels as biomaterials whose elastic properties dynamically respond to their environment.
I. Introduction
Attempting to mimic muscle tissue mechanics through the construction of artificial protein biomaterials is attractive due to myriad potential applications in tissue engineering. Such biomaterials can potentially substitute for damaged natural tissues, filling crucial roles in the treatment of cardiovascular and musculoskeletal diseases. Current exploratory efforts are grounded in the fact that muscle tissue mechanics originates from the mechanical properties of specific elastomeric proteins 1. For example, the giant protein titin generates the passive elasticity of striated muscle 2,3. As a result, titin elasticity is responsible for the passive force of cardiac muscle during diastolic relaxation of the ventricles 4. The central role of titin in the elasticity of cardiac muscle is further evidenced by the recent discovery that mutations in the titin gene lead to dilated cardiomyopathy, a pathology that entails aberrant changes in the elasticity of the heart 5.
The elastic properties of titin derive from its molecular architecture, which has been well characterized through the use of force-spectroscopy techniques, especially single molecule atomic force microscopy (AFM). Titin has two mechanically active structural components contributing to its elasticity: (i) random coil regions that act as entropic springs that are easy to extend, and (ii) tandem immunoglobulin (Ig) domains, which are able to unfold and refold under mechanical force 6. Folding and unfolding greatly affect the compliance of the whole titin molecule. Since unfolded domains behave as random coil polypeptides, unfolding of a protein domain increases the effective contour length of titin, resulting in a softer protein. Refolding stiffens titin because it reduces the number of amino acids belonging to random coil structure. Any variation in the folding/unfolding properties of the Ig domains changes the elasticity of titin.
Titin-based biomaterials could be designed following the same principles of tissue elasticity regulation that are found in muscle. For example, the length and constituent domains in a particular titin isoform determine the compliance of that particular titin molecule, which in turn defines the stiffness of muscle 2. Regulation of tissue elasticity can also be achieved by modulating the mechanical properties of titin in situ. With this regard, we have recently shown that S-glutathionylation, a physiological posttranslational modification, causes Ig domains in titin to be more sensitive to mechanical unfolding, ultimately resulting in substantial softening of heart tissue 7.
The link between single protein mechanics and larger scale biomechanics has increasingly been exploited in recent years to build macroscopic protein hydrogels with desirable mechanical properties 8,9. Protein hydrogels hold great promise in biomedical engineering as tissue-mimetic biomaterials because they offer unique control over mechanical properties through manipulation of amino acid sequences and accessory ligands 10-13. Elastic protein hydrogels that mimic muscle elastic properties have previously been created from engineered constructs of alternating GB1 domains, which are individually folded domains 14, and randomly coiled repeat sequences from the insect joint protein resilin 9,15. This was accomplished through photoactivated crosslinking of tyrosines in the presence of pyridyl ruthenium ([Ru(bpy)3]2+), creating a protein network of dityrosine crossbridges 9,16 . The photoactivated, pyridyl ruthenium strategy has been used to cast hydrogels from other proteins, including aforementioned resilin 8, gelatin 17, the extracellular matrix protein tenascin-C 18 and even de- novo-synthesized proteins 19. In contrast to purely resilin-based hydrogels 8, comprised entirely of entropic spring-like network proteins, the GB1-resilin hydrogels showed hysteresis at intermediate to high levels of strain, suggesting that the hydrogels dissipate energy through unfolding of GB1 domains. The hysteresis was observed to be reversible on a very short timescale (less than ~1 s), which is compatible with the fast refolding rate of GB1. Also, upon step extensions to a given strain GB1-resilin hydrogels display stress relaxation curves that increase with strain. Such relaxation behavior is proposed to derive at least partially from force-triggered unfolding of network GB1 domains9. Since the mechanical properties of protein hydrogels depend on the folding/unfolding dynamics of the network domains, it now becomes possible to engineer materials that respond to their environment through changes in the unfolding/refolding properties of the constituent domains, just as titin does in muscle.
A key limitation in the field of protein hydrogel tensile testing is the need for relatively large volumes of protein solution to prepare gels compatible with conventional tensile testing instrumentation. This problem is exacerbated by the need for highly concentrated protein solutions, often on the hundred mg/ml scale, as precursors of solid gels. Hence, obtaining sufficient quantities of protein to characterize hydrogel mechanics is a laborious, time-consuming process. Such requirements may be even impossible to meet when testing challenging-to-express proteins. Here, we propose reducing the volume of the hydrogel samples to accelerate the investigation of biomaterials for tissue engineering. We present a new tensile testing device that is capable of generating reliable data from the mechanical loading of ~1 cm long, fiber-like hydrogels made from volumes of protein solution as low as 3 µL, or ~30 times less than the current requirements 20. Our new system is inspired by technologies developed to study muscle fiber mechanics. It employs a voice coil actuator to apply extension protocols at precise speeds to these low volume gels, while a force transducer records the forces generated during the stretch protocol. A custom-fabricated cuvette maintains the hydrogels in solution through the duration of testing. We used this new tensile-tester system to examine the mechanical properties of biomaterials made from a multidomain polyprotein of the I91 domain of titin.
II. Tensile Tester Setup
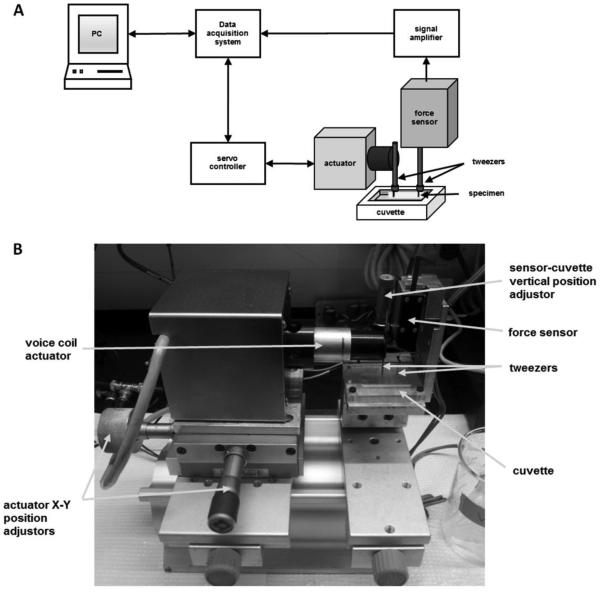
The tensile testing apparatus is represented in Figure 1. Custom programming in Wavemetrics Igor Pro was used to input extension protocols and output the force. The computer communicates with the servocontroller and receives input from the signal amplifier through a BNC-2090 connector block (National Instruments). The three basic components of the setup are described below:
Figure 1. Custom-built protein hydrogel tensile tester.
(A) Schematic drawing of the tensile testing system for low volume protein hydrogels. (B) Photograph identifying the various components of the tensile tester.
(i) An LFA-2010 Linear Focus voice coil actuator (Equipment Solutions) is used to stretch the biomaterial. The voice coil, which can move up to 10 mm in less than 3 ms, is PC-controlled through an SCA814 servocontroller (Equipment Solutions). The controller reports the actual position of the actuator through feedback communication with the computer. The extender arm of the actuator is attached to one end of the hydrogel fiber using stainless steel tweezers (SI-TM7 general-purpose mount, World Precision Instruments). To facilitate attachment and the subsequent positioning of the biomaterial, the coil is mounted on a stage that is adjustable in the X-Y directions using manipulators (Figure 1B).
(ii) An SI-KG4A force transducer (World Precision Instruments) records the force generated by the hydrogel. The transducer is attached to the end of the biomaterial with a second set of SI-TM7 tweezers. The current signal that is generated upon hydrogel loading is converted to voltage and then amplified by a BAM21 Optical Force Transducer Amplifier (World Precision Instruments). Force signal in Volts can be converted into force units through calibration using weights of known mass.
(iii) The custom-fabricated acrylic cuvette allows for the incubation of hydrogels in various reagents throughout the duration of tensile testing. Hence, it is possible to carry out day-long incubations between extension cycles with the attached hydrogel remaining in situ. This is useful in exploration of the relationship between hydrogel mechanics and the chemically sensitive conformations of network protein domains. Finally, although the transducer and cuvette are in a fixed spatial relationship, they are mounted on a vertically sliding stage, which can be adjusted relative to the voice coil to ensure the gel is positioned perpendicularly to both the actuator and the sensor (Figure 1B).
(iv) Advantages of the new instrument. Our custom-built device is highly versatile and can be easily adapted to carry out different stress/strain protocols. Thanks to the small response time of <3 milliseconds of the LFA-2010 voice coil actuator, it is also possible to conduct stress-clamp experiments by implementing feedback electronics.
III. Synthesis and Attachment of Hydrogels
Engineered modules of titin Ig domains are ideal building blocks for creating muscle-mimicking biomaterials because of the crucial physiological role of titin in determining muscle passive elasticity. To best correlate the properties of muscle tissue to the mechanics of the resulting biomaterials, we use polyproteins of the well characterized I91 domain of titin 7. To cast hydrogels, we used the Fancy and Kodadek photoactivated strategy, which results in crosslinks between exposed tyrosine residues 8,9,16. The eight I91 domains have tyrosine residues at position 9 (Figure 2A).
Figure 2. Synthesis and attachment of I918 muscle-mimetic hydrogels.
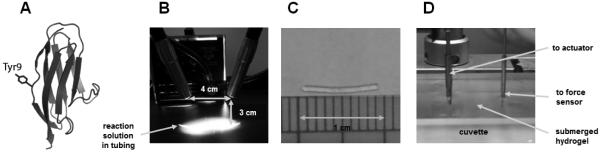
(A) Rendering of the I91 protein highlighting Tyr9, the residue targeted in the photocrosslinking strategy. PDB code: 1TIT. (B) Photograph of illuminator configuration while irradiating PTFE tubing loaded with photoactivated I918 concentrate. (C) A typically sized, translucent, ~1 cm I918 hydrogel. (D) Closeup of the tweezer attachment of the I918 hydrogel between the voice coil servoactuator and the force sensor.
Expression of the I918 polyprotein in E. coli was carried out according to described protocols starting from 1-2 liters of culture 21. The protein was purified using Ni-NTA affinity resin and, after washing with EW buffer (50 mM phosphate, 300 mM NaCl, pH 7) and 20 mM imidazole, was eluted into 1 mL fractions using EW buffer containing 250 mM imidazole. Fractions found to have suitably high concentrations of I918 were pooled together and dialyzed into PBS (50 mM phosphate, 150 mM NaCl, pH 7.5). The pool was further concentrated using 15 mL Millipore Centriplus (NMWL 50,000 kD; Amicon) and then 0.5 mL Millipore Microcon (NMWL 10,000 kD; Amicon) concentrators to final concentrations ranging from 100 to 150 mg/ml. To determine I918 concentration, we used a Beckman Coulter DU 720 spectrophotometer and an extinction coefficient of 0.685 cm−1·mL·mg−1 at 280 nm. This purification procedure rendered around 15 mg of highly concentrated protein per liter of culture.
To prepare the hydrogels, we added 0.2 µL of 25% ammonium persulfate and 0.5 µL of 6.67 mM pyridyl ruthenium (tris(2,2’-bipyridyl)-ruthenium(II) chloride hexahydrate) to 15 µL of concentrated I918 polyprotein. The photoactive reaction mixture was then loaded into PTFE tubing (0.022"ID x 0.042"OD, Cole-Parmer). This was done by fixing a 23 gauge hypodermic needle to a 1 mL syringe with a depressed plunger, and then inserting the tip into one end of a length of PTFE tubing. We immersed the free end of the tubing into the reaction solution before retracting the plunger, using negative pressure to load the tube. The needle was then removed, and the loaded tube was irradiated for 9 minutes with an EKE Lamp, 150 W, in a Fiber-Lite series 180 illuminator set to maximum intensity (Figure 2B). Subsequently, the gel was extruded into PBS with a blunted 25 gauge needle, and cut into lengths of about 1 cm with Vannas scissors (Figure 2C). To prevent degradation, samples were stored at 4 °C in PBS with 0.02% sodium azide. Before attachment of specimens into the tensile tester, the cuvette was loaded with 2-3 mL PBS to facilitate manipulation of the hydrogel. The ends of the hydrogel sample were then attached between the two pairs of tweezers leading to the voice coil actuator and the force sensor (Figure 2D). We did not detect any difference in the experimental results when we used cyanoacrylate-based glue to attach the gel. However, glue-based attachment is discouraged because it complicates cleaning and introduces uncertainties regarding potential chemical contaminations.
IV. Experimental Results and Discussion
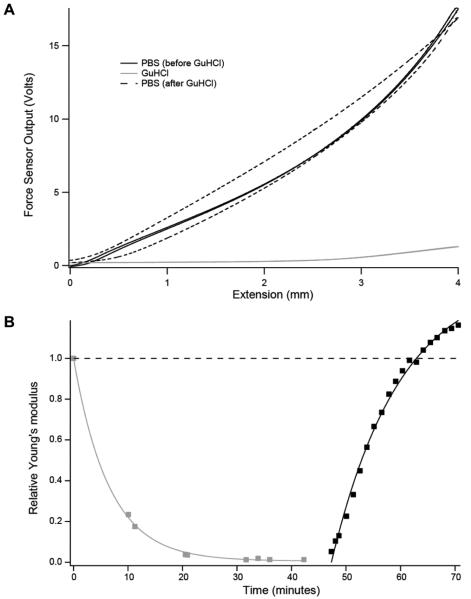
We used both force-extension and stress-relaxation protocols to characterize the properties of the I918 hydrogel. Force-extension protocols entailed one cycle of extension to a fixed length and relaxation at constant speed. These experiments generate force-length plots which allow determination of Young’s modulus (E) through the equation E = (F × L0) / (A0 × ΔL); where F is Force, L0 is the initial gel length, A0 is the initial gel cross-sectional area, and ΔL is the gel extension 9. With our new setup, we were able to show that the Young’s modulus of I91-based hydrogels decreases dramatically upon incubation in the protein denaturant guanidine hydrochloride (GuHCl) (Figure 3), supporting the view that network protein unfolding leads to the softening of the biomaterial. Similar observations have been made before for other protein hydrogels, suggesting that incubation with protein denaturant agents is a general method to soften protein hydrogels 9,19. We did 4 mm force-extension (0.667 mm/s) on a 7 mm gel in PBS, then replaced buffer with 6 M GuHCl, obtaining force-extensions traces for ~45 minutes. Then the denaturing solution was replaced with PBS to allow for protein refolding, and force-extension experiments were conducted for an additional period of ~25 minutes. Selected force-length plots are shown in Figure 3A. At 40% strain (2.8 mm stretch), the Young’s modulus of the gel decreases ~35 fold after 42 minutes in GuHCl. Incubation in PBS results in increased stiffness of the gel. After 23 minutes in PBS, the Young’s modulus increases to 1.14 times the starting value. We also observed an accompanying increase in hysteresis (Figure 3A), which reproduces observations from previous tensile experiments on hydrogels synthesized from de novo ferrodoxin-like domain construct 19. Such increase in hysteresis continued with longer incubation times in PBS (data not shown).
Figure 3. GuHCl reversibly modulates the elasticity of crosslinked I918 biomaterials.
(A) Results of force-extension cycles (0.667 mm/s) conducted on a 7 mm I918 gel in PBS (black, solid), after 42 minutes in GuHCl (gray), and 23 minutes after return to PBS (black, dashes). At 40% strain (2.8 mm stretch) during the extension, the Young’s modulus of the gel is reduced ~35 fold upon incubation in GuHCl. Subsequent incubation in PBS increases the Young’s modulus to 1.14 times the original value. We also observe a large increase in hysteresis. (B) Young’s modulus at 20% strain at different incubation times in GuHCl (gray) and after removal of GuHCl and incubation in PBS (black). Young’s moduli are given relative to the Young’s modulus before incubation in GuHCl. In this experiment, the hydrogel was attached to the force sensor and the voice coil using cyanoacrylate-based glue.
The kinetics of I91 protein unfolding induced by GuHCl have been extensively characterized before 22. To better understand the role of protein unfolding in the softening of I918 gels induced by GuHCl, we monitored the change in the Young’s modulus at 20% strain with incubation time in 6 M GuHCl (Figure 3B). Results are well described by a single exponential where the rate of softening is 0.15 min−1. Remarkably, the rate of unfolding of monomeric I91 molecules in 6M GuHCl is ~0.12 min−1 22. Coincidence between the rate of gel softening and the rate of I91 unfolding strongly supports a scenario where softening of the I91 hydrogels in GuHCl is caused primarily by protein unfolding 9.
We also followed the kinetics of stiffening of the gel upon removal of GuHCl, which can be fitted with a single exponential that gives a rate of stiffening of 0.08 min−1 (Figure 3B). This value is ~3 orders of magnitude slower than the rate of refolding of I91 measured by AFM 22. This result is compatible with two non-exclusive scenarios: (i) I91 refolding may be severely compromised in the gel, or (ii) some other effects may be causing stiffening of the gels. For instance, it is well known that rapid removal of GuHCl can trigger protein misfolding and aggregation 23, with unknown consequences for the mechanical properties of the hydrogel. Also, the diffusion of GuHCl out of the gel may be slow. A scenario where effects beyond protein folding contribute to the stiffening of the I91 gel upon GuHCl removal is supported by the increase in hysteresis we observed in force-extension experiments (Figure 3A), and also by the fact that the stiffness of the gels after GuHCl removal is higher than the original stiffness in PBS (Figure 3B).
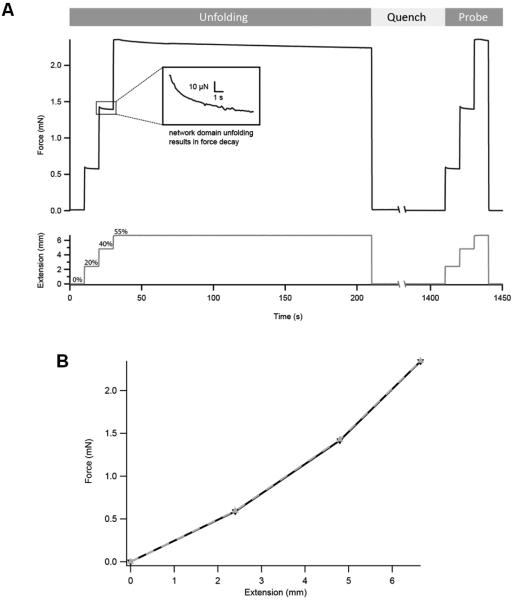
In the quest to engineer materials that mimic natural tissues, it is important to equip biomaterials with mechanisms of regulation similar to those found in vivo. A recent AFM study showed that mechanically induced S-glutathionylation of cryptic cysteines in titin leads to large-scale modulation of the elasticity of muscle tissue 7. This novel mechanism of regulation entails mechanical unfolding of titin’s Ig domains, which exposes cryptic cysteines and makes them reactive. The newly exposed cysteines can be modified by redox metabolites, resulting in persistent changes in elasticity through modification of the mechanical properties of the parent domains. Incorporating equivalent mechanisms to control the elasticity of protein hydrogels may produce smart gels with the ability to respond to physiological redox stimuli. Our I918 hydrogel is an excellent model system due to the presence of two cryptic cysteines in each I91 domain (Cys 47and Cys63, PDB code 1TIT). However, standard force-extension tensile tests are not well suited to study this form of mechanical regulation, since the conformation of the domains in the network dramatically changes during the test itself. A better alternative is to use a stress-relaxation configuration following an unfold-quench-probe scheme (Figure 4A). During the unfolding pulse, the hydrogel is pulled to high strain, triggering the unfolding of domains in the network. By leaving the gel in an extended state, modification of cryptic sites is favored. In the quench pulse, the force is relaxed and the proteins in the gel network are allowed to refold. Finally, the probe pulse measures variations in the stiffness of the gel caused by the experimental protocol. Any change in the elasticity of the gel caused by modification of cryptic sites becomes apparent in the probe pulse. In Figure 4A we show a representative force trace using the unfold-quench-probe protocol on a typical ~1.2 cm I918 biomaterial in PBS (in black). The extension is displayed below it, in gray. The unfolding pulse contained three extension steps, from 0% to 20%, and then to 40%, and 55% strain. The ramp interval between each extension step was 0.04 s. All steps lasted 10 seconds, except at 55% strain, where gels were held for 180 s to favor unfolding of protein domains in the hydrogel. After letting the gel recover for 20 minutes (1200 s) at 0% strain during the quench period, we ran a probe pulse with all steps lasting 10 seconds to characterize the recovery of the hydrogel from hysteresis 9. The declining force curves at each step of extension likely correspond to force-triggered unfolding of immunoglobulin domains within the gel network (Figure 4A, insert) 9. The gels show excellent recovery from hysteresis during the quenching period, as measured in the probe pulse (Figure 4B). We observed diminished recovery when the duration at high strain was increased, or the quench period was shortened (data not shown).
Figure 4. Stress relaxation experiments with I918 protein hydrogels.
(A) Force trace output (black), along with the corresponding extension protocol (gray). % strain values are indicated. Notice the sequential stress relaxation curves during the unfolding pulse, suggesting the force-triggered unfolding of domains forming the network of the gel. (B) Plot showing initial force generated at each extension step in both the unfolding (black, solid) and probe (gray, dashes) pulses. Both traces overlap to a large extent.
In the future, we plan to use our tensile testing scheme in the investigation of protein hydrogels that respond mechanically to a changing chemical environment in real-time. We speculate that I918 hydrogels may exhibit force-triggered redox activity, holding promise as a “smart” biomaterial that replicates muscle function through interfacing with redox signaling pathways in vivo. We also intend to exploit the feedback capability of the voice coil servo to add stress-clamp mode to our current position clamp tensile tester. This will allow a direct comparison with analogous force-clamp AFM procedures on single proteins, which is key to understand how the mechanical properties of the constituent proteins determines the macroscopic properties of protein hydrogels, including the effect of posttranslational modifications that mirror physiological situations.
V. Conclusion
We present a tensile testing device optimized for facile execution of complex loading protocols on low volume hydrogel fibers while maintaining a controlled chemical environment. We hope that this development will accelerate the pace of exploration and discovery of novel, protein-based tissue engineering biomaterials.
VI. Acknowledgements
This work was supported by NIH grants HL66030, HL61228, and NSF grant 1252857 to J.M.F. J.A.-C. was the recipient of an NIH K99 career development award (AI106072). F.S. acknowledges a Columbia University Summer Undergraduate Research Fellowship.
VII. References
- 1.Li H, Linke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernandez JM. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 2.Linke WA, Hamdani N. Gigantic business: titin properties and function through thick and thin. Circ Res. 2014;114:1052–1068. doi: 10.1161/CIRCRESAHA.114.301286. [DOI] [PubMed] [Google Scholar]
- 3.LeWinter MM, Granzier H. Cardiac titin: a multifunctional giant. Circulation. 2010;121:2137–2145. doi: 10.1161/CIRCULATIONAHA.109.860171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmes M, Trombitas K, Granzier H. Titin develops restoring force in rat cardiac myocytes. Circ Res. 1996;79:619–626. doi: 10.1161/01.res.79.3.619. [DOI] [PubMed] [Google Scholar]
- 5.Herman DS, Lam L, Taylor MRG, Wang LB, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJR, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of Titin Causing Dilated Cardiomyopathy. New Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 7.Alegre-Cebollada J, Kosuri P, Giganti D, Eckels E, Rivas-Pardo JA, Hamdani N, Warren CM, Solaro RJ, Linke WA, Fernandez JM. S-glutathionylation of cryptic cysteines enhances titin elasticity by blocking protein folding. Cell. 2014;156:1235–1246. doi: 10.1016/j.cell.2014.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou NE, Wong DC, Merritt DJ, Dixon NE. Synthesis and properties of crosslinked recombinant pro-resilin. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 9.Lv S, Dudek DM, Cao Y, Balamurali MM, Gosline J, Li H. Designed biomaterials to mimic the mechanical properties of muscles. Nature. 2010;465:69–73. doi: 10.1038/nature09024. [DOI] [PubMed] [Google Scholar]
- 10.Cao Y, Li YD, Li H. Enhancing the mechanical stability of proteins through a cocktail approach. Biophys J. 2011;100:1794–1799. doi: 10.1016/j.bpj.2011.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Carrion-Vazquez M, Oberhauser AF, Marszalek PE, Fernandez JM. Point mutations alter the mechanical stability of immunoglobulin modules. Nat Struct Biol. 2000;7:1117–1120. doi: 10.1038/81964. [DOI] [PubMed] [Google Scholar]
- 12.Li HB, Cao Y. Protein Mechanics: From Single Molecules to Functional Biomaterials. Accounts Chem Res. 2010;43:1331–1341. doi: 10.1021/ar100057a. [DOI] [PubMed] [Google Scholar]
- 13.Ainavarapu SR, Li L, Badilla CL, Fernandez JM. Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophysical journal. 2005;89:3337–3344. doi: 10.1529/biophysj.105.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y, Li HB. Polyprotein of GB1 is an ideal artificial elastomeric protein. Nat Mater. 2007;6:109–114. doi: 10.1038/nmat1825. [DOI] [PubMed] [Google Scholar]
- 15.Weis-Fogh T. A rubber-like protein in insect cuticle. J Exp Biol. 1960;37:889–907. [Google Scholar]
- 16.Fancy DA, Kodadek T. Chemistry for the analysis of protein-protein interactions: Rapid and efficient cross-linking triggered by long wavelength light. P Natl Acad Sci USA. 1999;96:6020–6024. doi: 10.1073/pnas.96.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elvin CM, Vuocolo T, Brownlee AG, Sando L, Huson MG, Liyou NE, Stockwell PR, Lyons RE, Kim M, Edwards GA, Johnson G, McFarland GA, Ramshaw JA, Werkmeister JA. A highly elastic tissue sealant based on photopolymerised gelatin. Biomaterials. 2010;31:8323–8331. doi: 10.1016/j.biomaterials.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Lv S, Bu T, Kayser J, Bausch A, Li H. Towards constructing extracellular matrix-mimetic hydrogels: an elastic hydrogel constructed from tandem modular proteins containing tenascin FnIII domains. Acta biomaterialia. 2013;9:6481–6491. doi: 10.1016/j.actbio.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Fang J, Mehlich A, Koga N, Huang J, Koga R, Gao X, Hu C, Jin C, Rief M, Kast J, Baker D, Li H. Forced protein unfolding leads to highly elastic and tough protein hydrogels. Nature communications. 2013;4:2974. doi: 10.1038/ncomms3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang J, Li H. A facile way to tune mechanical properties of artificial elastomeric proteins-based hydrogels. Langmuir : the ACS journal of surfaces and colloids. 2012;28:8260–8265. doi: 10.1021/la301225w. [DOI] [PubMed] [Google Scholar]
- 21.Alegre-Cebollada J, Kosuri P, Rivas-Pardo JA, Fernandez JM. Direct observation of disulfide isomerization in a single protein. Nat Chem. 2011;3:882–887. doi: 10.1038/nchem.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci U S A. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Y, Wetlaufer DB. Control of aggregation in protein refolding: the temperature-leap tactic. Protein Sci. 1996;5:517–523. doi: 10.1002/pro.5560050314. [DOI] [PMC free article] [PubMed] [Google Scholar]