Abstract
Background
This study aimed to have international experts converge on a harmonized definition of whole hippocampus boundaries and segmentation procedures, to define standard operating procedures for magnetic resonance (MR)-based manual hippocampal segmentation.
Methods
The panel received a questionnaire regarding whole hippocampus boundaries and segmentation procedures. Quantitative information was supplied to allow evidence-based answers. A recursive and anonymous Delphi procedure was used to achieve convergence. Significance of agreement among panelists was assessed by exact probability on Fisher’s and binomial tests.
Results
Agreement was significant on the inclusion of alveus/fimbria (P =.021), whole hippocampal tail (P =.013), medial border of the body according to visible morphology (P =.0006), and on this combined set of features (P =.001). This definition captures 100% of hippocampal tissue, 100% of Alzheimer’s disease-related atrophy, and demonstrated good reliability on preliminary intrarater (0.98) and inter-rater (0.94) estimates.
Discussion
Consensus was achieved among international experts with respect to hippocampal segmentation using MR resulting in a harmonized segmentation protocol.
Keywords: Hippocampus, Atrophy, Volumetry, Manual segmentation, Harmonization, Anatomical landmarks, Delphi procedure, Alzheimer’s disease, Medial temporal lobe, Hippocampal atrophy, Magnetic resonance, Neuroimaging, Standard operational procedures, Enrichment, MCI, Reliability
1. Introduction
Magnetic resonance (MR)-based hippocampal atrophy has been recognized as a supportive feature for the clinical diagnosis of Alzheimer’s disease (AD) [1–4] and disease tracking [5]. It has recently been qualified by the European Medicines Agency as an enrichment marker that can be used for subject enrolment in clinical trials on predementia AD [6]. However, a number of different protocols for manual segmentation exist [7,8], leading to highly heterogeneous volume estimates [7] and preventing its reliable use. The European Alzheimer’s Disease Consortium (EADC) and Alzheimer’s Disease Neuroimaging Initiative (ADNI) investigators, supported by the Alzheimer’s Association, have undertaken a project aimed to develop a Harmonized Protocol (HarP) for the manual segmentation of the boundaries of the whole hippocampus on MR scans [5,9] (www.hippocampal-protocol.net), and specifically on T1 weighted volumetric MR scans, the most frequently used sequence for hippocampal volumetry.
Earlier steps of the project have been described previously [10–12]. A survey of the available protocols was carried out: the 12 protocols most frequently used in the AD literature were examined, used to carry out segmentation using ADNI scans, and submitted to the authors of the protocols to certify the correct understanding and use of each protocol. Landmarks were extracted (www.hippocampal-protocol.net) [10], and operationalized into seven segmentation units (SUs): minimum hippocampal body (the common region included by all protocols), alveus/fimbria, subiculum-oblique, horizontal, and morphology, tail-crura, and whole-tail. Validity metrics of SUs were estimated such as intraclass correlation coefficients (ICC) test-retest and inter-rater reliability, and contribution to AD-related atrophy [11].
The present report describes how validity metrics of SUs were fed to a panel of international experts with substantial expertise on hippocampal segmentation and how, through a recursive and anonymous Delphi procedure, experts converged to a consensual definition on whole hippocampus boundaries segmentation. This consensual definition of the EADC-ADNI HarP for Manual Hippocampal Segmentation has subsequently been validated versus existing protocols [13] and pathology [14]. Benchmark images of hippocampi segmented according to the HarP have been produced to serve as reference for the qualification of naïve tracers [15] and automated segmentation algorithms [16].
2. Methods
2.1. Delphi voting procedure
We sought a consensual definition of the protocol for hippocampal segmentation through a Delphi panel procedure. Commonly used in biomedical research, this method facilitates convergence through recursive voting sessions [17]. After the first voting round, successive recursive rounds are run where panelists are informed about the decisions taken by the other panelists, and are then invited to vote again on the same issues, until convergence is achieved (Figure 1).
Fig. 1.
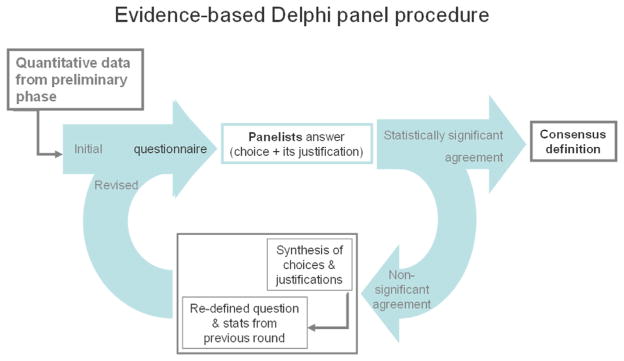
Evidence-based Delphi procedure used for defining the Harmonized Protocol for manual hippocampal segmentation.
In more detail, in round 1 panelists were asked to express their choice among alternatives for each issue of a questionnaire. In the subsequent rounds (rounds 2–5), for each item they were asked to quantify their agreement with the choices preferred by the majority in previous round through 9-level Likert scales. For these rounds, the questionnaire was integrated with both answers and related statistics from the previous round, and with a summary of the reasons why each choice was preferred. Reasons expressed for the less popular choices were also reported alongside with statistics. The complete original answers of all panelists were made available in anonymized forms.
Panelists were asked to answer the questionnaire through the dedicated web site www.hippocampal-protocol.net. They could access the same questionnaire with private credentials online at any time within a defined deadline, and provide their answers through their private account. They could choose among alternatives, justify their choices, see interactive 3D models illustrating the shape of hippocampus generated with their choices, revise and change their answers until they finally submitted the complete questionnaire. Final boxes were also made available for free comments relating any issue of the questionnaire, of the procedure, or of the project. These free comments, together with our answers, were also presented to all in the following round, allowing for a sharing of information and opinion, anonymously to participants.
A summary of the answered questionnaire was released to each participant at the end of each voting round.
2.1.1. Differences with the traditional Delphi method
We modified some aspects of the traditional Delphi method [17] to increase the relevance of quantitative information at each stage of the procedure, and define a consensual segmentation protocol through an evidence-based Delphi procedure.
First, for each issue raised in the questionnaire we collected and supplied all possible quantitative information that was pertinent to AD and helpful answering specific questions [11]. In this way, panelists could evaluate and integrate their preferences based on personal experience in manual hippocampal segmentation on (usually) the 2D coronal plane with the quantitative data collected and attributed to the segmentation units rendered in 3D. Secondly, we asked panelists to justify all of their choices. This allowed the other panelists to receive at the next round not just the statistics for the preferred choice, but also the reasons why these were preferred. Finally, we terminated the interrogation only when the number of panelists agreeing with an option was significantly greater than the number of those disagreeing at statistical testing. This differs from the traditional Delphi method, which terminates the interrogation when a median value greater than five in the 9-level Likert-scaled answer denotes agreement for the preferred choice.
2.2. Delphi panelists
A call for experts in hippocampal anatomy and manual segmentation was launched to the EADC and ADNI centers taking part in the project. The main authors of the protocols surveyed in the preliminary phase [10] were also invited to participate in the Delphi panel. Finally, one expert (HW) who asked to take part after having known of the project was admitted after evaluation of her expertise as documented by her scientific contribution in the field. Only one expert per laboratory was allowed to participate in the Delphi panel.
There were 17 panelists who registered for participation in the Delphi voting sessions. One panelist did not contribute to the Delphi at any stage. Participant panelists were from: David Geffen School of Medicine, University of California (Liana Apostolova and George Bartzokis), UCL Institute of Neurology, University College London (Josephine Barnes), University of California at Davis (Charles DeCarli), Rush University (Leyla deToledo-Morrell), Institute for Ageing and Health, Newcastle University, Wolfson Research Centre (Michael Firbank), Karolinska Institute (Lotte Gerritsen), VU University Medical Center (Wouter Hennemann), Mayo Clinic (Clifford Jack), Boston University School of Medicine (Ronald Killiany), Centre for Neuroscience, University of Alberta (Nikolai Malykhin), McGill Centre for Studies on Aging (Jens Pruessner), University of Eastern Finland (Hilkka Soininen), Northwestern University Feinberg School of Medicine (Lei Wang), University Health Center, St. Antoine (Craig Watson), and University of Zurich (Henrike Wolf).
The five Delphi voting rounds were run from April 2011 to March 2012.
2.2.1. Information fed to the Delphi panel
Delphi rounds were managed through a bespoke section of the project web site (http://www.hippocampal-protocol.net/SOPs/delphipanel.php).
An introduction page for each round briefly described the kind of information presented to the panelist and how to answer and submit.
Issues addressed in the questionnaires were divided into “Landmarks” and “Segmentation procedures”. The “Landmarks” section was aimed to provide the definition of the anatomical boundaries of the whole hippocampus for the HarP, choosing among the range observed in the 12 surveyed protocols. The selection of landmarks was handled through the SUs defined in Boccardi et al. [11]. Briefly, all landmark differences extracted from the most popular protocols in the AD literature [10] were operationalized into SUs, i.e., “pieces” of hippocampus that can be included or not in the segmentation, depending on the corresponding landmark definition. Some of these SUs were named as hippocampal subfields for their being roughly overlapping with them, but they only represent the variability of landmarks across the surveyed protocols. With this method, quantitative information could be obtained for each of these SUs, and the choice of inclusion or exclusion of SUs by the Delphi panel thus corresponded to landmarks definition. To facilitate communication with panelists, SUs were rendered as 3D objects by modeling the segmentations performed in the previous steps of this project [11]. In this way, panelists could evaluate the quantitative evidence while having a rather concrete idea of which part of the hippocampus was included or excluded with each landmark selection, and what did this mean in terms of size of the included or excluded tissue, of increase or decrease in segmentation reliability, and of informative power of each “piece” of hippocampus as to AD pathology. Panelists were accurately informed about how this information was collected. They were experts in hippocampal anatomy and segmentation, and they could thus evaluate each question on landmark definition considering both points of view of their personal experience in segmenting based on specific landmarks on the 2D coronal plane, and evaluate the quantitative evidence corresponding to those landmarks that we collected and rendered through the 3D segmentation units.
The “Segmentation procedures” section aimed to have panelists converge on a number of disputed or ambiguous technical segmentation issues.
2.2.2. Definition of anatomical landmarks
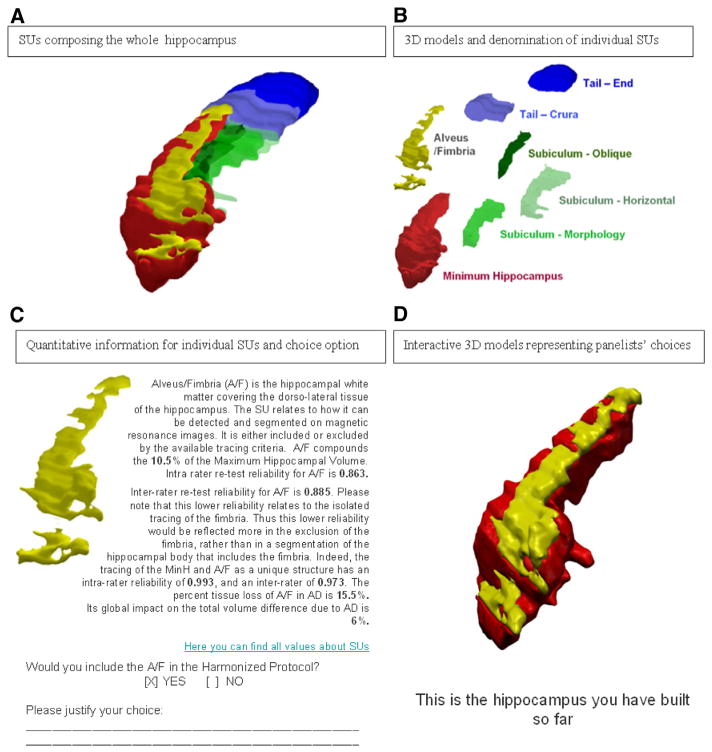
For their reference, panelists were presented the definition and 3D renderings of a union of (Figure 2A) and individual (Figure 2B, 2C) SUs, together with hyperlinks to the tables reporting the pertinent quantitative information published [11]. Briefly, these tables reported percent values of the SU volume relative to total hippocampal volume, ICC of intra- and inter-rater reliability in the segmentation of each SU, and percent tissue reduction in mild cognitive impairment (MCI) and AD compared with controls as computed from the preliminary phase of this project [11]. Interactive 3D models (Figure 2D) were built dynamically to help panelists evaluate the results of their choices (Figure 2C).
Fig. 2.
Example of information provided to panelists in the Delphi rounds for the definition of landmarks. Abbreviation: SUs, segmentation units.
Reports describing the previous steps of the project [10,11] were made available for the exclusive use of panelists in the context of this Delphi exercise.
Because the “minimum hippocampal body” (red SU in Figure 2D) was included by all of the protocols surveyed in Boccardi et al. [10], this SU was described, but it was included a priori in the protocol, and no decisions were taken with respect to its inclusion or borders. Therefore, the inclusion of the full hippocampal head and the definition of the ventral, lateral, and dorsal borders of the hippocampal gray matter in the head and body were defined based on the preexisting agreement among the most popular protocols in the AD literature [10,11]. Similarly, structures that were consistently excluded in the surveyed protocols (such as the entorhinal cortex) were not proposed as possible structures to include, but were a priori excluded from the HarP.
Questions fed to the Delphi panel covered the following:
Inclusion of the alveus/fimbria (dichotomous choice, proposed from round 1);
Definition of the medial border of the hippocampal body (proposed from round 1). Panelists were invited to choose one out of four options: (1) vertical line from the above CA1 to the parahippocampal white matter (no subiculum, as described in Convit et al. [18], (2) oblique line following the inclination of the medial parahippocampal white matter (“subiculum-oblique”), as described in refs. [19,20], and used to collapse different arbitrary oblique lines [11]), (3) horizontal line, obtained through a line drawn horizontally from the uppermost point of the medial parahippocampal white matter to the cistern cerebrospinal fluid (CSF), as described in Haller et al. [21] and Bartzokis et al. [22], (4) morphologic shape, consisting of following the boundary that can be identified from the morphology of the parahippocampal cortex, as in refs. [23–26];
Definition of the most caudal slice (proposed from round 1). The choice was proposed among three options: (1) “no tail”, terminating segmentation at the level where both the superior and inferior collicula are visible, as described in Bartzokis et al. [22], (2) “crus-crura”, terminating segmentation at the level where the crura of the fornices could first be seen in full profile [24,26], (3) “all tail”, segmenting until the last ovoid shape of hippocampal gray matter could be detected;
Inclusion of vestigial tissue (proposed from round 1). The issue was initially proposed to explore whether, in the opinion of panelists, the 3D tools currently used enable to segment reliably the Andrea Retzius and the fasciolar gyri. From round 2, when a first preference emerged from panelists for including the whole tail, the opportunity to include the vestigial tissue was proposed as a dichotomous item. (Table 1)
Table 1.
Questions asked during the Delphi rounds
| Questions landmarks | Round 1 | Round 2 | Round 3 | Round 4 | Round 5 |
|---|---|---|---|---|---|
| Alveus/fimbria | Inclusion/exclusion alveus/fimbria (Y/N) | Agreement on including the alveus/fimbria (9-level Likert) | Agreement on criterion for separating the fimbria from the fornix (9-level Likert) | ||
| Medial border of the hippocampal body | How to segment medial border of the body (four options) | Agreement on segmenting hippo medial border according to morphology criterion (9-level Likert) | How to segment the medial border of the body if no morphologic details can be found (two options) | ||
| Most caudal slice | How to segment the tail (three options) | Agreement on including the whole tail (9 level Likert) | Agreement on including the whole tail (9 level Likert) | ||
| Vestigial tissue | Possibility to separate vestigial tissue/hippo tail with 3D navigation (Y/N) | Possibility to separate vestigial tissue/hippo tail with 3D navigation (Y/N) Opportunity to exclude vestigial GM (9 level Likert) |
How to segment the hippocampal tail at the level of the vestigial tissue (Inclusion vs Exclusion) | ||
| Global model of harmonized hippocampus | Agreement on preliminary model of harmonized hippocampus (9 level Likert) | ||||
| Segmentation procedures | |||||
| Disambiguation of hippocampal head from amygdala with 3D navigation | Possibility to separate amygdala/hippo head with 3D navigation (Y/N) | Agreement on disambiguating the amygdala from hippo head through 3D navigation (9-level Likert) | |||
| Internal CSF pools | How to segment internal CSF pools (four options) | Agreement on excluding internal CSF pools based on connection with other hypointense voxels (9-level Likert) | Agreement on excluding internal CSF pools based on connection with other hypointense voxels in 3D navigation (9-level Likert) | ||
| Poorly visible structures (e.g.: subiculum in very atrophic subjects) | How to segment poorly visible structures (three options) | How to segment poorly visible structures (two options) | Agreement on definition on how to segment poorly visible structures (9-level Likert) | ||
| MRI orientation | Image orientation (two options) | Agreement on orienting MRIs along the hippo axis (9-level Likert) | Image orientation along which hippo long axis (three options) | Agreement on orienting images along the mean angle of the right and left hippo axes | Agreement on maintaining AC-PC based on the newly acquired data |
Abbreviations: Y/N, dichotomous answer allowed (Yes/No); AC-PC, anterior commissure–posterior commissure; GM, gray matter; CSF, cerebrospinal fluid; MRI, magnetic resonance imaging.
NOTE. Items for which agreement was not achieved were reproposed, in the same or modified way, in subsequent rounds.
An example of questions proposed to the panelists can be found in Figure 2C. All original questionnaires, summarized in Table 1, can be downloaded from: www.centroalzheimer.it/public/MB/SOPs/PaperConsensus/delphi-1.pdf www.centroalzheimer.it/public/MB/SOPs/PaperConsensus/delphi-2.pdf www.centroalzheimer.it/public/MB/SOPs/PaperConsensus/delphi-3.pdf www.centroalzheimer.it/public/MB/SOPs/PaperConsensus/delphi-4.pdf www.centroalzheimer.it/public/MB/SOPs/PaperConsensus/delphi-5.pdf
2.2.3. Segmentation procedures
The definition of segmentation procedures involved:
segmentation of the hippocampal head from the amygdala in the most rostral slices (proposed from round 1): panelists were asked whether, in their opinion, the currently available 3D navigation tools enable satisfactory discrimination of the boundaries between the amygdala and the hippocampal head;
segmentation of internal CSF pools (proposed from round 1): panelists were asked whether internal pools should be segmented, and using what criteria (e.g., only when connected to external CSF, or in every case);
segmentation of structures that can not always be clearly visualized on MR scans, but are expected in specific locations based on a priori anatomical knowledge, as it happens for the subiculum in very atrophic subjects (proposed from round 1);
MR image orientation (proposed from round 1) panelists were asked whether the MR scans should be oriented along the anterior commissure (AC) to the posterior commissure (PC) line, along the axis of the segmented hippocampus, or along the mean angle between the two hippocampal axes for each subject;
separation of the alveus/fimbria from the fornix (proposed from round 3, see Table 1, first line “alveus/fimbria”, column “round 3”).
Literature and anatomical information was made available whenever possible to improve the understanding of all nuances, and support informed decisions.
2.3. Statistics
A 9-level Likert scale was used to express level of agreement, 1 corresponding to minimum, 9 to maximum agreement. The median value was used as a central tendency measure. Besides the median value, we computed the statistical significance of the number of panelists agreeing versus the number of panelists disagreeing on the presented choices. P values from Fisher’s exact test were used to evaluate inhomogeneity of answers across levels of agreement. To evaluate the significance of agreement versus disagreement, the exact probability calculated using a binomial test of the choice was computed, after having dichotomized the answers into “disagreement” (Likert levels 1–4) and “agreement” (Likert levels 6–9). Here, level 5 was considered as neutral, “neither agreement nor disagreement”, and excluded from the test. Convergence was defined when both these tests were significant. If statistical significance was not achieved on both tests after a few rounds, the choice taken by the majority was accepted as a concordance index, as usually done in the Delphi procedure [17]. When not otherwise specified, P values reported in this article refer to the binomial test.
3. Results
3.1. Landmarks
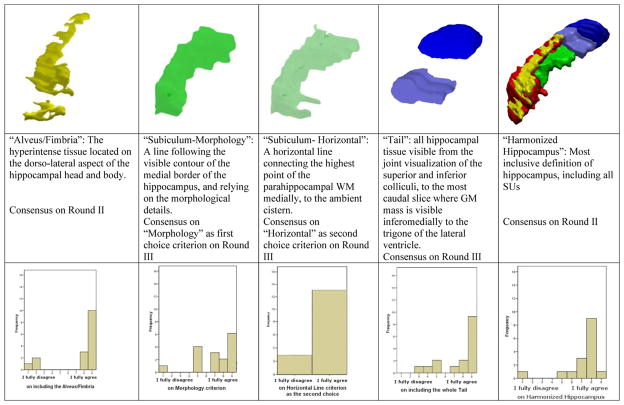
Statistically significant agreement was achieved on including alveus/fimbria, the whole visible hippocampal tail, and on segmenting the medial border of the body following visible morphology (Figure 3; Table 2). The Horizontal Line criterion for segmenting the subiculum at the level of the hippocampal body was significantly agreed on as a second choice criterion, for those slices where no morphologic details allow to clearly identify a medial boundary on the MR (Figures 2 and 3). The inclusion of the vestigial tissue in the hippocampal tail was agreed on by most panelists, although this is the only item for which agreement did not reach statistical significance on the binomial test (63% of agreement for inclusion, P (Fisher’s) = .022, (binomial) = .454).
Fig. 3.
Consensus on landmarks through the Delphi panel voting sessions. Agreement for the inclusion of each SU was expressed through a Likert scale, from 1 (“I fully disagree”) to 9 (“I fully agree”). Subiculum—horizontal line was voted as a “second choice” criterion for those slices where no morphologic details can be detected, that would allow for the adoption of the Subiculum—morphology criterion. The Minimum Hippocampus (red SU) was included a priori since it was the part included by all of the protocols surveyed in [9]. Abbreviation: SU, segmentation unit.
Table 2.
Overview of the decisions taken by the Delphi panel
| Round | Mdn | Likert scores
|
P | Synthesis of reasons for the most frequent choice | |||
|---|---|---|---|---|---|---|---|
| 9 8 7 6
|
5
|
4 3 2 1
|
|||||
| Agree
|
Neutral
|
Disagree
|
|||||
| N (%) | N (%) | N | |||||
| Segmentation unit selection | |||||||
| Inclusion of alveus/fimbria (A/F) | II | 9 | 13 (81%) | 0 (0%) | 3 (19%) | .021 |
|
| Segmentation of subiculum (morphology criterion) | III | 7.5 | 11 (69%) | 4 (25%) | 1 (6%) | .006 |
|
| Segmentation of subiculum (horizontal criterion—II choice) | III | — | 13 (81%) | — | 3 (19%) | .021 |
|
| Inclusion of tail end | III | 9 | 12 (75%) | 2 (12.5%) | 2 (12.5%) | .013 |
|
| Inclusion of vestigial tissue | III | — | 10 (63%) | — | 6 (37%) | n.s. |
|
| Harmonized Hippocampus | II | 8 | 14 (88%) | 1 (6%) | 1 (6%) | .001 |
|
| Segmentation procedures | |||||||
| Disambiguating amygdala with 3D navigation | II | 8 | 16 (100%) | 0 (0%) | 0 (0%) | <.0005 |
|
| CSF pools | III | 8 | 14 (88%) | 0 (0%) | 2 (12%) | .004 |
|
| Not visible structures (final definition) (e.g.: subiculum in very atrophic subjects) | III | 8 | 16 (100%) | 0 (0%) | 0 (0%) | <.0005 |
|
| Separating alveus/fimbria from fornix | IV | 8 | 12 (82%) | 0 (0%) | 1 (8%) | .003 |
|
| AC-PC image orientation* | V | 9 | 11 (79%) | 2 (14%) | 1 (7%) | .006 |
|
Abbreviations: Mdn, median value at Likert scale. ; n.s., not significant; AC-PC, anterior commissure–posterior commissure; CSF, cerebrospinal fluid.
NOTE. Data come from five voting rounds. “Round” denotes when the consensus was achieved. Two votes were missing in round V. No figures in the “Neutral” column indicates that the question was dichotomous (see Table 1).
NOTE. The quantitative data provided to panelists are described in [10].
NOTE. Synthesis of reasons for the most frequent choice is obtained from justifications for answers provided by panelists, with the exception of the AC-PC item*.
Reasons for agreement with AC-PC orientation relate to the solution proposed in the questionnaire, due to methodological requirements arisen relative to project consistency, and not to the spontaneous contribution of panelists as for the other items. See Results and Discussion.
The previously given set of landmarks (besides the horizontal line and the vestigial tissue definitions) was chosen by most panelists from the first round, and, as a whole, it was significantly agreed on in round 2 (88%, P =.001). However, the voting rounds had been carried out until statistically significant convergence was achieved for each individual item (except vestigial tissue).
Based on the quantitative investigation previously performed [11], we computed that the hippocampus as defined previously covers 100% of hippocampal tissue, captures 100% of AD-related hippocampal atrophy, and, measured across three experts from independent centers (Brescia, Mayo Clinic, LONI), has good intra-rater (Tracer 1: 0.99 (confidence interval or CI 95%: 0.96–1 Tracer 2: 0.98 (0.94–1), and Tracer 3: 0.95 (0.83–0.99) for the left, and 0.99 (0.97–1), 0.99 (0.96–1), and 0.97 (0.88–0.99) for the right hippocampus) and inter-rater (left: 0.94 (0.79–0.98); right: 0.94 (0.81–0.99) reliability [12].
3.2. Segmentation procedures
Panelists agreed that the 3D navigation, currently possible with most brain visualization and segmentation software, allows segmentation of the most rostral hippocampal tissue from the amygdala (Table 2) (100%, P =.0005). They agreed on the definition, fine-tuned based on other panelists’ comments and suggestions, to exclude the internal pools of CSF from the hippocampal segmentation when the hypointense voxels are connected to other hypointense voxels in 3D or in the rostro-caudal direction. Such stipulations regarding connection of hypointense voxels were made to ensure that these hypointense voxels likely constitute CSF (88%, P = .004). Panelists agreed on separating the fimbria from the fornices, in the caudal coronal view, at the level where the white matter tract changes inclination, diverging from hippocampal gray matter in its longitudinal extension (Table 2). Panelists also agreed on alerting tracers to attempt to see structures that are not visible on MR, but that may be expected based on a priori anatomical knowledge, such as the subiculum in very atrophic subjects. Methods which can be used to identify such structures include changing image contrast and 3D navigation. Panelists agreed not to segment such structures if, after these visualization attempts, they remain non-detectable (Table 2; Appendix II).
With respect to MR orientation, most panelists (62.5% versus 6.2%, P = .012) initially agreed on orienting MRs based on the long axis of the hippocampus, while 31.3% expressed neither agreement nor disagreement for image orientation, stating that both orientations would lead to good segmentation results. However, the issue was reproposed due to a methodological problem: all of the data made available to panelists were collected on AC-PC oriented images. These data, including segmentation reliability, influenced the panelists choices, but there was no guarantee that segmentation on a different orientation would be characterized by the same reliability and informative power features, based on which panelists took their decisions. To fully understand this problem, new quantitative data were collected [12], aimed to compare inter-rater reliability on images oriented along the hippocampal axis and along AC-PC. This additional information, that was not previously available in the literature, indicated nonsignificantly higher ICC values for AC-PC than hippocampal oriented images, but significantly greater overlapping reliability for segmentations in AC-PC oriented images [12]. As it can be appreciated in Figure 2 in ref. [12], the CSF inlet between the hippocampal head and the amygdala is more clearly visible in AC-PC images than in images oriented along the hippocampal axes. This provides a better visual separation of the two structures and of their boundaries, in a region that is typically a major source of segmentation mistakes and disagreement. Examples of performance and spatial overlapping between three tracers on AC-PC and on hippocampal axes oriented images can be visualized at the following links:
Worst overlapping among three tracers on images oriented along the hippocampal axis: http://www.centroalzheimer.it/public/MB/SOPs/PaperCheck4Axes/Subject_09_worseAxis_Right.mov
Worst overlapping among three tracers on AC-PC oriented images: http://www.centroalzheimer.it/public/MB/SOPs/PaperCheck4Axes/Subject_09_worseACPC_Left.mov
Best overlapping among three tracers on images oriented along the hippocampal axis: http://www.centroalzheimer.it/public/MB/SOPs/PaperCheck4Axes/Subject_04_bestAxis_Left.mov
Best overlapping among three tracers on AC-PC oriented images: http://www.centroalzheimer.it/public/MB/SOPs/PaperCheck4Axes/Subject_03_bestACPC_Right.mov
These data were thus supplied to the Delphi panel, together with the description of the methodological problem, and panelists converged on deciding that the AC-PC orientation was most suitable for segmentation (Table 2).
A detailed description of all landmarks and segmentation procedures is reported in Appendix II: “User Manual”.
4. Discussion
In this study, we achieved a consensus on landmarks and procedures to define a HarP for the manual segmentation of the boundaries of the whole hippocampus from high resolution T1 weighted MR scans. Panelists converged on the most inclusive definition of the hippocampus, including alveus and fimbria, the whole hippocampal tail including the vestigial gray matter, and the subiculum segmented following morphologic details. Until now, different laboratories have produced very heterogeneous measures mainly due to the use of different segmentation protocols [7,8]. This EADC-ADNI HarP provides standard procedures for manual hippocampal segmentation expected to obtain homogeneous measurements, facilitating direct comparisons across studies. Preliminary data showed very high inter-rater reliability across different laboratories [12]. Detailed validity metrics of the HarP have been specifically evaluated in the subsequent proper validation phases, comparing its reliability with that of currently used local protocols, and estimating variance due to different sources (tracer, side, time point, scanner, magnet field strength) [13].
The consensual definition was achieved through five Delphi rounds. The Delphi procedure was modified to accommodate empirical evidence. First, in a preliminary phase quantitative information was collected to allow panelists to make decisions with the aid of empirical evidence [10,11]. Secondly, panelists were asked to justify their choices, so that in the subsequent round provision of reasons for preferred choices, in addition to statistics was made. This enabled panelist to re-evaluate their positions and move toward a consensus based on a richer base of information. Finally, agreement was achieved when a statistically significant majority–rather than a simple majority–was obtained, with only one exception, where one out of two tests was not significant. Understandably, a few disagreements still remained to the last rounds, but this was confined to a maximum of 2 out of 16 panelists.
Quantitative information was supplied, to help panelists base decisions on evidence. Indeed, some criteria widely used in currently available segmentation protocols, such as the use of arbitrary lines, were based on pragmatic definitions that had never been quantitatively investigated in this way. For example, the segmentation of the medial border of the hippocampal body based on the visible morphology, or of the tail, was often mistrusted as possibly less reliable than segmentation based on arbitrary lines, with consequent exclusion of even wide portions of hippocampus, for the right sake of reliability. The data that we collected in the preliminary phase [11] gave reassuring information regarding the segmentation of visible shape from MR, and panelists could base their choice on empirical measurement of variables specifically pertinent to the definition of a segmentation protocol for AD, instead of basing their choices on legitimate but untested worries.
Panelists’s justifications for their answers denoted that they made wide use of the empirical information contained in the questionnaires (Table 2). It should be stressed however that answers were not uniquely guided by evidence; panelists’ mandate was in fact to use evidence to implement their personal experience. Moreover, panelists were asked to factor in their choices the possibility of reliably transferring information and segmentation instructions between different raters and laboratories.
For one item (image orientation) the questionnaire was re-submitted to panelists after consensus had been reached. This was due to the fact that the chosen orientation along the long hippocampal axis was not consistent with the whole set of data that panelists examined while formulating their choices, which were collected on AC-PC oriented images. Before raising this issue to the panel, new data were collected evaluating the reliability of segmentation on the long hippocampal axis and AC-PC line [12], something so far unexplored. This investigation showed that segmentations on AC-PC images lead to higher inter-rater reliability. When reproposed to the Del-phi panel, the AC-PC line orientation was chosen based both on methodological consistency within the project and evidence of higher overlap of segmented labels across tracers.
On the whole, this “evidence-based Delphi panel”, to our knowledge the first of its kind, seems to have efficiently guided panelists toward convergence. The very initial prediction that the HP could have been defined as a sort of average between the protocols defined or preferred by those participating to the Delphi panel was actually contradicted by results, indicating that the product of this work is an entirely new definition, not uniquely based on possible personal bias of participants, but emerged as the result of a complex procedure that could keep into account a large set of pertinent variables.
4.1. Limitations
The heterogeneities among hippocampal volume estimates across different laboratories using different protocols might arise from sources of variability that were not addressed in the present study. A theoretically relevant source of variability is represented by the different software and settings used for manual segmentation, visualization and volume computation. To date, it is agreed that the available 3D visualization tools greatly improve hippocampal segmentation, and the check of landmarks and morphology in all of the three planes is required for proper segmentation [10]. However, this is allowed through different tools and in different modalities across the available tools. Some software interpolates MR information to provide a clearer rendering of the brain, thus appearing more similar to the real brain than to the voxelized MR. However, there is no evidence on how such interpolation may interfere with appropriate segmentation, nor has any quantitative information ever been collected about reliability of segmentation carried out in these different visualization conditions. In the same way, the visualization of the segmented region is rendered differently by different tools, with some tools showing only the position of the cursor in the 3D planes, and outlining the trace only in the coronal plane, and others filling all of the voxels included in the segmentation with bold color in all three planes, thus making very clearly apparent not only the landmarks, but also the segmented tissue, in all planes. As well, some tools allow not just visualization, but also the editing of segmentation in the 3 planes. The different contribution of these heterogeneous tools to proper hippocampal segmentation should be investigated, and a standard procedure making use of the most helpful tool should be defined on the basis of such evidence that, to our knowledge, has not yet been collected for the specific field of hippocampal volumetry.
In this study, and in the whole project, we have kept all variables constant. In this way, we ruled out discrepancies due to different segmentation tools, but have not been able to estimate any different contribution that they may provide, a task that needs being carried out to set proper standard operating procedures for all steps involved in hippocampal volumetry. More precisely, we chose a freely available software allowing subvoxel segmentation and subvoxel volume computation (i.e., http://www.loni.ucla.edu/Software/MultiTracer). The free distribution of MultiTracer and the accuracy of segmentation allowed by the subvoxel brush are highly desirable features that may facilitate widespread use of this tool for standard hippocampal volumetry. However, this software has other disadvantages, particularly the very limited visualization of the segmented tissue on the sagittal and axial planes (when segmentation is carried out in coronal). Based on all of the above considerations, we must thus underline that hippocampal volumetry as defined by the HarP landmarks and procedures (/centroalzheimer.it/public/SOPs/online/HarmonizedProtocol_ACPC_UserManual_Biblio.Pdf) is not necessarily bound to the methods used in this project. Many features of the different segmentation tools must be carefully evaluated before any final decision is taken about which one should be adopted in the complete standard procedure. In this study we only aimed to provide a standard definition of landmarks and on how to handle specific issues in hippocampal segmentation from MR. This provides a protocol that would prove reliable in AD clinic and research, independently on any other confounding variable, that will need to be addressed in subsequent steps.
Analogous problems arise regarding the computation of total intracranial volume, used to correct hippocampal volumes. We never corrected hippocampal volumes in the different steps of this project, so we did not introduce this sort of variability. This value, however, will be required when using the HarP for both clinical and research aims, and proper investigation aimed to identify the optimal estimate needs to be carried out before defining a standard procedure.
Moreover, individual variability of course affects landmarks, and may affect segmentation variability based on different landmarks. We were not able to exactly quantify this effect, however, the reliability estimates of segmentation units used to help panelists in their decisions were computed on a sample of 77 subjects [11], therefore our estimates may be considered relatively stable on this regard. Future steps of this project will involve the quantification of the variability due to subject in the validation of the HarP [13], and the production of benchmark HarP segmentations on a set of 135 different subjects, to allow training on a wide range of morphologic variability for future tracers and for automated algorithms [16].
Different definitions for hippocampal segmentation may be useful for different aims, especially in areas of research other than AD. However, the definition of the HarP is inclusive enough that it covers most needs, at least as long as one is interested in measuring the whole hippocampus.
On the other hand, the inclusive definition of the HarP does include some structures that may not be considered as hippocampus proper, like the alveus and fimbria and the subiculum. These structures were included because their boundaries with the proper hippocampal gray matter can not be segmented reliably on ordinarily used MR scans. Because they also show a similar degree of atrophy as found in the rest of the hippocampal tissue in AD [11], their inclusion has been considered acceptable for AD studies. However, the recently initiated project of harmonization of hippocampal subfields segmentation may provide evidence and consensus regarding subfield delineation (http://www.hippocampalsubfields.com/).
If the use of the HarP may be regarded as adequate also in different settings than the dementia-related, we should consider that its adoption by centers that are not expert in hippocampal volumetry may be challenging under some respects. The main issue consists in the fact that many users, unfamiliar to hippocampal segmentation, will undergo training a remoto, through a standard system. So far, the rather complex task of hippocampal segmentation was typically taught to new tracers in training sessions carried out personally by experts. The standard use of the HarP will require that sufficiently clear instructions are provided, allowing people in the world to familiarize with the segmentation tool, the preprocessing of the MR, and the segmentation of the hippocampus as prescribed. Within our harmonization project, we have created and tested a standard web-platform [27] devised to train people unfamiliar to the HarP to segment based on the consensual landmarks described in this article. Preliminary data suggest that this system works also for trainees who are not expert of hippocampal segmentation. However, tutorials explaining the whole procedure through a narrated video, reporting performance of segmentation examples with special attention to regions that are frequent sources of mistakes should be produced to help filling the gap of remote communication. We can also anticipate that the large amount of work foreseen for the standard use of hippocampal segmentation for Alzheimer’s disease will soon be performed by automated algorithms, due to the long time and effort required by manual segmentation. Therefore, even greater efforts will be deployed to implement automated segmentation. These would easily be used in both expert and non expert centers. However, implementation of their use will require the development of user-friendly interfaces, and, more importantly and upstream, of accurate and evidence-based criteria enabling their certification as medical devices.
The subsequent steps of this project consist in the validation of the protocol [13] and the production of a large set of benchmark images reflecting appropriate hippocampal segmentation [16]. This set of actions will define a reliable and standard procedure for hippocampal segmentation, as a first step to the use of standard hippocampal volumetry in routine clinical and research activities.
Clinicians and researcher will be able to use hippocampal volumetry to apply, together with the panel of biomarkers recently defined for AD, the International Working Group/National Institute on Aging-Alzheimer’s Association groups and European Medicines Agency criteria [1–4] for diagnostic or enrichment aims in the evaluation of patients with cognitive impairment; developers will be provided with a gold standard against which to validate their automated segmentation algorithms.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: Hippocampal volumetry is a useful biomarker for Alzheimer’s disease (AD) and for enrichment in mild cognitive impairment clinical trials, but the wide heterogeneities among different protocols provides exceedingly different volume estimates across studies. The definition of standard operating procedures for hippocampal volumetry is required for its concrete use as a biomarker. In this work, a consensually HarP for hippocampal volumetry has been defined by a panel of international experts in the field of AD. Interpretation: The use of this protocol will enable to compare the results of different studies directly, and to pool results from different laboratories. This will increase the power of future experiments using hippocampal volumetry and speed up clinical trials for disease-modifying drugs for AD. Future directions: Subsequent steps of this study consist in validating this protocol, and implementing its concrete use by setting systems for remote human and automated algorithms training.
Acknowledgments
The project “Harmonized Protocol For Manual Hippocampal Segmentation: An EADC-ADNI Effort” has been funded thanks to an unrestricted grant from the Alzheimer’s Association, Chicago, US (grant number IIRG-10-174022). The project PI is Giovanni B Frisoni, IRCCS Fatebenefratelli, Brescia, Italy; the co-PI is Clifford R. Jack, Mayo Clinic, Rochester, MN; the Statistical Working Group is led by Simon Duchesne, Laval University, Quebec City, Canada; project Coordinator is Marina Boccardi, IRCCS Fatebenefratelli, Brescia, Italy. EADC Centres (local PI) are: IRCCS Fatebenefratelli, Brescia, Italy (GB Frisoni); University of Kuopio and Kuopio University Hospital, Kuopio, Finland (H Soininen); Höpital Salpètriere, Paris, France (B Dubois and S Lehericy); University of Frankfurt, Frankfurt, Germany (H Hampel); University Rostock, Rostock, Germany (S Teipel); Karolinska institutet, Stockholm, Sweden (L-O Wahlund); Department of Psychiatry Research, Zurich, Switzerland (C Hock); Alzheimer Centre, Vrije Univ Medical Centre, Amsterdam, The Netherlands (F Barkhof and P Scheltens); Dementia Research Group Institute of Neurology, London, UK (N Fox); Neuromed, Centre for Neuroimaging Sciences, London, UK (A Simmons). ADNI Centres are: Mayo Clinic, Rochester, MN (CR Jack); University of California Davis, CA (C DeCarli and C Watson); University of California, Los Angeles (UCLA), CA (G Bartzokis); University of California San Francisco (UCSF), CA (M Weiner and S Mueller); Laboratory of NeuroImaging (LoNI), University of California, Los Angeles (UCLA), CA (PM Thompson and LG Apostolova); Rush University Medical Center, Chicago, IL (L deToledo-Morrell); Rush Alzheimer’s Disease Center, Chicago, IL (D Bennet); Northwestern University, IL (J Csernansky); Boston University School of Medicine, MA (R Killiany); John Hopkins University, Baltimore, MD (M Albert); Center for Brain Health, New York, NY (M De Leon); Oregon Health & Science University, Portland, OR (J Kaye). Other Centres are: McGill University, Montreal, Quebec, Canada (J Pruessner); University of Alberta, Edmonton, Alberta, Canada (R Camicioli and N Malykhin); Department of Psychiatry, Psychosomatic, Medicine & Psychotherapy, Johann, Wolfgang Goethe-University, Frankfurt, Germany (J Pantel); Institute for Ageing and Health, Wolfson Research Centre, Newcastle General Hospital, Newcastle, UK (J O’Brien). Population-based studies: PATH through life, Australia (P Sachdev and JJ Maller); SMART-Medea Study, The Netherlands (MI Geerlings); Rotterdam Scan Study, The Netherlands (T denHeijer). Statistical Working Group: AFAR (Fatebene-fratelli Association for Biomedical Research) San Giovanni Calibita – Fatebenefratelli Hospital – Rome, Italy (P Pasqualetti); Laval University, Quebec City, Canada (S Duchesne); MNI, McGill University, Montreal, Canada (L Collins). Advisors: Clinical issues: PJ Visser, Department of Psychiatry and Neuropsychology, Maastricht University, Maastricht, The Netherlands; EADC PIs: B Winbald, Karolinska Institute, Sweden and L Froelich, Central Institute of Mental Health, Mannheim, Germany; Dissemination & Education: G Waldemar, Copenhagen University Hospital, Copenhagen, Denmark; ADNI PI: M Weiner, University of California San Francisco (UCSF), CA; Population Studies: L Launer, National Institute on Aging (NIA), Bethesda and W Jagust, University of California, Berkeley, CA.
Preparatory work (reported in Boccardi et al., J Alzheimers Dis, 2011; 26:61–75 and in Boccardi et al, Alzheimer’s and Dementia 2013, doi:pii: S1552-5260(13)00078-2) has been funded thanks to unrestricted grants from Lilly International (grant n. 10012941) and Wyeth International (a part of the Pfizer group).
The Statistical Working Group is co-funded through an international collaborative grant from the Ministère du Développement Economique, de l’Innovation et de l’Exportation of Quebec, grant n. PSR-SIIRI 547.
JB is an Alzheimer’s Research UK senior research fellow based at the Dementia Research Centre, Department of Neurodegenerative Disease, UCL, Institute of Neurology. The Dementia Research Centre is an Alzheimer’s Research UK Coordinating Centre and has also received equipment funded by Alzheimer’s Research UK and Brain Research Trust. Part of this work was supported by the NIHR Queen Square Dementia BRU.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.jalz.2014.02.009.
References
- 1.Albert MS, DeKosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–9. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR, Jr, Kawas CH, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–9. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:280–92. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubois B, Feldman HH, Jacova C, Dekosky ST, Barberger-Gateau P, Cummings J, et al. Research criteria for the diagnosis of Alzheimer’s disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6:734–46. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 5.Frisoni GB, Jack CR. Harmonization of magnetic resonance-based manual hippocampal segmentation: a mandatory step for wide clinical use. Alzheimers Dement. 2011;7:171–4. doi: 10.1016/j.jalz.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Committee for Medicinal Products for Human Use - CHMP. Qualification opinion of low hippocampal volume(atrophy) by MRI for use in clinical trials for regulatory purpose in pre-dementia stage of Alzheimer’s disease. EMA/CHMP/SAWP/809208/2011. 2011 Nov 17; [Google Scholar]
- 7.Geuze E, Vermetten E, Bremner JD. MR-based in vivo hippocampal volumetrics: 1. Review of methodologies currently employed. Mol Psychiatry. 2005;10:147–59. doi: 10.1038/sj.mp.4001580. [DOI] [PubMed] [Google Scholar]
- 8.Konrad C, Ukas T, Nebel C, Arolt V, Toga AW, Narr KL. Defining the human hippocampus in cerebral magnetic resonance images–an overview of current segmentation protocols. Neuroimage. 2009;47:1185–95. doi: 10.1016/j.neuroimage.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack CR, Jr, Barkhof F, Bernstein MA, Cantillon M, Cole PE, Decarli C, et al. Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criterion for Alzheimer’s disease. Alzheimers Dement. 2011;7:474–85. e4. doi: 10.1016/j.jalz.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boccardi M, Ganzola R, Bocchetta M, Pievani M, Redolfi A, Bartzokis G, et al. Survey of protocols for the manual segmentation of the hippocampus: preparatory steps towards a joint EADC-ADNI harmonized protocol. J Alzheimers Dis. 2011;26:61–75. doi: 10.3233/JAD-2011-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boccardi M, Bocchetta M, Ganzola R, Robitaille N, Redolfi A, Duchesne S, et al. Operationalizing protocol differences for EADC-ADNI manual hippocampal segmentation. Alzheimers Dement. 2013 doi: 10.1016/j.jalz.2013.03.001. pii: S1552–5260(13) 00078–2. [DOI] [PubMed] [Google Scholar]
- 12.Boccardi M, Bocchetta M, Apostolova LG, Preboske G, Robitaille N, Pasqualetti P, et al. Establishing magnetic resonance images orientation for the EADC-ADNI manual hippocampal segmentation protocol. J Neuroimaging. 2013 doi: 10.1111/jon.12065. http://dx.doi.org/10.1111/jon.12065. [DOI] [PubMed]
- 13.Frisoni G, Jack CR, Bocchetta M, Bauer C, Fredriksen K, Liu Y, et al. The EADC-ADNI Harmonized Protocol for Hippocampal Segmentation on Magnetic Resonance: Evidence of Validity. Alzheimers Dement. doi: 10.1016/j.jalz.2014.05.1756. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Apostolova L, Zarow C, Biado K, Babakchanian S, Boccardi M, Somme J, et al. Pathologic validation of the EADC-ADNI Harmonized Hippocampal Protocol. Neurology. 2014;82(10 Supplement P6):331. [Google Scholar]
- 15.Bocchetta M, Boccardi M, Ganzola R, Apostolova LG, Preboske G, Wolf D, et al. Harmonized benchmark labels of the hippocampus on MR: the EADC-ADNI project. Alzheimers Dement. doi: 10.1016/j.jalz.2013.12.019. in press. [DOI] [PubMed] [Google Scholar]
- 16.Boccardi M, Bocchetta M, Nishikawa M, Ganzola R, Grothe M, Wolf D, et al. Providing standardized labels of the EADC-ADNI harmonized hippocampal protocol for automated algorithm training. Alzheimer’s Association International Conference; Boston. 2013; Congress Presentation. [Google Scholar]
- 17.Murphy M, Black N, Lamping D, McKee C, Sanderson C, Askham J, et al. Consensus development methods, and their use in clinical guideline development. Health Technol Assess. 1998;2:1–88. [PubMed] [Google Scholar]
- 18.Convit A, De Leon MJ, Tarshish C, De Santi S, Tsui W, Rusinek H, et al. Specific hippocampal volume reductions in individuals at risk for Alzheimer’s disease. Neurobiol Aging. 1997;18:131–8. doi: 10.1016/s0197-4580(97)00001-8. [DOI] [PubMed] [Google Scholar]
- 19.Killiany RJ, Moss MB, Albert MS, Sandor T, Tieman J, Jolesz F. Temporal lobe regions on magnetic resonance imaging identify patients with early Alzheimer’s disease. Arch Neurol. 1993;50:949–54. doi: 10.1001/archneur.1993.00540090052010. [DOI] [PubMed] [Google Scholar]
- 20.Malykhin NV, Bouchard TP, Ogilvie CJ, Coupland NJ, Seres P, Camicioli R. Three-dimensional volumetric analysis and reconstruction of amygdala and hippocampal head, body and tail. Psychiatry Res. 2007;155:155–65. doi: 10.1016/j.pscychresns.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 21.Haller JW, Banerjee A, Christensen GE, Gado M, Joshi S, Miller MI, et al. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202:504–10. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- 22.Bartzokis G, Altshuler LL, Greider T, Curran J, Keen B, Dixon WJ. Reliability of medial temporal lobe volume measurements using reformatted 3D images. Psychiatry Res. 1998;82:11–24. doi: 10.1016/s0925-4927(98)00007-9. [DOI] [PubMed] [Google Scholar]
- 23.deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, et al. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging. 2004;25:1197–203. doi: 10.1016/j.neurobiolaging.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Jack CR., Jr MRI-based hippocampal volume measurements in epilepsy. Epilepsia. 1994;35(Suppl 6):S21–9. doi: 10.1111/j.1528-1157.1994.tb05986.x. [DOI] [PubMed] [Google Scholar]
- 25.Pantel J, O’Leary DS, Cretsinger K, Bockholt HJ, Keefe H, Magnotta VA, et al. A new method for the in vivo volumetric measurement of the human hippocampus with high neuroanatomical accuracy. Hippocampus. 2000;10:752–8. doi: 10.1002/1098-1063(2000)10:6<752::AID-HIPO1012>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 26.Soininen HS, Partanen K, Pitkanen A, Vainio P, Hanninen T, Hallikainen M, et al. Volumetric MRI analysis of the amygdala and the hippocampus in subjects with age-associated memory impairment: correlation to visual and verbal memory. Neurology. 1994;44:1660–8. doi: 10.1212/wnl.44.9.1660. [DOI] [PubMed] [Google Scholar]
- 27.Duchesne S, Valdivia F, Robitaille N, Abiel Valdivia F, Bocchetta MM, Boccardi M, et al. Manual segmentation certification platform. IEEE; 2013. pp. 35–9. Medical Measurements and Applications Proceedings (MeMeA) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.