Abstract
With the rapid growth of electronic medical records (EMR), there is an increasing need of automatically extract patterns or rules from EMR data with machine learning and data mining technqiues. In this work, we applied unsupervised statistical model, latent Dirichlet allocations (LDA), to cluster patient diagnoics groups from Rochester Epidemiology Projects (REP). The initial results show that LDA holds the potential for broad application in epidemiogloy as well as other biomedical studies due to its unsupervised nature and great interpretive power.
Introduction
With the rapid growth of electronic medical records (EMRs), it becomes more and more essential to develop methods to automatically mine information from EMRs with machine learning and data mining techniques in a timely and accurate manner [1, 2].
Recently, Latent Dirichlet Allocations (LDA) [3] has gained popularity in diverse fields due to the fact that it holds great promise as a means of gleaning actionable insight from the text or image datasets. In natural langauge processing (NLP), LDA clusters both words and documents into topics by approximating word or term distributions [4]. As an unsuperivsed statistical model, LDA makes use of Bayseisan inference to update the probability estimates for a hypothesis.
As LDA does not require a priori knowledge but can generate good interpretative models, enjoy good portability [5] and meanwhile it has the flexibility of adding implicit as well as explict priors to build diverse models [6–9], it thus holds the potential for broad applications, such as comorbidity studies, drug repurposing, biological connections among diseases and so on in biomedical research [10]. In this paper, we propose to use LDA to identify associations among diagnosis code groups utilizing an epidemiology cohort, Rochester Epidemiology Projects (REP) [11], and aim to understand the comorbidities. The paper starts with the introduction of background and related work in section 2; it then presents experimental methods in section 3 where the experiment data is introduced and adapted topic modeling for diagnosis group associations and topic analysis approaches are illustrated respectively. Section 4 presents the results and what can be found from those topics. Finally, in section 5, we discuss potential expansions, existing limitations and how we can make more improvements.
Background and Related Work
Disease classification and grouping in epidemiology studies
In epidemiology, the three Cs (cause, contribute and correlate) in studying disease etiology proposed by Green [12] have long been the principle. However, diseases can be related biologically or phenotypically. There are different approaches to group diseases. The first approach defines disease groups by the symptoms of the affected organ. This kind of grouping derives from observational correlation between pathological analysis and clincal syndromes [13]. With the development of novel quantitaitive approaches to network analysis and the explosion of currently avaiable genomic, transcrptomic, proteomic and metabolomic data sets, biological systems based on network has been applied to disease classifications [14].
The most popular disease classification systems used is the International Classification of Disease (ICD) [15, 16], which classifies diseases systematically based on the analysis of the general health situation of population groups. It is used to monitor the incidence and prevalence of diseases and other health problem and has become the standard diagnosis tool for epidemiology.
However, ICD classification can be too finer granualarity for clinical practice since the number of ICD codes is too large and the distinctions among some codes are not clear. The large number of ICD-9-CM (the 9th version, Clinical Modification) codes also makes statistical analysis and reporting difficult and time-consuming. the Agency for Healthcare Research and Quality (AHRQ) introduces Clinical Classification Software [17] (CCS) to cluster patient diagnosis and procedures into a manageable number of clinically meaningful categories. This way, 14,000 diagnosis codes are reduced to 279 groups.
Topic modeling in boimedical informatics Specifications
In biomedical informatics, probabilistic topic modeling has been applied to patients’ notes to discover relevant clinical concepts and relations between patients [18]. Angues et al. [19] applied unsupervised LDA to primary clinical dialogues for visualizing shared content in communication. Wang et al. developed BioLDA [20] to find complex biological relationships in recent PubMed articles. Wu and Xu [21] made use of LDA to rank gene-drug relationships in biomedical literatures based on Kullback-Leibler (KL) distance between topics derived from LDA. Bisgin et al. [12, 22] mined FDA drug labels using topic modeling. Fifty-two unique topics, each containing a set of terms, were identified and then the probabilistic topic associations were used to measure the similarity between drugs. Bian et al. [23] utilized the topic features to categorize the collections tweets into latent topics and those topics are used as features to train SVM prediction models for mining adverse effects labels. Newman et al. [24] and Bundachus and Tresp [25] employed topic models to interpret MeSH terms. Chen et al. [26] proposed to use LDA to promote ranking diversity for genomics information retrieval and they claimed that topic distributions of retrieval passages can help identify aspects more accurately. Chen et al. [27] extended LDA by including background distribution to study microbial samples. Under their setting, each microbial sample is a document and each functional element is a word. They found that estimating the probabilistic topic model can uncover the configuration of functional groups. All of those studies have shown the potentiality of topic modeling.
Experimental Methods
In this study, our main goal is to investigate the effectiveness of topic modeling in discvering assocations among disease groups. We first generate topic distribution for selected medical records for certain population and then the connections among disease groups are analyzed.
Rochester epidemiology project (REP) and data inclusion
The Rochester Epidemiology Project (REP) is a collaboration between health care providers in southeaster Minnesota, which involves Olmsted Medical Center, Mayo Clinic, Rochester Family Medicine Clinic and other medical care providers in southeastern MN. The REP is a unique records-linkage research infrastructure that has existed since 1966. It includes the medical records of all persons who have ever lived in Olmsted County, Minnesota between January 1, 1966 and the present, and who have given permission for their medical information to be used for research. Those persons comprise more than 500,000 unique individuals and more 6 million person years of follow-up through 2010. Historically, the Olmsted County population is less racially diverse then the US as a whole [11, 28] and similar to the state of Minnesota and surrounding states [29]. The REP data we use has been processed and saved as a matrix with rows being the patient ID and columns the diagnosis code group defined by AHRQ. There are 256 diagnosis code groups in total in our data. As an initial study, we only select 4644 patients who are above 65 and paid 80 visits over the chosen set of years for this study.
Topic modeling
Topic modeling is originally a tool for text analysis. Now, we adapt it to the association analysis of diagnosis group. In text analysis, LDA represents a document as a mixture of fixed topics. Under the context of our data, LDA represents a collection of patients as a mixture of fixed topics. Each topic z has the weight in a patient p and each topic is a distribution over a finite vocabulary of diagnosis code groups, and each code group c has a probability ϕ in topic z. Placing symmetric Dirichlet priors on θ and ϕ, with θ ~ Dirichlet(α) and ϕz ~ Dirichlet(β), where, α and β are hyper-parameters to control the sparsity of distributions, the generative model is given by:
where K is the total number of topics, C is the total number of diagnosis code groups in the patient collection, and pi and zi are the passage and the topic of the ith code group ci respectively. Each code group in the vocabulary ci ∈ V = [c1, c2, …,Cc] is assigned to each latent topic variable zi. Given a topic zi = k, the expected posterior probability of topic mixings of a given patient p and the expected posterior probabilities of code group ci are calculated as below.
where is the count of ci in topic k, and np,k is the count of topic k in patient p.
In this study, we used the LDA approach to obtain the parameter ϕ for every diagnosis code group. The topics were extracted by using the R package topicmodels, which is based on Blei et al [3].
1.1. Associations discovery of diagnosis group
The topic distributions over diagnosis code group measures the connection (or relatednedss) of a disease with a specific topic (i.e. the conditional probability of topic for a given disease as shown in Figure 1. As shown in the previous section, in our work, the document is the patient while the term is the diagnosis code group. Therefore, the posterior distribution would determine the probability of a patentient given a topic and would determine the probability of a diagonosis code group given a topic. More specifically, some patients were assigned to the most probable topics and some diagnosis code groups were assigned to the most probable topics.
Figure 1.
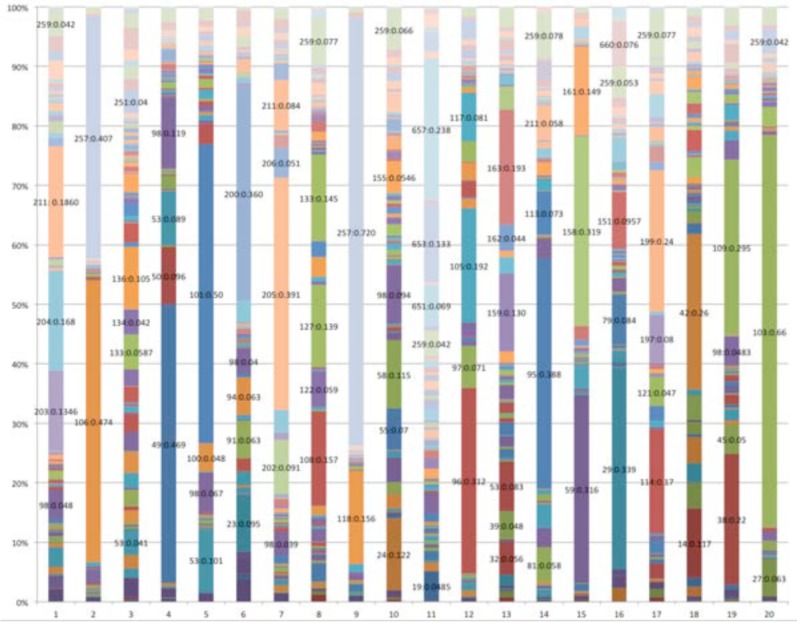
Diagnosis code group proportion for 20 topics where x-axis is the topic and y-axis is the proportion of each code group in that topic
Results and Analysis
There are a total of 4644 patients with their diagnosis code groups obtained with simeple exclusion criterias described above. LDA was employed to generate topic distributions for both the patients and the diagnosis code groups. We tested diverse topic numbers ranging from 20 to 147 and compared the resultant topics with respect to loglikelihood distributions and perplexities. Similar results were obtained when the number of topics is between 20 to 35. We chose the number of topics to be 20 and analyzed the common properties shared by the diseases with proportion higher than 0.05 in each topic.
Topic analasis in terms of disease relations
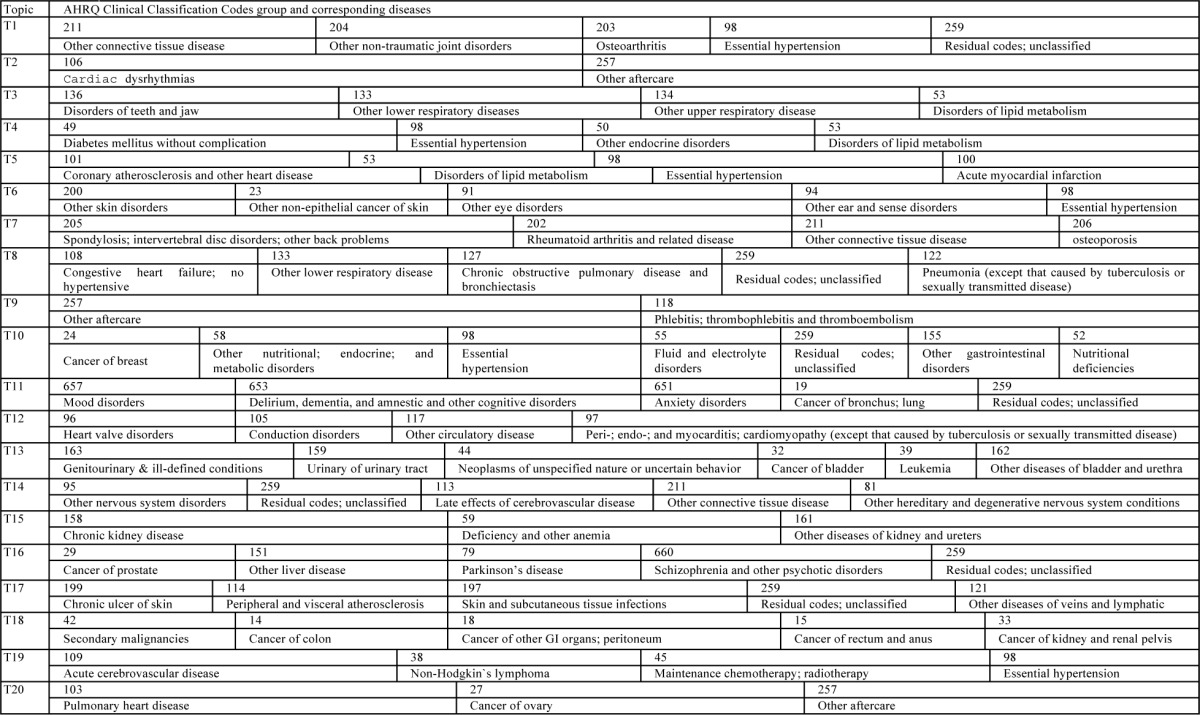
In Figure 1, the proportion of diagnosis code group for each of the topics is drawn with sample results when topic each topic is dominated by a few code groups which involve much larger ratios than remainings. Namely, each topic is represented by a few key diognosis code groups. In Table 1, the interpreations of those dominant diagnosis code groups are given. The five diagnosis code groups in T1 are almost relatedto joint disorders except the last two, 98 and 259. T7 is also about joints, but it focuses more infections. The last two, are found in many topics. In fact, they two can be thought related to diverse diseases. That is why they have high proportions in many topics. Two components occupy 0.88 of T2 and T9. Both of them invovle the code aftercare while the other one for T2 is related to heart rhythm and the one for T9 is to infections in intravenes. It seems that these two topics are not clustered very well. But if we think from the perspective that aftercare plays imporant parts in quite a few severe diseases, especially diseases related to heart, it is quite reasonable for them two to co-occur often. T3 is obviously about respiratory diseases, with the four main codes nearly evenly distributed. Diseases in T4 seem to all related to fat-induced diseases since diabetes, hypertension, lipid metabolism may all be causes by eating too much high-calory food. T8 all involves repiratory. Congestive heart failure and repiratory problems may be related. T5 and T12 are about heart diseases. number is 20. As can be seen, although each topic is composed of some proportion of all 253 diagnosis code group,
Table 1.
Corresponding diagnosis code group for each topic in Figure 1
|
Nonetheless, T5 is more about heart organ itself while T12 is more about the circulation. T6, T11, T13, T14, T15, T17 and T18 have strong category features as sense, mental, nervous, urinary, system, kidney, skin and gastrointestinal diseases. T10 can be classified as internal secretion diseases. T16 seems more about dieseases seen among old people although data we used is in fact about patients who are older than 65. The results indicated that topic modeling can yield statistically significant topics that group and identify diseases sharing some commonality. Basically, what we have discovered about diseases, is consistent with what is shown by topic modeling for other domains, like text mining, natural langauge processing or image processing.
Topic analysis in terms of patient grouping
Figure 2 shows the distributions of patients’ topic assignemtns. T1, T2 and T3 occupy about 0.3 among all topics and T4, T5, T6 and T7 also share about 0.06 respectivlye while T18, T19 and T20 only occupies about 0.017, 0.015 and 0.008 respectively. This is natural since the first seven diseases are all about heart diseases, respirative, tissue or joint disease which are quite common ones among old people. In contrast, the last three are about some rare diesease such as colon cancers, cerebrovascular or cancer of overy.
Figure 2.
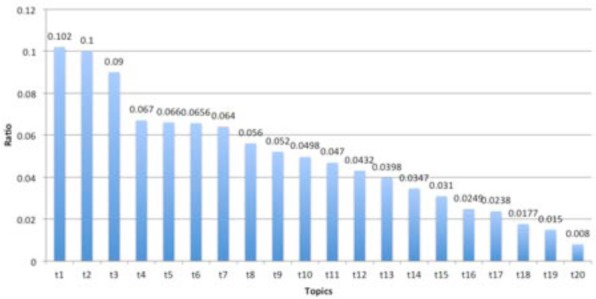
Patient ratio among topics
The actual counts of diagnosis code groups for each topic are somewhat different from the corresponding proportions. The former is based on the maximum probability of some topics for the given patients while the proportions are calculated with the summation of posterior probabilities for each topic. This difference shows that some diagnosis code groups have more counts than others. Namely, for some diseases, patients have to pay more visits than other diseases. Hence, the patient topic distribution analyses can reveal the subtle nature of diseases.
Discussion and Limitations
Although many techniques, such as principle component analysis (PCA) [30], factor analysis (FA) [31] or probabilistic latent semantic indexing (pLSI) [32] have been used in clustering medical data, topic modeling has been proved to be a model with distinct advantages. One of them is to group semantically related documents as well as terms together. In this work, LDA groups related diagnosis code groups into clusters. This provides strong interpretive potential in making phenotyping analysis or designing clinical decision support systems. Secondly, in contrast to PCA, FA or pLSI, LDA assume that each document may involve multiple components or topics and the generative process is based on Bayesian nature. Therefore, it is suitable for hierarchical analysis. Thirdly, the Dirichlet prior enables LDA can smooth its topic distribution, thus overcoming the overfitting problem of other models.
Another advantage is the unsupervised nature of LDA and its flexibilities. LDA itself does not require any training data or a priori knowledge about diseases. However, it doesn’t prevent LDA to incorporate supervised information or external knowledge as prior or even as supervised labels. In our on-going work, one of our goals is to use section headers, physicians comments or labels on clinical notes as observed side information to train supervised or semi-supervised topic models for prediction tasks. LDA is designed for document analysis mainly because it is good at doing heterogeneous data analysis. Hence, it is now broadly used in image processing, bioinformatics and information retrieval. That is the main reason that we applied LDA in diagnosis code analysis.
Undoubtedly, there are limitations for the unsupervised LDA. The first limitation is the inconsistent mapping between the topics and the actual common properties of disease group. This can be found from the 20 topics generated. Some topics cluster some diagnosis code group together without much similarity. For example, cancer of prostate and other liver diseases in topic 16 seem not so related but they are the two highest code groups in it. Yet, we cannot say there is no reason for them to cluster together. They may be related due to some uncovered comorbidity. Finding out the exact cause requires addition information and domain knowledge. If we can add some supervised information, we may have a better control on the model generation and prediction. This may also imply that a topic is not necessarily associated with only one concept, and it could be related to several commonalities shared by diagnosis code group. The third limitation is that in this work, we didn’t do much on the evaluations though we review and measure whether topics generated fit classification standard in AHRQ. It is still necessary to evaluate topics from other standards, such as similarity measurements, human judgments and so on.
In addition, there may be inconsistency for the results of each sampling. A common problem existing among sampling methods is its stability. LDA, starting with Dirichlet distributions, generates topic distributions. Next, it generates topics and diagnosis code groups in turn via a series of multinomial distributions. Although the conjugate nature between Dirichlet and multinomial distributions guarantee the theoretically soundness and the simplicity of the model, the results, after a few hundreds of iterations via Gibbs sampling, yielded are usually slightly different each time. Although we cannot fully control the stability of Gibbs sampling, Sato et al. [33] and Asuncion et al. [34] have proved that the collapsed variational Bayes inference with a zero-order Taylor expansion approximation, called CVB0 inference can get better performance than Gibbs sampling methods. Replacing the current inference methods with CVB0 can be one solution to explore in the future.
In this work, we identified 20 topics that could almost be connected with some group of diseases. However, we also observe that the same diagnosis code group might fall into different topics. For example, Residual codes; unclassified has been seen to share above 5% among 7 different topics. Based on the AHRQ definition, such codes cannot exactly be classified. This may be partially the reason that such codes are assigned to different topics. Such phenomenon is very popular in human languages considering the polysemy natures of words. But in diagnostic code grouping analysis, this may lead to confusions on the topic grouped together if we cannot find strong reasons for them. The phenomenon may need domain experts to interpret. Further distance assessment, like KL divergence or mutual information, may help find clearer demarks between each group.
Conclusions and Future Work
This study ivestigates the efficacy of topic modeling for the discovery of hidden patterns from a large epidemiology cohort. The results demonstrate that disease groups based on topic modeling do have statistically significance and also can reveal semantic commonalities among diseases. In our future work, we would add other patient information, such as drug, lab, procedure events and temporality to the analysis. In addition, temporal trends plays important roles in any epidemiological study. In addition, we would focus on an “interesting subpopulation” (e.g., a very complex or poorly understood disorder) to explore whether topic modeling help to unravel a complex disorder. The construction of temporal topic modeling on an epidemiology cohort may also lead to interesting discovery.
Acknowledgments
This work was made possible by joint funding from National Institute of Health R01LM009959A1 and National Science Foundation ABI:0845
References
- 1.Smith CA. ElectronicHealth Records. 2003 [Google Scholar]
- 2.Li J-s, Yu H-y, Zhang X-g. Data Mining in Hospital Information System.
- 3.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. Journal of Machine Learning Research. 2003;3:993–1022. [Google Scholar]
- 4.Griffiths TL, Steyvers M. Finding scientific topics; Proceedings of the National academy of Sciences of the United States of America; 2004. pp. 5228–5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisgin H, et al. Mining FDA drug labels using an unsupervised learning technique-topic modeling. BMC bioinformatics. 2011;12(Suppl 10):S11. doi: 10.1186/1471-2105-12-S10-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.AlSumait L, Barbará D, Domeniconi C. On-line lda: Adaptive topic models for mining text streams with applications to topic detection and tracking. in Data Mining, 2008. ICDM′08; Eighth IEEE International Conference on; IEEE; 2008. [Google Scholar]
- 7.Xu G, Zhang Y, Yi X. Modelling user behaviour for web recommendation using lda model. in Web Intelligence and Intelligent Agent Technology, 2008. WI-IAT'08; IEEE/WIC/ACM International Conference on; IEEE; 2008. [Google Scholar]
- 8.Phan X-H, Nguyen L-M, Horiguchi S. Learning to classify short and sparse text & web with hidden topics from large-scale data collections; Proceedings of the 17th international conference on World Wide Web; ACM; 2008. [Google Scholar]
- 9.Nguyen C-T, et al. Web search clustering and labeling with hidden topics; ACM Transactions on Asian Language Information Processing (TALIP); 2009. p. 12. [Google Scholar]
- 10.Bisgin H, et al. Investigating drug repositioning opportunities in FDA drug labels through topic modeling. BMC bioinformatics. 2012;13(Suppl 15):S6. doi: 10.1186/1471-2105-13-S15-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauver JLS, et al. Use of a medical records linkage system to enumerate a dynamic population over time: the Rochester epidemiology project. American journal of epidemiology. 2011;173(9):1059–1068. doi: 10.1093/aje/kwq482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green J. The three C’s of etiology. Wide Smiles; 1996. [Google Scholar]
- 13.Loscalzo J, Kohane I, Barabasi A-L. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Molecular systems biology. 2007;3(1) doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bugrim A, Nikolskaya T, Nikolsky Y. Early prediction of drug metabolism and toxicity: systems biology approach and modeling. Drug discovery today. 2004;9(3):127–135. doi: 10.1016/S1359-6446(03)02971-4. [DOI] [PubMed] [Google Scholar]
- 15.Cherkin DC, et al. Use of the International Classification of Diseases (ICD-9-CM) to identify hospitalizations for mechanical low back problems in administrative databases. Spine. 1992;17(7):817–825. doi: 10.1097/00007632-199207000-00015. [DOI] [PubMed] [Google Scholar]
- 16.Found EC. ICD-9-CM Codes.
- 17.Elixhauser A, Steiner C, Palmer L. Clinical classifications software (CCS) Book Clinical Classifications Software (CCS); 2008. (Editor ed^ eds) [Google Scholar]
- 18.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic acids research. 2004;32(suppl 1):D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hersh WR, et al. TREC 2006 Genomics Track Overview. TREC. 2006 [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, et al. Finding complex biological relationships in recent PubMed articles using Bio-LDA. PLoS One. 2011;6(3):e17243. doi: 10.1371/journal.pone.0017243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellaachia A, Guven E. Predicting breast cancer survivability using data mining techniques. Age. 2006;58(13):10–110. [Google Scholar]
- 22.Jackson S, et al. Bacillus cereus and Bacillus thuringiensis isolated in a gastroenteritis outbreak investigation. Letters in Applied Microbiology. 1995;21(2):103–105. doi: 10.1111/j.1472-765x.1995.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 23.Ogilvie MM, Tearne CF. Spontaneous abortion after hand-foot-and-mouth disease caused by Coxsackie virus A16. British medical journal. 1980;281(6254):1527. doi: 10.1136/bmj.281.6254.1527-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman D, Karimi S, Cavedon L. Using topic models to interpret MEDLINE’s medical subject headings; AI 2009: Advances in Artificial Intelligence; Springer; 2009. pp. 270–279. [Google Scholar]
- 25.Bimboim H, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic acids research. 1979;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Y, et al. A LDA-based approach to promoting ranking diversity for genomics information retrieval. BMC genomics. 2012;13(Suppl 3):S2. doi: 10.1186/1471-2164-13-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, et al. Bioinformatics and Biomedicine (BIBM), 2011 IEEE International Conference on. IEEE; 2011. Inferring functional groups from microbial gene catalogue with probabilistic topic models. [Google Scholar]
- 28.St Sauver JL, et al. Data resource profile: the Rochester Epidemiology Project (REP) medical records-linkage system. International journal of epidemiology. 2012;41(6):1614–1624. doi: 10.1093/ije/dys195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.St Sauver JL, et al. Mayo Clinic Proceedings. Elsevier; 2012. Generalizability of epidemiological findings and public health decisions: an illustration from the Rochester Epidemiology Project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolliffe I. Principal component analysis. Wiley; Online Library: 2005. [Google Scholar]
- 31.Kline P. An easy guide to factor analysis. 1993 [Google Scholar]
- 32.Hofmann T. Proceedings of the 22nd annual international ACM SIGIR conference on Research and development in information retrieval. ACM; 1999. Probabilistic latent semantic indexing. [Google Scholar]
- 33.Sato I, Nakagawa H. Rethinking collapsed variational Bayes inference for LDA. 2012. arXiv preprint arXiv: 1206.6435.
- 34.Asuncion A, et al. On smoothing and inference for topic models; Proceedings of the Twenty-Fifth Conference on Uncertainty in Artificial Intelligence; AUAI Press; 2009. [Google Scholar]


