Abstract
Electron crystallography is well suited for studying the structure of membrane proteins in their native lipid bilayer environment. This technique relies on electron cryomicroscopy of two-dimensional (2D) crystals, grown generally by reconstitution of purified membrane proteins into proteoliposomes under conditions favoring the formation of well-ordered lattices. Growing these crystals presents one of the major hurdles in the application of this technique. To identify conditions favoring crystallization a wide range of factors that can lead to a vast matrix of possible reagent combinations must be screened. However, in 2D crystallization these factors have traditionally been surveyed in a relatively limited fashion. To address this problem we carried out a detailed analysis of published 2D crystallization conditions for 12 β-barrel and 138 α-helical membrane proteins. From this analysis we identified the most successful conditions and applied them in the design of new sparse and incomplete factorial matrices to screen membrane protein 2D crystallization. Using these matrices we have run 19 crystallization screens for 16 different membrane proteins totaling over 1,300 individual crystallization conditions. Six membrane proteins have yielded diffracting 2D crystals suitable for structure determination, indicating that these new matrices show promise to accelerate the success rate of membrane protein 2D crystallization.
Keywords: Two-dimensional (2D) crystal, membrane protein, electron crystallography, high-throughput screening, membrane protein reconstitution, electron cryomicroscopy, 96-well format
1. Introduction
Membrane protein electron crystallography was pioneered in the 1970s by Henderson and Unwin through their studies of bacteriorhodopsin (Henderson and Unwin, 1975), and relies on electron cryomicroscopy (cryo-EM) of two-dimensional (2D) crystalline specimens of membrane proteins in a lipid bilayer. This method is thus ideal for studying the structure of membrane proteins in their natural membrane environment (Ubarretxena-Belandia and Stokes, 2010;Ubarretxena-Belandia and Stokes, 2012). As in X-ray crystallography, growing suitable crystals represents one of the major bottlenecks in the application of this technique. 2D crystals are typically grown by reconstitution of purified, detergent-solubilized membrane proteins into lipid bilayers at a high enough density to favor the formation of a regular array (Jap et al., 1992;Kühlbrandt, 1992;Mosser, 2001). Several methods, including dialysis (Kühlbrandt, 1992), controlled dilution (Remigy et al., 2003), adsorption onto a hydrophobic resin (Rigaud et al., 1997) or complexation with cyclodextrins (Signorell et al., 2007) are generally employed for detergent removal and reconstitution of the protein into proteoliposomes. Identifying the conditions for growing 2D crystals requires screening over a wide range of factors including pH, temperature, lipid composition, lipid-to-protein ratio, detergent, amphiphiles, mono- and divalent-ions, inhibitors and ligands. A systematic screen over all of these factors generates a huge matrix of possible reagent combinations, which should ideally be sampled to cover the majority of 2D crystallization space. For 3D crystallization, a vast portion of crystallization space can be screened efficiently and rapidly using sparse (Jancarik et al., 1991;Rupp and Wang, 2004) and incomplete factorial crystallization matrices (Carter Jr, 1990;Gorrec et al., 2011) in combination with high-throughput approaches. In contrast, factors relevant for 2D crystallization have traditionally been surveyed in a relatively limited fashion, potentially missing truly optimal conditions, or in some cases failing to even obtain crystals. However, the recent development of high-throughput tools for 2D crystallization (Cheng et al., 2007;Vink et al., 2007;Coudray et al., 2008;Hu et al., 2010;Iacovache et al., 2010;Karathanou et al., 2010;Kim et al., 2010;Coudray et al., 2011) make it now possible to conduct 2D crystallization trials at a higher pace and reproducibility, and moreover, sufficient amount of data is now available on membrane protein 2D crystallization (reviewed in (Abeyrathne et al., 2012)) to allow the rational design of new and more comprehensive 2D crystallization screens. To this end we first built a 2D crystallization database with information mined from successful 2D crystallization conditions reported in the literature. We analyzed this information to evaluate the effect of the different crystallization factors, and from this analysis we designed new sparse and incomplete factorial matrices to screen membrane protein 2D crystallization. Using these matrices we have been able to grow 2D crystals suitable for structure determination for several membrane proteins.
2. Materials and methods
2.1 2D crystallization database
To construct a database of 2D crystallization experiments we mined the successful conditions from ~250 2D crystallization screens published in ~200 journal articles. To guide us in our literature search we used a recent review by Abeyrathne et al. (Abeyrathne et al., 2012), which tabulated all the membrane proteins studied by electron crystallography up to the year 2012. We tabulated all crystallization conditions according to different factors including pH, temperature, lipid composition, lipid-to-protein ratio, detergent, amphiphiles, and mono- and divalent-ions, along with their respective concentrations. We completed this database with additional fields to describe particular properties of the membrane protein, detergent, and lipid that constitute the initial ternary mixture in a typical 2D crystallization experiment. The 2D crystallization conditions were analyzed by constructing a series of bar charts showing the number of entries in the database as a function of individual crystallization factors.
2.2 Design of a sparse matrix 2D crystallization screen
To design the screen in an unbiased manner we applied the k-means algorithm to form 10 groups using 94 successful 2D crystallization conditions from 57 unique membrane proteins. These conditions were chosen to be as non-redundant as possible. The input fields for the algorithm were: phospholipids characterized by their alkyl chain length and headgroup composition (in percentages of Phosphatidylcholine (PC), Phosphatidylethanolamine (PE), Phosphatidylglycerol (PG), Phosphatidic acid (PA), Phosphatidylserine (PS), Cardiolipin (CA) and E.coli polar lipid extract), pH, NaCl and MgCl2 concentration, and temperature characterized by its median value and variation (which is non-zero when temperature cycling is used).
2.3 Design of an incomplete factorial 2D crystallization screen
The design of so called “incomplete factorial screens” or “grid screens” relies on three major rules: 1) the level of the different factors are assigned randomly; 2) first-order interactions of the level should be balanced; 3) redundancy should be avoided so setups are as different as possible (Carter and Carter, 1979). In order to design a comprehensive incomplete factorial 2D crystallization screen we first identified the ten main factors affecting 2D crystallization, each one of which represents an axis in a ten-dimensional crystallization space. Second, we assigned discrete levels for each factor that then represent points along the corresponding axis. We note that the low occurrence of some of these levels in the database may not reflect the fact that they are not successful, but rather that they have been rarely used. Nevertheless, these levels have been included in the design of the incomplete factorial matrix. For instance, lipid composition is represented by one axis and the levels include commonly used lipids, such as DMPC, DOPC and E.coli lipid extracts, as well as seldom used lipids like DOPG. This approach ensures coverage of a wide range of lipid headgroups, chain lengths and degree of unsaturation. To be compatible with the conventional high-throughput format of 96 conditions per run, our incomplete factorial matrix consisted of 90 conditions (corresponding to points in the ten-dimensional space) plus 6 spots for controls. The 90 points were randomly selected with two constraints: each level should be represented the same number of times and second-order interactions were balanced to avoid redundancy. For instance, the second constraint forced a given lipid to be combined with a multitude of pHs. In this manner over 20,000 trial matrices were generated. In order to identify one matrix covering the largest amount of ten-dimensional space we computed the standard deviation (SD) of the nearest neighbor-distance (nnd). More specifically, for each of the 90 conditions we computed nnd values relative to the remaining 89 conditions. We note that, because we minimized redundancy during generation of the trial matrices, none of the conditions were identical and thus the nnd values were always > 0. As a consequence, if the conditions were equidistant from each other in ten-dimensional space, then the nnd values would be equivalent and the SD(nnd) would be zero. Following this argument, the matrix with the lowest SD(nnd) represents the most dispersed matrix, i.e., the matrix that most effectively samples the ten-dimensional crystallization space.
2.4 Lipids, detergents and Proteins for 2D crystallization screens
Lipids supplied as powder were purchased from Avanti Polar Lipids (Alabaster, AL) and detergents were bought from Anatrace (Maumee, OH). Detergent solubilized lipid stocks, at a final lipid concentration of 2mg/ml, were prepared in distilled water by first resuspending the lipid at 10mg/ml, and then mixing a 200µl aliquot from this suspension with 800µl of aqueous solution containing detergent. As previously described (Kim et al., 2010), turbidity measurements were employed to determine the minimal detergent concentration needed to solubilize each lipid species. These detergent solubilized lipid stocks could be stored at 4°C for up to four days, or frozen at −80°C for long-term storage.
Sixteen different membrane proteins were expressed and purified either in our own laboratories or in collaborating laboratories. These proteins were usually expressed with either N- or C-terminal affinity-tags to allow purification by affinity chromatography. Generally, the linker region between the tag and the target protein included a TEV or thrombin proteolytic site and, when possible, the tag was removed by proteolysis. The stability, polydispersity, and oligomeric state in detergent for some of these membrane proteins was determined using size-exclusion chromatography performed in combination with analyses of static light scattering, ultraviolet absorbance, and refractive index using protocols described in Slotboom et al. (Slotboom et al., 2008). Each purified protein, at a final concentration of 0.5-1.5mg/ml, was clarified by centrifugation at 200,000xg for 30 minutes before crystallization.
2.5 Set up of the 96-well micro-volume dialysis crystallization plate
A protein concentration of 0.3-0.6mg/ml and a volume of 15–25µl were employed per crystallization condition. Thus ~0.4-1mg of a membrane protein target was needed to set up a complete 96-condition screen. Aliquots of 15–25µl of the purified membrane protein at different lipid-to-protein ratios were prepared in 96-well disposable microtiter plates, adding the protein solution last. These samples were incubated for 1h at room temperature to ensure a homogeneous population of protein/detergent/lipid micelles. Thereafter, each sample was transferred to individual sample wells of the microfluidic dialysis plate. This commercially available (XZ-HT-96) 96-well micro-volume dialysis crystallization plate was designed by GN Biosystems (Santa Clara, CA) and can accommodate sample and buffer volumes of 5–28µl and up to 500µl, respectively. The MWCO of the dialysis membrane ranged from 12,000 to 14,000Da. Following the addition of the protein and buffer, the plates were film sealed and incubated at the desired dialysis temperature. Buffer was exchanged twice a day for a period of up to two weeks. After removing the sealing film the samples were recovered through slits at the top of each dialysis chamber and transferred to a 96-well disposable microtiter plate. As an alternative to the 96-well micro-volume dialysis crystallization plate a fraction of the conditions were also set up in 20µl dialysis buttons against 50ml dialysis buffer for detergent removal (Stokes et al., 2010).
2.6 Automated EM analysis of crystallization trials
Our automated pipeline to screen membrane protein 2D crystallization has been described elsewhere (Kim et al., 2010), and more details can be found at http://temimps.nysbc.org/technology.htm., or at the Structural Biology Knowledgebase (http://www.sbkb.org). In brief, batches of 100 carbon coated EM grids were prepared as described in Vink et al. (Vink et al., 2007). For automated negative staining, the EM grids were manually transferred onto a magnetic support platform (SPRI plate 384 Post Magnet Plate, Agencourt, Beverly MA) that was placed on a liquid-handling robot (Biomek FX, Beckman Coulter, Fullerton, CA). The staining of 2µl of each crystallization condition using a solution of 0.25% uranyl acetate in water was carried out by the liquid-handling robot using a 96-pipette head as previously described (Kim et al., 2010). The specimen grids were then manually transferred to a custom-made storage tray for automated grid insertion into the electron microscope and image acquisition (Hu et al., 2010). The specimen grids were inserted with a two-part robot consisting of a selective compliance articulated robot arm (SCARA) for loading samples into a microscope holder containing a modified tip, and a Cartesian robot for placing the holder into the electron microscope. A standard 120 keV JEOL-1230 transmission electron microscope (JEOL USA Inc.) modified with a rewired toggle switch controlling the airlock to allow robot control was used. A computer program for controlling the robots was integrated with the Leginon program (Potter et al., 1999;Suloway et al., 2005), which provides a module for automated imaging of individual samples. Images for each grid corresponding to an individual crystallization condition were taken in an unattended manner from regions of interest selected by the ANIMATED-TEM algorithms developed by Coudray et al. (Coudray et al., 2011). The resulting images are uploaded into the Sesame laboratory information management system (Zolnai et al., 2003), where they are associated with other data relevant to the crystallization screen (Hu et al., 2010).
3. Results and Discussion
All together we used ~250 screens from ~200 journal articles to construct a database of successful 2D crystallization conditions, which is freely accessible at http://www.sesame.wisc.edu/ using the Jafar module. Figure 1A depicts the resolution of the 2D crystals in the database as a function of their type. With 47% of the entries planar 2D crystals or sheets produced the highest resolution. These crystals, characterized by a planar bilayer with a coherent 2D array of proteins, are relatively large in both dimensions and are favored for a 3D analysis of their structure. Related vesicular crystals, arising from flattened lipid vesicles containing two overlapping 2D lattices, constitute 17% of the entries, and tend to produce lower resolution than sheets since their rectangular shape and smaller size affects tilted data collection. Finally with 36% of the entries, tubular crystals, characterized by an array of proteins with helical symmetry in a cylindrical lipid vesicle that provides many different views of the molecules and thus do not need to be tilted, can also produce high-resolution 3D structures. With 70% of entries the vast majority of the 2D crystals in the database were produced by detergent dialysis, followed by detergent adsorption by bio-beads with 15% of entries, and with the lipid monolayer technique, complexation by cyclodextrin, dilution, or a combination of these methods accounting for the remaining 15% of entries.
Figure 1. Types of 2D crystals and membrane protein families.
A. The type, distribution and resolution of crystals in the 2D crystallization database. Depending on the publication resolution may refer to the final resolution of a 3D structure, 2D projection map, or diffraction pattern. B. The functional classification and distribution of the different membrane protein families in the 2D crystallization database.
3.1 Membrane proteins used in the analysis
The database included 2D crystallization experiments from a total of 12 β-barrel and 138 α-helical membrane proteins. Approximately 50% of these proteins were from bacteria, 45% from eukaryotes, and 5% from archaea. Two thirds of all the α-helical membrane proteins had 4–12 transmembrane domains, while the largest protein complexes contained 20–60 transmembrane domains and up to 5,000 amino acid residues. We grouped these proteins into seven different functional families (Fig. 1B). The largest families are the transporters and the channels, followed by photosynthetic proteins, ATPases and respiratory proteins. GPCRs and membrane enzymes are also present, but account for less than 10% of the entries in each case.
3.2 Detergent selection for 2D crystallization
Detergents are amphipathic molecules, consisting of a polar head group and a hydrophobic chain, capable of solubilizing membrane proteins by creating a mimic of a lipid bilayer environment. In general the choice of detergent is a critical factor for the initial solubilization and subsequent purification, and even functional characterization of a membrane protein. In addition, the detergent plays a prominent role in the formation and stabilization of the detergent/protein/lipid ternary complexes that constitute the starting point in a 2D crystallization experiment. The detergent is also critical during the reconstitution of the membrane protein into lipid bilayers, as its interactions with lipids and protein play an important role during this process (Rigaud et al., 2000). However, after reconstitution of the lipid bilayer, the role of detergent becomes less prominent as protein-protein and protein-lipid interactions dominate in the formation of well-ordered lattices. Indeed, detergent is only a minor component of the final 2D crystal. Despite a great deal of effort to understand the effect of detergent on membrane proteins, there is no single detergent that can be generally and reliably applied. Indeed, in numerous cases, the detergent used for the initial extraction and purification will not be effective in preserving function or in promoting the formation of well diffracting 2D crystals. Thus the choice of detergent must be made empirically.
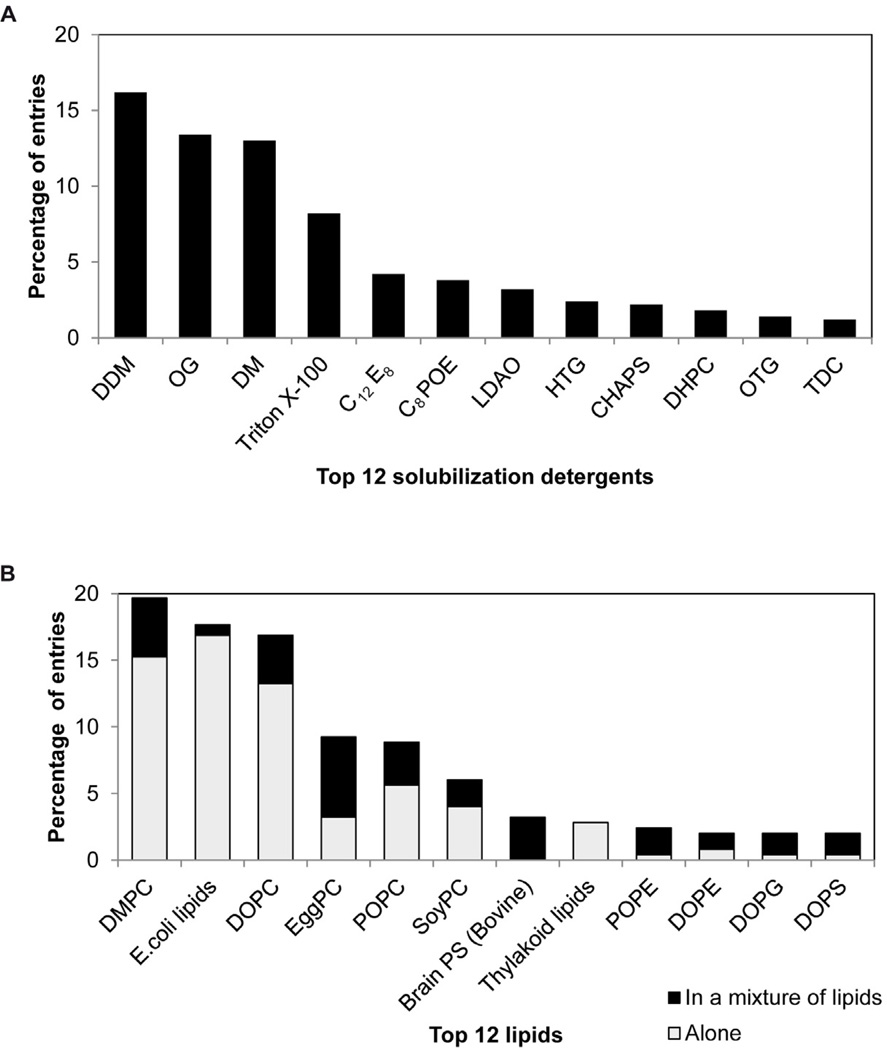
Figure 2A shows the top twelve detergents that to date have been successfully used in 2D crystallization. These data show a clear preference for the alkyl maltopyranoside detergents, accounting for ~30% of the entries in the database, followed by the alkyl glucopyranoside detergents with ~18% of the entries. Of the alkyl maltopyranoside detergents, n-Dodecyl-β-D-maltopyranoside (DDM) is as successful as n-Decyl-β-D-maltopyranoside (DM). Regarding the alkyl glucopyranosides, n-Octyl-β-D-glucopyranoside (OG) is clearly the most successful, whereas n-Hexyl-β-D-glucopyranoside (HG), n-Heptyl-β-D-thioglucopyranoside (HTG), and n-Octyl-β-D-thioglucopyranoside (OTG) have been seldom used. The alkyl polyoxyethylene detergents were also successful with ~18% of the entries in the database. Of these detergents, α-[4-(1,1,3,3-Tetramethylbutyl)phenyl]-ω-hydroxy-poly(oxy-1,2-ethanediyl) (Triton X-100) appears to be the most successful. We note that although the detergents OG, DDM and DM are also commonly used in 3D crystallization, the detergent Triton X-100 is seldom employed, as it is polydisperse and has a rather large micelle that tends to interfere with lattice formation. Likely, its prominence in 2D crystallization reflects the fact that Triton X-100 is very efficient at solubilization, forming mixed micelles of detergent and lipid at relatively low concentrations without the formation of complex intermediate aggregates (López et al., 1998). Because the detergent is ultimately removed, the properties of the micelle are not as important to the outcome of 2D crystallization. With regard to detergent concentration, a relatively wide range has been used for 2D crystallization with a median value of 3.4x the critical micelle concentration (CMC). In rare cases, the detergent concentration can be as high as 130xCMC, though more typical values are 2xCMC for high CMC-detergents, such as OG and DM, and up to 5.7xCMC for low-CMC detergents such as DDM and Triton X-100.
Figure 2. Detergent and lipid.
A. The twelve most frequent detergents in the 2D crystallization database. In approximately 66% of the entries, both membrane protein and lipid were solubilized in the same detergent, whereas in the remaining entries a different detergent was used. B. The twelve most frequent lipids in the 2D crystallization database. E. coli* denotes E. coli polar lipid extract.
Our analysis did not yield clear correlations between physicochemical properties of the proteins and the detergents that could help predict outcomes for future 2D crystallization trials. This is not surprising, as previous attempts by X-ray crystallographers to establish meaningful correlations between membrane proteins and detergents in 3D crystallization have also not been successful (Newstead et al., 2008). However, we noted several interesting trends. The detergent OG has been successful in 2D crystallization primarily with membrane proteins displaying an average negative electrical charge at pH 7.0, while the opposite is true for DM. The rational for this observation is unclear as both detergents are nonionic. We also note that 75% of proteins in our database that were successfully crystallized from OG or tetraethylene glycol monooctyl ether (C8E4) have less than 8 transmembrane helices. In addition, 70% of the proteins crystallized from either of these two detergents have a molecular weight below 40 kDa, while only 27% of the proteins crystallized from other detergents have such low molecular weight. One possible explanation for these observations is that the relatively short alkyl chain of both OG and C8E4 affects the ability of these detergents to solubilize membrane proteins that are very hydrophobic or that contain a large number of transmembrane helices. In support of this idea, a recent study (White et al., 2008) tested the extraction of 122 yeast membrane proteins in 6 detergents - n-Dodecyl-N,N-Dimethylamine-N-Oxide (LDAO), DDM, OG, and C8E4, 1-Dodecanoyl-2-Hydroxy-sn-Glycero-3-Phosphocholine (FC12) and Triton X-100 - and found that the short-chain detergents were, in general, the least effective. They also reported that as the number of transmembrane domains and the molecular weight of the proteins increased, the more difficult it was to solubilize them in OG or C8E4. Finally, the authors also underlined that OG and C8E4 were less effective as the percentage of charged and polar amino acids (EDKRHNQST) in the transmembrane region increased. Interestingly, we have neither found in our database any protein successfully crystallized from these two detergents when the percentage of charged and polar amino acids is above 20%. An alternative explanation is that although OG and C8E4 may solubilize the larger or more highly charges proteins, they are unstable in the micelles and thus aggregate during purification.
3.3 Lipid selection for 2D crystallization
The choice of lipid is another key factor in 2D crystallization. Lipids can have two distinct roles. Arranged in a bilayer, phospholipids provide a physical environment for reconstitution. Even as bilayers, phospholipids can adopt a variety of morphologies determined by the structure of the lipid, the nature of the lipid headgroup and its degree of hydration, the degree of unsaturation of the fatty acid chains, lateral pressure, temperature, ionic strength, and pH. As a consequence, the physical chemistry that governs the reconstitution of a membrane protein in a bilayer is complex, and thus the choice of lipid must ultimately be made empirically. However, we do know that it is energetically unfavorable for membrane proteins to expose their hydrophobic regions to water, or to embed hydrophilic regions in the hydrocarbon core of the lipid bilayer; thus, the hydrophobic regions of phospholipid and proteins need to be matched (Mouritsen and Bloom, 1993). Lipids can also fulfill an additional role as specific ligands. Indeed, individual lipids are often found bound to specific sites in membrane proteins (Wiener, 2006;Hunte and Richers, 2008), being essential for their structural integrity and functional activity (Dowhan, 1997).
Figure 2B shows the top twelve lipids that to date have been successfully used in 2D crystallization. These data show a clear preference for phosphatidylcholine lipids, accounting for over 65 % of the entries in the database. Polar and total lipid extracts from E. coli, composed mainly of phosphatidylethanolamine and phosphatidylglycerol lipids in a 8:2 ratio, were the next most successful with 20% of the entries. Of the phosphatidylcholine lipids, dimyristoylphosphatidylcholine (DMPC: 14:0) had most entries in the database, followed by dioleoylphosphatidylcholine (DOPC: 18:1) and palmytyl-oleoylphosphatidylcholine (POPC: 16:0/18:1). Natural lipid extracts rich in DOPC and POPC, such as egg-yolkPC and soybeanPC have also been used successfully in 10% and 7% of the cases, respectively.
As was the case with the detergents discussed above, we did not observe clear correlations between physicochemical properties of proteins and lipids in 2D crystallization. We note, however, that the vast majority of β-barrel membrane proteins - found only in prokaryotic membranes and in mitochondria - have been crystallized in either DMPC or E. coli lipid extracts. In contrast, DOPC a lipid often used to crystallize α-helical membrane proteins has not been used successfully with β-barrel membrane proteins. We also note that thylakoid lipids have been used exclusively in the crystallization of PSII complexes. Indeed, lipids used for 2D crystallization generally reflect the composition of native membranes, though given the relatively limited number of proteins represented in this database and the general lack of high-throughput methodologies employed for 2D crystallization, we believe it would be interesting to explore a broader range of lipids (e.g., cardiolipin and sphingomyelin) in future screens.
3.4 The lipid to protein ratio in 2D crystallization
The lipid to protein ratio (LPR) is another critical variable in 2D crystallization. If the lipid content is too high, then the protein will not be sufficiently concentrated in the plane of the lipid bilayer to induce 2D crystallization. Alternatively, if the lipid concentration is too low, the membrane protein target will aggregate and denature upon detergent removal. In principle, one should be able to precisely control the LPR in the initial protein/detergent/lipid ternary mixture. In reality, however, the purified membrane protein carries unpredictable amounts of bound lipids derived from the host membrane (Zhao et al., 2010;De Zorzi et al., 2013). Therefore, most laboratories will initially screen a broad range of LPRs followed by a more exhaustive sampling once crystallization hits are identified.
On a weight per weight basis the LPRs that have been successfully used in 2D crystallization vary from 0.1 to 1.5 without a clear peak or specific distribution. However, the LPR expressed on a mol per mol basis is more informative as it shows a unimodal distribution with a peak around a LPR of 25 (Fig. 3A). Despite the fact that the LPR generally increases as the number of transmembrane domains increases (Fig. 3B), the peak at a LPR of ~25 remains valid for membrane proteins with 6–14 transmembrane domains. When the LPR is estimated from the dimensions of the unit cell of a 2D crystal - by using a cross section area of 1nm2 for a α-helix and 0.6nm2 for a phospholipid - a correlation between LPR and the number of transmembrane domains is even more striking with distinct peaks at LPRs of ~25 and ~40 for membrane proteins with 8 and 12 transmembrane domains, respectively (Fig. 3C). Given that two thirds of all the α-helical membrane proteins in the database had 4–12 transmembrane domains these observations are broadly relevant to future crystallization screens.
Figure 3. Lipid to protein ratio.
A. The distribution of lipid to protein ratios (LPRs) in mol per mol in the 2D crystallization database. B. The distribution of LPRs as a function of the number of number of transmembrane helices. C. The distribution of LPRs calculated from the dimensions of the unit cell of a 2D crystal (using a cross section area of 1nm2 for a α-helix and 0.6nm2 for a phospholipid) as a function of the number transmembrane helices.
What could be the role of these lipids? According to the insight on protein-lipid interactions derived from high-resolution electron crystallography structures, approximately half of these lipids will likely play a role as annular lipids to form a shell around the protein and engage in transient and relatively nonspecific interactions with the protein. The remaining half will constitute the bulk of the bilayer. Indeed, in the structure of bacteriorhodopsin at 3.5Å resolution (Grigorieff et al., 1996), 10 lipids were visible at the perimeter of each monomer (bacteriorhodopsin has 7 transmembrane helices), while additional lipids outside this shell could not be quantified. The structure of aquaporin-0 from eye lens at 1.9Å resolution revealed a belt of nine well-defined lipid molecules at the perimeter of each protein monomer (aquaporin-0 has 6 transmembrane helices) (Gonen et al., 2005;Hite et al., 2008). The aquaporin-0 structure also revealed lipids outside the shell of annular lipids, which lacked direct interactions with the protein and thus constituted the bulk of the bilayer.
3.5 pH in 2D crystallization
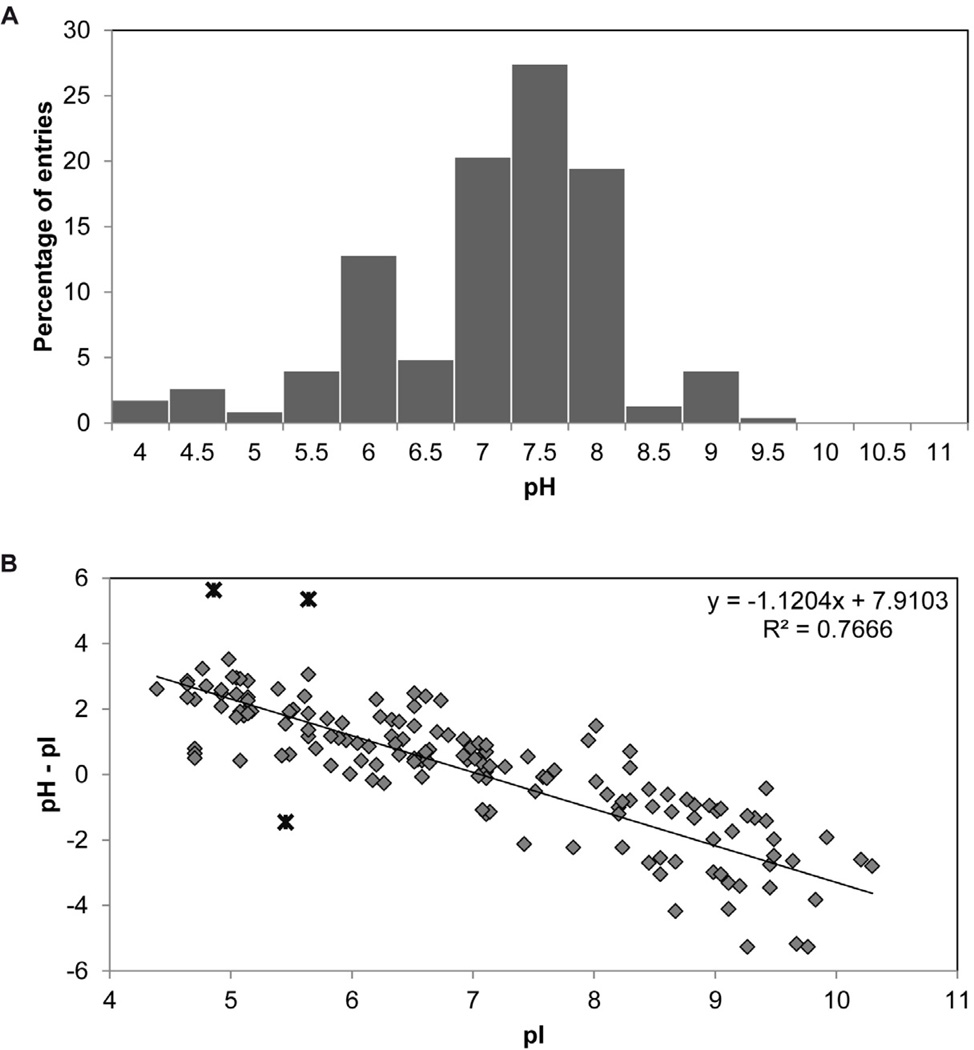
As with 3D crystallization the pH can have a significant impact on 2D crystallization. Figure 4A shows the success rate in 2D crystallization as a function of pH, with 60% of the entries in the database falling within the range of pH 7–8, and 13% of the entries clustering at around pH 6. Further analysis of the data revealed a positive correlation between the isoelectric point (pI) of the protein and the pH (Fig. 4B), in line with previous observations made by Kantardjieff at al. (Kantardjieff and Rupp, 2004) in their analysis of 3D crystallization factors for soluble proteins reported in the Protein Data Bank.
Figure 4. pH.
A. Distribution of pHs in the 2D crystallization database. B. Positive correlation between the pI of the membrane proteins in the database and the pH used in 2D crystallization.
3.6 Type and concentration of salts in 2D crystallization
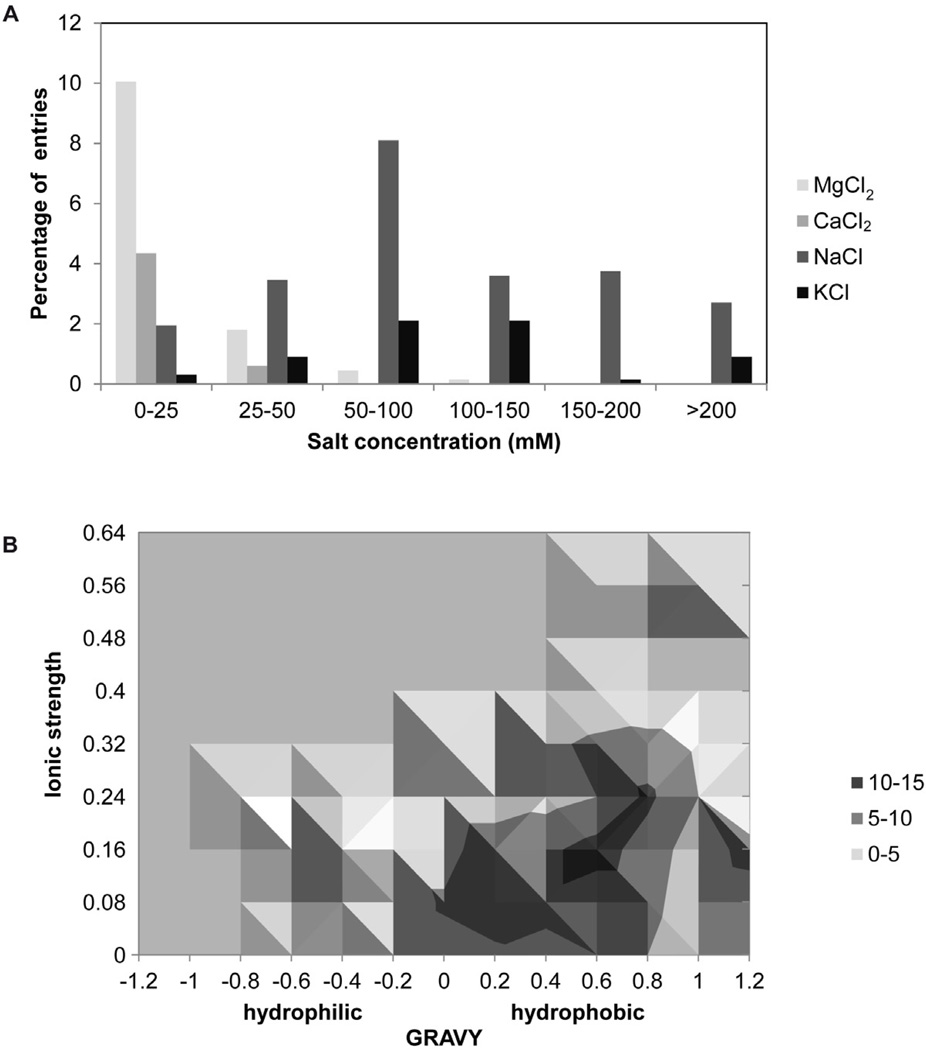
Ions can often be essential for 3D crystallization as they can affect protein conformation, protein-protein interactions, and protein solubility. In the context of 2D crystallization, divalent cations are known to promote the stability of the phospholipid bilayer. Specifically, because they intercalate between the polar head group regions of phospholipids, divalent cations shield electrostatic repulsions between negatively charged phosphate regions. Of the various salts represented in the database NaCl, KCl, MgCl2, and CaCl2 have been the most successful (Fig. 5A). NaCl in combination with MgCl2 accounts for over 60% of entries. In general, divalent salts are used at a lower concentration than monovalent salts. Polyvalent cations and ions can also serve as substrates and cofactors for transporters, channels and enzymes. In these cases, they can stabilize a particular protein intermediate and thus lock in a conformation that is amenable for 2D crystallization. In several examples, including PS II core (da Fonseca et al., 2002), BetP (Tsai et al., 2007), bovine lens connexin (Lampe et al., 1991), cyt b6f (Mosser et al., 1997), NanC (Signorell et al., 2007), GalP (Zheng et al., 2010), and aquaporin-2 (Schenk et al., 2005) the addition of specific divalent cations was crucial for 2D crystallization.
Figure 5. Salts.
A. The distribution of the concentrations of the prevalent salts (in the crystallization buffers) in the 2D crystallization database. B. The ionic strength (IS) in 2D crystallization as a function of the grand average of hydropathicity (GRAVY). The data suggests a positive correlation in 2D crystallization between helical membrane proteins with a positive GRAVY value and a high IS.
As part of our analysis we also explored a possible correlation between the ionic strength (IS) of a crystallization condition and the grand average of hydropathicity (GRAVY) for the protein. As shown in Figure 5B our analysis suggests a positive correlation between α-helical membrane proteins with a positive GRAVY and a high IS during 2D crystallization. Several specific examples illustrate this observation. For instance, 50 mM NaCl (IS = 0.05) yielded optimal crystals of CtrA3 (GRAVY = 0.137) (Chintalapati et al., 2008). In the case of aquaporin-8 (GRAVY = 0.67) a 100 mM NaCl (IS = 0.1) was necessary (Agemark et al., 2012), whereas GalP (GRAVY = 0.6) crystallized in a combination of 150 mM NaCl, 25 mM CaCl2 and 25 mM MgCl2 (IS = 0.23) (Zheng et al., 2010). Finally, the Enzyme IIC mannitol (GRAVY = 0.87) yielded the best crystals when the IS was 0.3 or above, while at lower IS the crystals deteriorated (Stuart et al., 2004).
3.7 Temperature in 2D crystallization
The predominant lipid bilayer form in biological membranes is the lamellar liquid crystalline phase (Lα which is characterized by considerable disorder in the acyl chains. Below a transition temperature characteristic of the particular lipid, the acyl chains are packed more tightly together giving rise to the lamellar gel phase (Lβ) and a higher overall lipid density within the bilayer. Our analysis shows that the majority of membrane protein 2D crystallization has been carried out at constant temperatures above this transition temperature.
Room temperature (20–27°C) has been successful in 46% of the entries in the database (51% if we consider the 20–30°C range), followed by low temperature (4°C) with 16% and high temperature (37°C) with 8% of the entries. Cycling the temperature above and below the transition temperature has also been shown to be effective in 17% of the entries. In rare cases, other temperatures (8°C, 12°C and 50°C) or cycling between 4°C and room temperature have also been used. Aside from its effect on the phase transition of the lipid bilayer, temperature can also affect the stability of the target protein, and the rate of detergent removal through dialysis. At low temperatures the protein of interest might be more stable during 2D crystallization, which can take several days and up to two weeks, but the phase transition of lipids containing DMPC might be affected and the dialysis rates of detergent slowed. In contrast, at high temperatures not all proteins might be stable, but it is possible to shorten the experiment duration by increasing the detergent removal rate and favoring crystallization by increasing Brownian motion.
3.8 Design of sparse and incomplete factorial 2D crystallization matrices
Based on our analysis of successful 2D crystallization conditions reported in the literature, we have designed a targeted sparse matrix crystallization screen. We applied the k-means algorithm to form 10 groups derived from 94 successful 2D crystallization conditions from 57 unique membrane proteins. The resulting matrix comprises 10 conditions, which is intended to be screened at 3 to 6 different LPRs, depending on the amount of protein available (Table 1). While the three most successful lipids (E.coli polar extract, DMPC and DOPC) are well represented in the screen, it also includes a few other species. The pH range of 6–8 covers ~75% of the entries in the database, the divalent salt concentration is kept low, as in ~80% of the entries, and the NaCl concentrations cover the most successful 50–300 mM range. Thus with 30 conditions, this targeted sparse matrix screen allows one to survey a significant portion of the published 2D crystallization space.
Table 1.
Sparse matrix 2D crystallization screen.
| Condition # | Lipids | Temperature | pH | MgCl2 [mM] | NaCl [mM] |
|---|---|---|---|---|---|
| 1 | E.coli polar | cycle | 6 | 50 | 100 |
| 2 | E.coli polar | RT | 7.5 | 2 | 150 |
| 3 | DOPC | cycle | 7.5 | 0 | 100 |
| 4 | DOPC | 4°C | 7 | 10 | 100 |
| 5 | DMPC | RT | 7 | 0 | 300 |
| 6 | DMPC:DOPC (1:4) | cycle | 7 | 40 | 150 |
| 7 | DMPC:DMPS (8.5:1.5) | RT | 8 | 2 | 50 |
| 8 | DOPC:DOPA (9:1) | RT | 7.5 | 0 | 200 |
| 9 | EggPC | RT | 6 | 2 | 50 |
| 10 | DOPE | RT | 7.5 | 0 | 100 |
RT = room temperature; cycle = temperature cycling between 12 h at 27°C and 12 h at 37°C. The lipid ratios are given in weight.
Incomplete factorial or grid screens offer an alternative to maximize the exploration of crystallization space. Designed to optimize the screening of a large amount of factors in as few experiments as possible, these screens have become increasingly popular in 3D crystallization (DeLucas et al., 2003;Luft et al., 2011). Using the most important factors affecting 2D crystallization, we designed an incomplete factorial 2D crystallization screen of 90 conditions (Table 2). In combination with 6 control conditions, this screen is compatible with the conventional 96-well high-throughput format.
Table 2.
Incomplete factorial matrix 2D crystallization screen.
| Buffer | Lipid | Co-factor | LP R |
Monovalent Salt | Divalent Salt | Detergent | Detergent removal |
Additive | Tem p |
|---|---|---|---|---|---|---|---|---|---|
| µdia+5mM β- | |||||||||
| pH6.5 MES | EggPC/DOPS | none | 1.5 | 250mM Na2SO4 | 5mM CaCl2 | C12E8 | CD | 2% glycerol | 4°C |
| pH7.5 | µdia+5mM β- | ||||||||
| HEPES | DOPG | 10% cholesterol | 0.1 | 250mM KCl | 5mM MgCl2 | DDM | CD | 2% glycerol | 4°C |
| µdia+5mM β- | |||||||||
| pH5.5 Citrate | DMPC | 10% brain extract | 0.1 | 150mM Na2HPO4 | 50mM CaCl2 | DDM | CD | none | 4°C |
| TritonX10 | µdia+5mM β- | ||||||||
| pH6.5 MES | E.coli polar | 10% cholesterol | 0.3 | 150mM NaCl | 5mM MgCl2 | 0 | CD | 2% glycerol | 4°C |
| µdia+1 mM β- | |||||||||
| pH8.5 Tris | DOPG | 10% cholesterol | 0.5 | 250mM NaCl | 5mM CaCl2 | C12E8 | CD | none | 4°C |
| µdia+1 mM β- | |||||||||
| pH5.0 Citrate | SoyPC | none | 1.3 | 50mM NaCl | none | DM | CD | none | 4°C |
| µdia+1 mM β- | 5mM Β- | ||||||||
| pH5.5 Citrate | E.coli polar | 10% cholesterol | 1.1 | 150mM KCl | 50mM MgCl2 | OG | CD | MEA | 4°C |
| pH7.0 | EggPC/EggP | µdia+1 mM β- | |||||||
| HEPES | A | 10% brain extract | 0.3 | 150mM Na2HPO4 | 5mM MgCl2 | OG | CD | none | 4°C |
| TritonX10 | µdia+1 mM β- | ||||||||
| pH4.5 Citrate | DMPC | 10% cholesterol | 0.3 | 150mM Na2SO4 | 5mM CaCl2 | 0 | CD | none | 4°C |
| 5mM Β- | |||||||||
| pH6.5 MES | SoyPC | 10% brain extract | 0.5 | 150mM Na2SO4 | 50mM MgCl2 | DDM | µdia | MEA | 4°C |
| pH5.5 Citrate | EggPC/DOPS | 10% cholesterol | 0.7 | 50mM KCl | 5mM CaCl2 | DM | µdia | 2% glycerol | 4°C |
| pH6.0 MES | DMPC | none | 0.1 | 50mM Na2HPO4 | 5mM CaCl2 | OG | µdia | 2% glycerol | 4°C |
| TritonX10 | |||||||||
| pH9.0 Tris | POPC | none | 1.3 | 150mM KCl | 50mM CaCl2 | 0 | µdia | 2% glycerol | 4°C |
| pH7.0 | EggPC/EggP | TritonX10 | |||||||
| HEPES | A | 10% cholesterol | 0.9 | 250mM mM NaCl | 50mM MgCl2 | 0 | µdia | 2% glycerol | 4°C |
| pH7.0 | µdia+5mM β- | 5mM Β- | |||||||
| HEPES | POPC | 10% cholesterol | 1.3 | 50mM Na2HPO4 | 5mM CaCl2 | DDM | CD | MEA | 20°C |
| µdia+5mM β- | |||||||||
| pH5.0 Citrate | EggPC/DOPS | 10% cholesterol | 1.5 | 50mM Na2SO4 | 5mM CaCl2 | DM | CD | 2% glycerol | 20°C |
| µdia+5mM β - | |||||||||
| pH4.5 Citrate | SoyPC | none | 0.9 | 50mM Na2HPO4 | 5mM MgCl2 | DM | CD | none | 20°C |
| pH7.0 | TritonX10 | µdia+5mM β- | 5mM Β- | ||||||
| HEPES | EggPC/DOPS | none | 0.1 | 250mM Na2HPO4 | 50mM MgCl2 | 0 | CD | MEA | 20°C |
| TritonX10 | µdia+5mM β- | ||||||||
| pH8.0 Tris | DOPC | 10% brain extract | 0.5 | 50mM KCl | 50mM MgCl2 | 0 | CD | 2% glycerol | 20°C |
| µdia+1 mM β- | |||||||||
| pH6.0 MES | DOPG | 10% brain extract | 0.9 | 250mM Na2SO4 | 50mM MgCl2 | DDM | CD | none | 20°C |
| EggPC/EggP | µdia+1 mM β- | ||||||||
| pH6.5 MES | A | 10% cholesterol | 1.1 | 250mM Na2HPO4 | none | DM | CD | 2% glycerol | 20°C |
| EggPC/EggP | µdia+1 mM β- | 5mM Β- | |||||||
| pH8.0 Tris | A | none | 1.5 | 50mM Na2SO4 | 50mM CaCl2 | OG | CD | MEA | 20°C |
| µdia+1 mM β- | |||||||||
| pH5.5 Citrate | DOPC | 10% brain extract | 0.1 | 50mM NaCl | 5mM MgCl2 | OG | CD | 2% glycerol | 20°C |
| 5mM Β- | |||||||||
| pH8.5 Tris | DMPC | 10% cholesterol | 1.3 | 50mM Na2SO4 | 5mM MgCl2 | C12E8 | µdia | MEA | 20°C |
| 5mM Β- | |||||||||
| pH9.0 Tris | SoyPC | 10% brain extract | 0.5 | 150mM NaCl | none | C12E8 | µdia | MEA | 20°C |
| pH6.5 MES | POPC | none | 0.9 | 50mM KCl | 50mM CaCl2 | OG | µdia | none | 20°C |
| TritonX10 | |||||||||
| pH6.0 MES | DOPC | 10% cholesterol | 1.5 | 150mM KCl | none | 0 | µdia | none | 20°C |
| EggPC/EggP | µdia+5mM β- | 5mM Β- | |||||||
| pH9.0 Tris | A | 10% brain extract | 0.7 | 50mM NaCl | 5mM CaCl2 | C12E8 | CD | MEA | 27°C |
| µdia+5mM β- | |||||||||
| pH8.0 Tris | EggPC/DOPS | 10% cholesterol | 0.9 | 150mM Na2SO4 | none | C12E8 | CD | 2% glycerol | 27°C |
| µdia+5mM β- | 27°C | ||||||||
| pH5.0 Citrate | POPC | 10% brain extract | 0.1 | 150mM KCl | 5mM MgCl2 | C12E8 | CD | none | |
| µdia+5mM β- | 5mM Β- | 27°C | |||||||
| pH8.5 Tris | E.coli polar | 10% brain extract | 1.5 | 150mM Na2SO4 | 50mM CaCl2 | DDM | CD | MEA | |
| µdia+5mM β- | 27°C | ||||||||
| pH8.0 Tris | SoyPC | 10% cholesterol | 1.3 | 250mM KCl | 50mM MgCl2 | OG | CD | none | |
| µdia+1 mM β- | 5mM Β- | 27°C | |||||||
| pH8.0 Tris | E.coli polar | none | 0.1 | 250mM NaCl | 5mM CaCl2 | DDM | CD | MEA | |
| µdia+1 mM β- | 27°C | ||||||||
| pH4.5 Citrate | POPC | 10% cholesterol | 0.7 | 250mM NaCl | 50mM MgCl2 | DM | CD | 2% glycerol | |
| µdia+1 mM β- | 27°C | ||||||||
| pH9.0 Tris | EggPC/DOPS | none | 0.5 | 50mM Na2SO4 | 5mM MgCl2 | DM | CD | none | |
| pH5.0 Citrate | DOPC | none | 0.3 | 250mM NaCl | 50mM CaCl2 | DDM | µdia | 2% glycerol | 27°C |
| 5mM Β- | 27°C | ||||||||
| pH5.0 Citrate | EggPC/DOPS | 10% brain extract | 0.7 | 150mM NaCl | 5mM CaCl2 | OG | µdia | MEA | |
| pH6.5 MES | DOPG | none | 1.5 | 150mM Na2HPO4 | 50mM MgCl2 | OG | µdia | none | 27°C |
| pH7.5 | TritonX10 | 5mM Β- | 27°C | ||||||
| HEPES | DMPC | none | 0.9 | 50mM NaCl | 50mM MgCl2 | 0 | µdia | MEA | |
| pH7.5 | EggPC/EggP | TritonX10 | 27°C | ||||||
| HEPES | A | 10% cholesterol | 0.1 | 250mM Na2SO4 | none | 0 | µdia | none | |
| µdia+5mM β- | 5mM Β- | 37°C | |||||||
| pH4.5 Citrate | DOPC | none | 1.1 | 250mM NaCl | 5mM CaCl2 | C12E8 | CD | MEA | |
| µdia+5mM β- | 37°C | ||||||||
| pH8.5 Tris | DOPC | none | 0.7 | 150mM Na2HPO4 | none | DDM | CD | none | |
| pH7.0 | µdia+5mM β- | 5mM Β- | 37°C | ||||||
| HEPES | DMPC | 10% cholesterol | 0.5 | 250mM Na2SO4 | 50mM CaCl2 | DM | CD | MEA | |
| µdia+1 mM β- | 5mM Β- | 37°C | |||||||
| pH6.0 MES | SoyPC | 10% cholesterol | 1.1 | 150mM Na2HPO4 | 50mM CaCl2 | C12E8 | CD | MEA | |
| µdia+1 mM β- | 37°C | ||||||||
| pH8.0 Tris | DMPC | none | 1.5 | 150mM NaCl | 50mM MgCl2 | C12E8 | CD | 2% glycerol | |
| pH7.0 | µdia+1 mM β- | 37°C | |||||||
| HEPES | SoyPC | 10% cholesterol | 0.7 | 150mM KCl | 5mM MgCl2 | DDM | CD | 2% glycerol | |
| pH7.5 | µdia+1 mM β- | 37°C | |||||||
| HEPES | POPC | 10% cholesterol | 1.5 | 250mM Na2HPO4 | 5mM CaCl2 | OG | CD | 2% glycerol | |
| TritonX10 | µdia+1 mM β- | 5mM Β- | 37°C | ||||||
| pH4.5 Citrate | E.coli polar | 10% brain extract | 1.3 | 50mM KCl | none | 0 | CD | MEA | |
| EggPC/EggP | TritonX10 | µdia+1 mM β- | 37°C | ||||||
| pH5.0 Citrate | A | none | 0.5 | 50mM Na2HPO4 | 50mM CaCl2 | 0 | CD | none | |
| pH4.5 Citrate | DOPC | 10% cholesterol | 0.1 | 50mM Na2SO4 | 50mM CaCl2 | C12E8 | µdia | none | 37°C |
| pH9.0 Tris | DOPG | none | 1.1 | 150mM Na2SO4 | 5mM MgCl2 | DDM | µdia | 2% glycerol | 37°C |
| pH6.0 MES | POPC | 10% brain extract | 0.5 | 150mM Na2HPO4 | 5mM MgCl2 | DM | µdia | 2% glycerol | 37°C |
| pH7.0 | 37°C | ||||||||
| HEPES | DOPC | 10% brain extract | 1.1 | 150mM NaCl | 5mM CaCl2 | DM | µdia | none | |
| 5mM Β- | 37°C | ||||||||
| pH5.5 Citrate | DOPG | 10% brain extract | 0.9 | 50mM Na2HPO4 | none | OG | µdia | MEA | |
| TritonX10 | 37°C | ||||||||
| pH8.5 Tris | E.coli polar | 10% brain extract | 0.9 | 250mM KCl | 5mM CaCl2 | 0 | µdia | 2% glycerol | |
| µdia+5mM β- | cycl | ||||||||
| pH4.5 Citrate | EggPC/DOPS | none | 0.9 | 150mM Na2HPO4 | none | DDM | CD | 2% glycerol | e |
| µdia+5mM β- | 5mM Β- | cycl | |||||||
| pH9.0 Tris | SoyPC | 10% brain extract | 1.5 | 250mM Na2SO4 | none | DM | CD | MEA | e |
| EggPC/EggP | µdia+5mM β- | cycl | |||||||
| pH8.5 Tris | A | 10% brain extract | 0.9 | 150mM KCl | 50mM MgCl2 | DM | CD | 2% glycerol | e |
| pH7.0 | µdia+5mM β- | 5mM Β- | cycl | ||||||
| HEPES | DOPG | none | 0.3 | 50mM KCl | none | OG | CD | MEA | e |
| µdia+5mM β- | cycl | ||||||||
| pH6.0 MES | E.coli polar | none | 0.7 | 250mM Na2HPO4 | 5mM MgCl2 | OG | CD | none | e |
| TritonX10 | µdia+5mM β- | cycl | |||||||
| pH9.0 Tris | DMPC | 10% brain extract | 1.1 | 250mM NaCl | 50mM CaCl2 | 0 | CD | none | e |
| µdia+1 mM β- | cycl | ||||||||
| pH6.0 MES | EggPC/DOPS | 10% brain extract | 1.5 | 150mM Na2SO4 | 50mM CaCl2 | C12E8 | CD | none | e |
| µdia+1 mM β- | 5mM Β- | cycl | |||||||
| pH5.5 Citrate | POPC | none | 0.5 | 250mM KCl | none | DDM | CD | MEA | e |
| µdia+1 mM β- | 5mM Β- | cycl | |||||||
| pH6.5 MES | DOPC | 10% cholesterol | 1.3 | 250mM Na2SO4 | 50mM CaCl2 | OG | CD | MEA | e |
| pH7.5 | TritonX10 | µdia+1 mM β- | cycl | ||||||
| HEPES | SoyPC | 10% brain extract | 0.3 | 50mM Na2SO4 | 5mM CaCl2 | 0 | CD | 2% glycerol | e |
| EggPC/EggP | 5mM Β- | cycl | |||||||
| pH4.5 Citrate | A | none | 1.1 | 250mM KCl | 5mM MgCl2 | C12E8 | µdia | MEA | e |
| cycl | |||||||||
| pH9.0 Tris | E.coli polar | 10% cholesterol | 0.3 | 50mM Na2HPO4 | 50mM MgCl2 | C12E8 | µdia | none | e |
| 5mM Β- | cycl | ||||||||
| pH8.0 Tris | DOPG | 10% cholesterol | 0.7 | 50mM NaCl | 50mM CaCl2 | DM | µdia | MEA | e |
| 5mM Β- | 4°C | ||||||||
| pH8.0 Tris | POPC | 10% brain extract | 0.3 | 250mM Na2HPO4 | none | C12E8 | Button | MEA | |
| 5mM Β- | 4°C | ||||||||
| pH6.0 MES | DOPC | none | 0.3 | 250mM KCl | none | DM | Button | MEA | |
| pH7.0 | 4°C | ||||||||
| HEPES | E.coli polar | 10% brain extract | 1.5 | 250mM KCl | 50mM CaCl2 | DM | Button | none | |
| TritonX10 | 5mM Β- | 4°C | |||||||
| pH8.5 Tris | POPC | none | 1.1 | 250mM Na2SO4 | 5mM MgCl2 | 0 | Button | MEA | |
| pH7.5 | 20°C | ||||||||
| HEPES | E.coli polar | none | 0.5 | 150mM KCl | 5mM CaCl2 | C12E8 | Button | none | |
| 5mM Β- | 20°C | ||||||||
| pH8.0 Tris | EggPC/DOPS | 10% brain extract | 1.1 | 150mM KCl | 50mM CaCl2 | DDM | Button | MEA | |
| pH5.0 Citrate | DOPG | 10% brain extract | 1.1 | 50mM Na2SO4 | none | DDM | Button | 2% glycerol | 20°C |
| pH7.5 | 5mM Β- | 20°C | |||||||
| HEPES | DOPC | none | 0.7 | 150mM Na2SO4 | 50mM MgCl2 | DM | Button | MEA | |
| pH5.0 Citrate | DMPC | 10% cholesterol | 0.5 | 250mM KCl | none | OG | Button | 2% glycerol | 20°C |
| pH6.5 MES | DMPC | 10% brain extract | 0.7 | 50mM Na2HPO4 | none | C12E8 | Button | 2% glycerol | 27°C |
| pH4.5 Citrate | DOPG | 10% brain extract | 1.3 | 250mM Na2HPO4 | 50mM MgCl2 | C12E8 | Button | 2% glycerol | 27°C |
| pH5.5 Citrate | SoyPC | 10% cholesterol | 1.5 | 50mM KCl | 5mM MgCl2 | DDM | Button | none | 27°C |
| pH8.5 Tris | SoyPC | none | 0.1 | 150mM NaCl | 50mM CaCl2 | OG | Button | 2% glycerol | 27°C |
| TritonX10 | 5mM Β- | 27°C | |||||||
| pH5.0 Citrate | E.coli polar | 10% cholesterol | 0.9 | 150mM Na2HPO4 | 5mM MgCl2 | 0 | Button | MEA | |
| EggPC/EggP | 37°C | ||||||||
| pH5.5 Citrate | A | none | 0.3 | 150mM Na2SO4 | 5mM CaCl2 | DM | Button | 2% glycerol | |
| pH7.5 | 5mM Β- | 37°C | |||||||
| HEPES | EggPC/DOPS | 10% brain extract | 1.3 | 250mM NaCl | 5mM MgCl2 | OG | Button | MEA |
LPR = Lipid-to-Protein Ratio (w/w); RT = room temperature; cycle = temperature cycling between 12 h at 27°C and 12 h at 37°C; detergent = detergent used for lipid solubilization; µdia = microdialysis plate; Button = dialysis buttons; β-CD = cyclodextrin; B-MEA: β-mercaptoethanol; The lipid ratios are given in weight. Ratio of EggPC/EggPA and EggPC/D.
The incomplete factorial screen covers a large variety of lipid polar head groups, acyl chain lengths and levels of unsaturation. In addition, mixtures of lipids are well represented in the screen, either as the principal constituents of the bilayer or as more specific protein co-factors. For instance, we included cholesterol in the screen as it has been used as a co-factor to improve the quality and reduce the aggregation of rhodopsin 2D crystals (Mielke et al., 2002), and is known to stabilize other membrane proteins. Brain lipid extract, rich in sphingolipids and gangliosides, is also included as a co-factor in several of the conditions. The distribution of LPRs efficiently cover a wide range of observed values, and pHs between 4.5–9 cover ~100% of the entries in the database and the majority of the physiological pH range available for protein and lipids. The concentrations of divalent salts (Ca2+ and Mg2+) is kept in the 5–50 mM range, and the monovalent salt type and concentration range explores known 2D crystallization space. In addition, additives known to stabilize proteins such as glycerol and (β-mercaptoethanol (β-ME) are also sampled as they have been found to influence the size and order of BetP (Tsai et al., 2007) and glutathione transferase (Schmidt-Krey et al., 1998) 2D crystals. Finally, in this screen we have also tried to sample various kinetics of detergent removal. In some cases, 2D crystallization can occur concomitantly with reconstitution of the membrane protein into a lipid bilayer. However, in other cases, crystallization occurs after reconstitution has finished. This entire process depends on the relative kinetics of detergent removal and crystal formation, as well as on the physical nature of the intermolecular interactions that stabilize the crystal lattice (Rigaud et al., 2000;Rigaud and Lévy, 2003). The CMC of a detergent is a key factor affecting the kinetics of detergent removal. Detergents having a CMC > 0.1% (w/w) can be eliminated by dialysis in less than a week, whereas low CMC (< 0.05%) detergents require significantly longer dialysis times. The low CMC detergents are also good candidates for adsorption by hydrophobic resins such as Biobeads (Rigaud et al., 1997), or complexation by cyclodextrins (Signorell et al., 2007). Indeed, α-, β- or γ-cyclodextrins can be used to remove low CMC detergents from lipid/detergent/protein ternary mixtures to facilitate the reconstitution and crystallization of membrane proteins, and we have therefore included cyclodextrin in the dialysis buffer of some of the conditions. In addition, our matrix also incorporates microdialysis buttons, as the larger area of their dialysis membrane relative to the crystallization plate as well as the higher buffer to sample volume ratio can also influence the kinetics of detergent removal. Finally, we explored the effect of detergent mixtures in 2D crystallization by virtue of using different detergents for membrane protein and lipid solubilization.
3.9 Screening results using the sparse and incomplete factorial 2D crystallization matrices
Crystallization was carried out by detergent dialysis in parallel on a 96-well format. Typically the screening of 2D crystallization trials involves four key steps: crystallization, specimen preparation for electron microscopy, image acquisition, and evaluation (Dreaden et al., 2013). We have incorporated all these steps into an automated pipeline that greatly improves throughput and reproducibility of the 2D crystallization process (Kim et al., 2010). For the implementation of the new sparse and incomplete factorial 2D crystallization matrices we have incorporated a new 96-well microfluidic dialysis plate into our crystallization pipeline. Our original 96-well dialysis block was built with standard 96-well SBS microplate dimensions and permitted the dialysis of 96 unique samples against 96 different buffers (Vink et al., 2007). This block could accommodate sample and buffer volumes of up to 53µl and 1ml respectively, and thus - at a typical protein concentration of 0.5 mg/ml - up to 2.5mg of protein were required for a full 96-condition 2D crystallization screen. In order to reduce the sample volume we have collaborated with GN Biosystems (Santa Clara, CA) to develop a microfluidic dialysis system for low-volume, high-throughput crystallization trials. This microfluidic dialysis plate can accommodate 5–28µl of sample per well and up to 500µl per buffer chamber. In our case, we have used 15–25µl of sample per well, the total amount of protein needed for a 96-condition screen would be reduced to approximately 0.4-1mg. Following dialysis for periods of up to two weeks, negatively stained specimens were prepared in parallel using a 96-position magnetic platform to hold EM grids and a liquid handling robot (Kim et al., 2010). Finally, robotic grid insertion into the electron microscope and computerized image acquisition (Hu et al., 2010) from regions of interest selected automatically (Coudray et al., 2011) ensured the rapid and reliable evaluation of the crystallization screen. The resulting images were uploaded into the Sesame laboratory information management system (Zolnai et al., 2003). Currently, an experienced microscopist performs data analysis and evaluation, but efforts to automate these steps using algorithms for crystal detection and automated collection of diffraction data are underway.
To date, 19 2D crystallization screens have been performed on 16 different target membrane proteins (Table 3). This corresponds to over 1,300 individual crystallization experiments evaluated by over 50,000 EM images. Six membrane protein targets have yielded diffracting crystals (Table 3, Fig. 6). In all these successful cases, initial hits were observed in either the sparse matrix or incomplete factorial crystallization screens, which provided the basis for designing more focused screens to improve crystal quality and abundance. Out of the 12 membrane protein targets tested with the sparse matrix screen, 5 have produced crystals. Crystals for 4 out of 6 membrane protein targets have been obtained with the incomplete factorial screen. Some of the targets have yielded crystals with both screens, whereas other targets crystals could only be obtained with one of the screens. This relatively high success rate suggests that the new matrices presented here have good potential to accelerate the process of membrane protein 2D crystallization.
Table 3.
Summary of 2D crystallization trials.
| Function | Organism | MW (kDa) | pI | TMD | TM (%) | Sparse | I.F. | Best outcome |
|---|---|---|---|---|---|---|---|---|
| ATPase | E.coli | 154.7 | 7.2 | 21 | 31 | Yes | No | Vesicles packed with protein |
| Channel | Rabbit | 565.2 | 5.2 | 8 | 4 | No | Yes | Striated vesicles |
| Channel | E.coli | 49.7 | 9.7 | 12 | 54 | No | Yes | Non-diffracting tubes |
| Channel | E.coli | 37.1 | 4.6 | 17 | 55 | Yes | Yes | Diffracting helical tubes and crystalline sheets |
| Enzyme | M.arvoryzae | 33.6 | 8.9 | 7 | 52 | Yes | No | Rare striated sheets and non-diffracting tubes |
| Enzyme | B.subtilis | 24.0 | 8.9 | 7 | 69 | Yes | No | Diffracting helical tubes |
| Other | Guinea pig | 66.5 | 5.5 | 2 | 7 | Yes | No | Sheet-like structure |
| Other | S.aureus | 49.8 | 5.3 | 6 | 29 | Yes | No | Vesicles, aggregates |
| Other | Mus musculus | 24.8 | 8.1 | 4 | 38 | Yes | No | Vesicles, aggregates |
| Other | Homo sapiens | 22.0 | 8.8 | 4 | 42 | Yes | No | Vesicles, aggregates |
| Transporter | I.loihiensis | 28.1 | 9.5 | 4 | 37 | No | Yes | Diffracting helical tubes |
| Transporter | S.oneidensis | 32.5 | 5.4 | 6 | 43 | Yes | Yes | Diffracting helical tubes |
| Transporter | E.coli | 32.5 | 6.0 | 6 | 43 | Yes | No | Diffracting helical tubes |
| Transporter | S.oralis | 46.0 | 9.4 | 11 | 55 | No | Yes | Rare non-diffracting filaments |
| Transporter | S.typhimurium | 44.8 | 10.3 | 12 | 61 | Yes | Yes | Diffracting helical tubes, and some sheets |
| Transporter | L.monocytogenes | 20.4 | 9.9 | 6 | 65 | Yes | No | Non-diffracting tubes |
MW = molecular weight; pI: isoelectric point; TMD: number of predicted transmembrane domains α-helices or β-strands); %TM: % of transmembrane sequence as predicted using the prediction software TMHMM (http://www.cbs.dtu.dk/services/TMHMM/); Sparse = Sparse matrix; I.F. = incomplete factorial matrix. I.F. Yes/No = indicates if a matrix has been run or not.
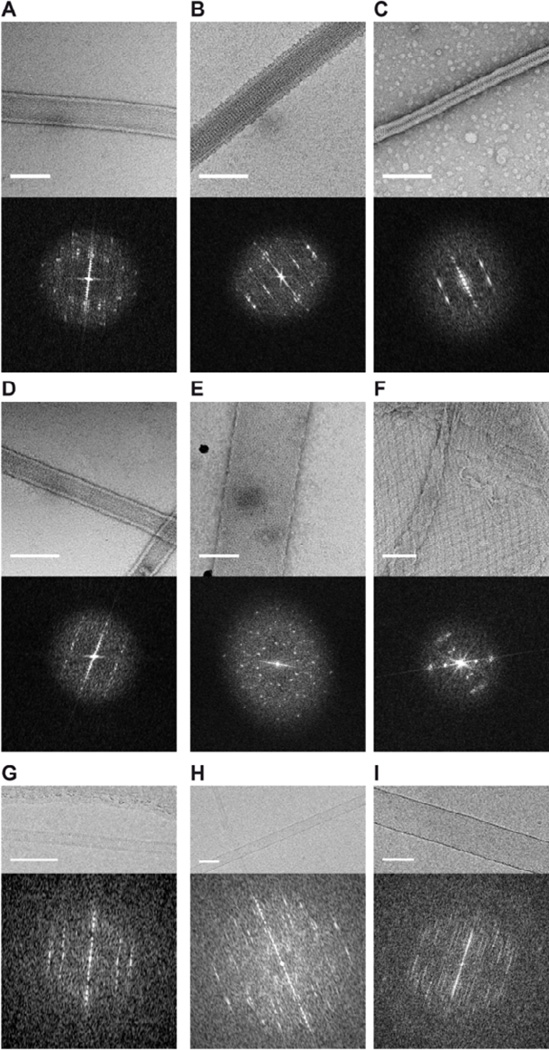
Figure 6. Gallery of 2D crystals obtained using the sparse and incomplete factorial matrices.
Images show negatively stained helical crystals of three different transporters (A, B, C) one enzyme (D) and one channel (E), and a crystalline vesicle of a channel (F). Cryo images of some of the targets are also shown (G, H, I). The scale bars are 100 nm.
We note that the majority of 2D crystals produced by our screens had a tubular morphology. The helical symmetry underlying this tubular morphology offers an advantage for structure determination, because a full 3D dataset can be obtained without the need to tilt the crystals and because new iterative helical real-space refinement (IHRSR) procedures have proven effective in producing high resolution structures (Behrmann et al., 2012;Wu et al., 2014). Despite screening a wide-range of conditions, we were unable to obtain either vesicular or sheet crystals for all but one of our protein targets. This preference for tubular 2D crystals likely reflects constraints of the crystal lattice due either to the topology or to the intermolecular protein contacts of the crystallized targets. Specific inter-molecular associations within the detergent depleted protein-lipid aggregates may also govern the resulting crystal symmetry and morphology. In some cases, a given protein will form both tubular and planar crystals (Jap et al., 1992), indicating that there are alternative sets of crystal contacts that are differentially selected by the conditions during reconstitution and crystal growth. Presumably, the curvature of tubular crystals is the result of the differences in the distance of the protein-protein contacts on either side of the membrane that arise when membrane proteins are incorporated into the lipid bilayer with a unique orientation. In contrast, membrane proteins inserted in an "upside-down" orientation can have a two-fold symmetry axis oriented parallel to the membrane plane and often produce extended, planar crystals (e.g., light harvesting complex II (Kühlbrandt and Wang, 1991), cytochrome oxidase (Valpuesta et al., 1990), glutathione transferase (Holm et al., 2006)). By analyzing only the primary or predicted secondary structures of the screened proteins, it is not easy to foresee their propensity to form tubular crystals. The six proteins that, so far, have yielded tubular crystals have different topologies (4–17 transmembrane helices), hydrophobicities (37–69% intramembranous), and isoelectric points (4.6–10.3), as predicted through www.cbs.dtu.dk/services/TMHMM/ and au.expasy.org/tools/protparam.html. Thus, additional proteins need to be screened, and more ambitious bioinformatics studies conducted, in order to draw more generalized conclusions.
4. Conclusions
This communication presents an attempt to rationalize membrane protein 2D crystallization. Using information derived from our analysis we have designed two new 2D crystallization matrices that complement each other and combined allow the efficient sampling of a broad range of the multifactorial 2D crystallization space. In combination with available procedures to automate the steps of crystallization, specimen preparation for electron microscopy, image acquisition, and evaluation, these matrices can improve the odds for producing well-ordered 2D crystals of membrane proteins. Once initial hits are identified with the sparse and/or incomplete factorial crystallization matrices it may take multiple rounds of optimization to improve the quality and abundance of 2D crystals suitable for data collection by cryo-EM and structure determination. Optimization is certainly case-specific, however some general rules include: (1) fine-tuning of the crystallization factors around the initial hits; (2) varying the protein concentration; (3) addition of known protein ligands, substrates or inhibitors; (4) modifying the chromatographic steps for protein purification or removing purification tags; and (5) altering the kinetics of detergent removal by exploring alternative reconstitution techniques such as dilution (Dolder et al., 1996), adsorption by Biobeads (Rigaud et al., 1997) or complexation by cyclodextrins (Signorell et al., 2007).
From a technical perspective there have been recent important advances for high-throughput screening of 2D crystallization trials and for automated imaging of membrane crystals with the electron microscope (Cheng et al., 2007;Vink et al., 2007;Coudray et al., 2008;Hu et al., 2010;Iacovache et al., 2010;Karathanou et al., 2010;Kim et al., 2010;Coudray et al., 2011;Glover et al., 2011). In addition, there have also been very recent key developments in detector technologies capable of recording EM images with unprecedented signal-to-noise ratio (Liao et al., 2013). Finally, there have also been key developments in software to facilitate the process of structure determination (Hirai et al., 1999;Philippsen et al., 2003;Gipson et al., 2007;Philippsen et al., 2007;Diaz et al., 2010), including new IHRSR procedures for helical 2D crystals (Egelman, 2010;Behrmann et al., 2012) and a semi-automated structure determination pipeline with the software package 2dx for flat 2D crystals (Scherer et al., 2014). In combination with all these advances, the 2D crystallization matrices presented here will be valuable to affirm electron crystallography as a viable tool for routine structure determination of membrane proteins along side NMR spectroscopy and X-ray crystallography.
Acknowledgements
We are grateful to the laboratories of Dr. Tom Walz, Dr. Wayne Hendrickson, Dr. Tamir Gonen, and Dr. Robert Stroud, and the Center for Eukaryotic Structural Genomics for supplying some of the proteins used for 2D crystallization. Funding for this work was provided by grants U54GM094598 and R01GM095747 from the National Institutes of Health and grant MCB-1244236 from the National Science Foundation.
Abbreviations
- β-CD
methyl-β-cyclodextrin
- DMPC
dimyristoylphosphatidylcholine
- DMPS
dimyristoylphosphatidylserine
- DOPA
dioleoylphosphatidic acid
- DOPC
dioleoylphosphatidylcholine
- DOPE
dioleoylphosphatidylethanolamine
- DOPG
dioleoylphosphatidylglycerol
- DOPS
dioleoylphosphoserine
- EggPA
egg-yolk phosphatidic acid
- EggPC
egg-yolk phosphatidylcholine
- POPC
palmytyl-oleoylphosphatidylcholine
- POPE
palmytyl-oleoylphosphatidylethanolamine
- soyPC
soybean phosphatidylcholine
- C8E4
tetraethylene glycol monooctyl ether
- C12E8
octaethyleneglycol mono-n-dodecylether
- C8POE
octyl- polyoxyethylene
- CHAPS
3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DDM
n-Dodecyl-b-D-maltopyranoside
- DHPC
diheptanoylphosphocholine
- DM
n-Decyl-b-D-maltopyranoside
- FC12
1-Dodecanoyl-2-Hydroxy-sn-Glycero-3-Phosphocholine
- HG
n-hexyl-beta-D-glucopyranoside
- HTG
n-Heptyl beta-Dthioglucopyranoside
- LDAO
n-Dodecyl-N,NDimethylamine-N-Oxide
- OG
n-Octyl-beta-D-glucopyranoside
- OTG
n-Octyl-beta-D-thioglucopyranoside
- TDC
taurodeoxycholate
- Triton X-100
α-[4-(1,1,3,3-Tetramethylbutyl)phenyl]-ω-hydroxy-poly(oxy-1,2-ethanediyl)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeyrathne PD, Chami M, Pantelic RS, Goldie KN, Stahlberg H. Preparation of 2D crystals of membrane proteins for high-resolution electron crystallography data collection. Methods in Enzymology. 2012;481:25–43. doi: 10.1016/S0076-6879(10)81001-8. [DOI] [PubMed] [Google Scholar]
- Agemark M, Kowal J, Kukulski W, Nordén K, Gustavsson N, Johanson U, Engel A, Kjellbom P. Reconstitution of water channel function and 2D-crystallization of human aquaporin 8. Biochimica et Biophysica Acta - Biomembranes. 2012;1818:839–850. doi: 10.1016/j.bbamem.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Behrmann E, Tao G, Stokes DL, Egelman EH, Raunser S, Penczek PA. Real-space processing of helical filaments in SPARX. Journal of Structural Biology. 2012;177:302–313. doi: 10.1016/j.jsb.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CW, Jr, Carter CW. Protein crystallization using incomplete factorial experiments. J Biol Chem. 1979;254:12219–12223. [PubMed] [Google Scholar]
- Carter C., Jr Efficient factorial designs and the analysis of macromolecular crystal growth conditions. Methods. 1990;1:12–24. [Google Scholar]
- Cheng A, Leung A, Fellmann D, Quispe J, Suloway C, Pulokas J, Abeyrathne P, Lam J, Carragher B, Potter C. Towards automated screening of two-dimensional crystals. Journal of Structural Biology. 2007;160:324–331. doi: 10.1016/j.jsb.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintalapati S, Alkurdi R, Terwisschavanscheltinga A, Kuhlbrandt W. Membrane Structure of CtrA3, a Copper-transporting P-type-ATPase from Aquifex aeolicus. J Mol Biol. 2008;378:581–595. doi: 10.1016/j.jmb.2008.01.094. [DOI] [PubMed] [Google Scholar]
- Coudray N, Beck F, Buessler J, Korinek A, Karathanou A, Remigy H, Kihl H, Engel A, Plizko J, Urban J. Automatic Acquisition and Image Analysis of 2D Crystals. Microscopy Today. 2008;16:48–49. [Google Scholar]
- Coudray N, Hermann G, Caujolle-Bert D, Karathanou A, Erne-Brand F, Buessler JL, Daum P, Plitzko JM, Chami M, Mueller U, Kihl H, Urban JP, Engel A, Remigy HW. Automated screening of 2D crystallization trials using transmission electron microscopy: a high-throughput tool-chain for sample preparation and microscopic analysis. J Struct Biol. 2011;173:365–374. doi: 10.1016/j.jsb.2010.09.019. [DOI] [PubMed] [Google Scholar]
- da Fonseca P, Morris EP, Hankamer B, Barber J. Electron crystallographic study of photosystem II of the cyanobacterium Synechococcus elongatus. Biochemistry. 2002;41:5163–5167. doi: 10.1021/bi0120650. [DOI] [PubMed] [Google Scholar]
- De Zorzi R, Nicholson WV, Guigner JM, Erne-Brand F, Venien-Bryan C. Growth of large and highly ordered 2D crystals of a K(+) channel, structural role of lipidic environment. Biophys J. 2013;105:398–408. doi: 10.1016/j.bpj.2013.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucas LJ, Bray TL, Nagy L, McCombs D, Chernov N, Hamrick D, Cosenza L, Belgovskiy A, Stoops B, Chait A. Efficient protein crystallization. J Struct Biol. 2003;142:188–206. doi: 10.1016/s1047-8477(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Diaz R, Rice WJ, Stokes DL. Chapter Five - Fourier-Bessel Reconstruction of Helical Assemblies. Methods in Enzymology. 2010;482:131–165. doi: 10.1016/S0076-6879(10)82005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolder M, Engel A, Zulauf M. The micelle to vesicle transition of lipids and detergents in the presence of a membrane protein: towards a rationale for 2D crystallization. FEBS Lett. 1996;382:203–208. doi: 10.1016/0014-5793(96)00180-9. [DOI] [PubMed] [Google Scholar]
- Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- Dreaden TM, Metcalfe M, Kim LY, Johnson MC, Barry BA, Schmidt-Krey I. Screening for Two-Dimensional Crystals by Transmission Electron Microscopy of Negatively Stained Samples. In: Ingeborg YC, editor. Electron Crystallography of Soluble and Membrane Proteins : Methods and Protocols. Vol. 955. Schmidt-Krey: Humana Press; 2013. pp. 73–101. [DOI] [PubMed] [Google Scholar]
- Egelman EH. Reconstruction of helical filaments and tubes. Methods Enzymol. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson B, Zeng X, Zhang Z, Stahlberg H. 2dx—User-friendly image processing for 2D crystals. Journal of Structural Biology. 2007;157:64–72. doi: 10.1016/j.jsb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Glover CA, Postis VL, Charalambous K, Tzokov SB, Booth WI, Deacon SE, Wallace BA, Baldwin SA, Bullough PA. AcrB contamination in 2-D crystallization of membrane proteins: lessons from a sodium channel and a putative monovalent cation/proton antiporter. J Struct Biol. 2011;176:419–424. doi: 10.1016/j.jsb.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrec F, Palmer CM, Lebon G, Warne T. Pi sampling: a methodical and flexible approach to initial macromolecular crystallization screening. Acta Crystallogr D Biol Crystallogr. 2011;67:463–470. doi: 10.1107/S0907444911008754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- Henderson R, Unwin PN. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Hirai T, Murata K, Mitsuoka K, Kimura Y, Fujiyoshi Y. Trehalose embedding technique for high-resolution electron crystallography: application to structural study on bacteriorhodopsin. J Electron Microsc (Tokyo) 1999;48:653–658. doi: 10.1093/oxfordjournals.jmicro.a023731. [DOI] [PubMed] [Google Scholar]
- Hite RK, Gonen T, Harrison SC, Walz T. Interactions of lipids with aquaporin-0 and other membrane proteins. Pflugers Arch. 2008;456:651–661. doi: 10.1007/s00424-007-0353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm P, Bhakat P, Jegerschold C, Gyobu N, Mitsuoka K, Fujiyoshi Y, Morgenstern R, Hebert H. Structural Basis for Detoxification and Oxidative Stress Protection in Membranes. J Mol Biol. 2006;360:934–945. doi: 10.1016/j.jmb.2006.05.056. [DOI] [PubMed] [Google Scholar]
- Hu M, Vink M, Kim C, Derr K, Koss J, D'Amico K, Cheng A, Pulokas J, Ubarretxena-Belandia I, Stokes D. Automated electron microscopy for evaluating two-dimensional crystallization of membrane proteins. J Struct Biol. 2010;171:102–110. doi: 10.1016/j.jsb.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Richers S. Lipids and membrane protein structures. Curr Opin Struct Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Iacovache I, Biasini M, Kowal J, Kukulski W, Chami M, van der Goot FG, Engel A, Rémigy HW. The 2DX robot: a membrane protein 2D crystallization Swiss Army knife. Journal of Structural Biology. 2010;169:370–378. doi: 10.1016/j.jsb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Jancarik J, Scott WG, Milligan DL, Koshland DE, Jr, Kim SH. Crystallization and preliminary X-ray diffraction study of the ligand-binding domain of the bacterial chemotaxis-mediating aspartate receptor of Salmonella typhimurium. J Mol Biol. 1991;221:31–34. doi: 10.1016/0022-2836(91)80198-4. [DOI] [PubMed] [Google Scholar]
- Jap BK, Zulauf M, Scheybani T, Hefti A, Baumeister W, Aebi U, Engel A. 2D crystallization: from art to science. Ultramicroscopy. 1992;46:45–84. doi: 10.1016/0304-3991(92)90007-7. [DOI] [PubMed] [Google Scholar]
- Kantardjieff KA, Rupp B. Protein isoelectric point as a predictor for increased crystallization screening efficiency. Bioinformatics. 2004;20:2162–2168. doi: 10.1093/bioinformatics/bth066. [DOI] [PubMed] [Google Scholar]
- Karathanou A, Coudray N, Hermann G, Buessler JL, Urban JP. Automatic TEM image analysis of membranes for 2D crystal detection. Adv Exp Med Biol. 2010;680:327–333. doi: 10.1007/978-1-4419-5913-3_37. [DOI] [PubMed] [Google Scholar]
- Kim C, Vink M, Hu M, Love J, Stokes DL, Ubarretxena-Belandia I. An automated pipeline to screen membrane protein 2D crystallization. J Struct Funct Genomics. 2010;11:155–166. doi: 10.1007/s10969-010-9088-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühlbrandt W. Two-dimensional crystallization of membrane proteins. Q Rev Biophys. 1992;25:1–49. doi: 10.1017/s0033583500004716. [DOI] [PubMed] [Google Scholar]
- Kühlbrandt W, Wang DN. Three-dimensional structure of plant light-harvesting complex determined by electron crystallography. Nature. 1991;350:130–134. doi: 10.1038/350130a0. [DOI] [PubMed] [Google Scholar]
- Lampe PD, Kistler J, Hefti A, Bond J, Muller S, Johnson RG, Engel A. In vitro assembly of gap junctions. J Struct Biol. 1991;107:281–290. doi: 10.1016/1047-8477(91)90053-y. [DOI] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López O, de la Maza A, Coderch L, López-Iglesias C, Wehrli E, Parra JL. Direct formation of mixed micelles in the solubilization of phospholipid liposomes by Triton X-100. FEBS Letters. 1998;426:314–418. doi: 10.1016/s0014-5793(98)00363-9. [DOI] [PubMed] [Google Scholar]
- Luft JR, Wolfley JR, Snell EH. What's in a drop? Correlating observations and outcomes to guide macromolecular crystallization experiments. Cryst Growth Des. 2011;11:651–663. doi: 10.1021/cg1013945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke T, Villa C, Edwards PC, Schertler GFX, Heyn MP. X-ray diffraction of heavy-atom labelled two-dimensional crystals of rhodopsin identifies the position of cysteine 140 in helix 3 and cysteine 316 in helix 8. J Mol Biol. 2002;316:693–709. doi: 10.1006/jmbi.2001.5352. [DOI] [PubMed] [Google Scholar]
- Mosser G. Two-dimensional crystallogenesis of transmembrane proteins. Micron. 2001;32:517–540. doi: 10.1016/s0968-4328(00)00047-0. [DOI] [PubMed] [Google Scholar]
- Mosser G, Breyton C, Olofsson A, Popot JL, Rigaud JL. Projection map of cytochrome b6 f complex at 8 A resolution. Journal of Biological Chemistry. 1997;272:20263–20268. doi: 10.1074/jbc.272.32.20263. [DOI] [PubMed] [Google Scholar]
- Mouritsen OG, Bloom M. Models of lipid-protein interactions in membranes. Annu Rev Biophys Biomol Struct. 1993;22:145–171. doi: 10.1146/annurev.bb.22.060193.001045. [DOI] [PubMed] [Google Scholar]
- Newstead S, Ferrandon S, Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Science. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen A, Schenk AD, Signorell GA, Mariani V, Berneche S, Engel A. Collaborative EM image processing with the IPLT image processing library and toolbox. J Struct Biol. 2007;157:28–37. doi: 10.1016/j.jsb.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Philippsen A, Schenk AD, Stahlberg H, Engel A. Iplt--image processing library and toolkit for the electron microscopy community. J Struct Biol. 2003;144:4–12. doi: 10.1016/j.jsb.2003.09.032. [DOI] [PubMed] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A, Tcheng D, Weber D, Carragher B. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. doi: 10.1016/s0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Remigy HW, Caujolle-Bert D, Suda K, Schenk A, Chami M, Engel A. Membrane protein reconstitution and crystallization by controlled dilution. FEBS Lett. 2003;555:160–169. doi: 10.1016/s0014-5793(03)01105-0. [DOI] [PubMed] [Google Scholar]
- Rigaud J, Chami M, Lambert O, Levy D, Ranck J. Use of detergents in two-dimensional crystallization of membrane proteins. Biochim Biophys Acta. 2000;1508:112–128. doi: 10.1016/s0005-2736(00)00307-2. [DOI] [PubMed] [Google Scholar]
- Rigaud J-L, Lévy D. Reconstitution of membrane proteins into liposomes. Meth Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- Rigaud JL, Mosser G, Lacapere JJ, Olofsson A, Levy D, Ranck JL. Bio-Beads: an efficient strategy for two-dimensional crystallization of membrane proteins. J Struct Biol. 1997;118:226–235. doi: 10.1006/jsbi.1997.3848. [DOI] [PubMed] [Google Scholar]
- Rupp B, Wang J. Predictive models for protein crystallization. Methods. 2004;34:390–407. doi: 10.1016/j.ymeth.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Schenk AD, Werten PJ, Scheuring S, de Groot BL, Muller SA, Stahlberg H, Philippsen A, Engel A. The 4.5 A structure of human AQP2. J Mol Biol. 2005;350:278–289. doi: 10.1016/j.jmb.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Scherer S, Kowal J, Chami M, Dandey V, Arheit M, Ringler P, Stahlberg H. 2dx_automator: implementation of a semiautomatic high-throughput high-resolution cryo-electron crystallography pipeline. J Struct Biol. 2014;186:302–307. doi: 10.1016/j.jsb.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Schmidt-Krey I, Lundqvist G, Morgenstern R, Hebert H. Parameters for the two-dimensional crystallization of the membrane protein microsomal glutathione transferase. J Struct Biol. 1998;123:87–96. doi: 10.1006/jsbi.1998.4018. [DOI] [PubMed] [Google Scholar]
- Signorell GA, Chami M, Condemine G, Schenk AD, Philippsen A, Engel A, Remigy HW. Projection maps of three members of the KdgM outer membrane protein family. J Struct Biol. 2007;160:395–403. doi: 10.1016/j.jsb.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Signorell GA, Kaufmann TC, Kukulski W, Engel A, Remigy HW. Controlled 2D crystallization of membrane proteins using methyl-beta-cyclodextrin. J Struct Biol. 2007;157:321–328. doi: 10.1016/j.jsb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Slotboom DJ, Duurkens RH, Olieman K, Erkens GB. Static light scattering to characterize membrane proteins in detergent solution. Methods. 2008;46:73–82. doi: 10.1016/j.ymeth.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Stokes DL, Rice WJ, Hu M, Kim C, Ubarretxena-Belandia I. Two-dimensional crystallization of integral membrane proteins for electron crystallography. Methods Mol Biol. 2010;654:187–205. doi: 10.1007/978-1-60761-762-4_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart MCA, Koning RI, Oostergetel GT, Brisson A. Mechanism of formation of multilayered 2D crystals of the Enzyme IIC-mannitol transporter. Biochimica et Biophysica Acta - Biomembranes. 2004;1663:108–116. doi: 10.1016/j.bbamem.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tsai C-J, Ejsing CS, Shevchenko A, Ziegler C. The role of lipids and salts in two-dimensional crystallization of the glycine-betaine transporter BetP from Corynebacterium glutamicum. Journal of Structural Biology. 2007;160:275–286. doi: 10.1016/j.jsb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Ubarretxena-Belandia I, Stokes DL. Present and future of membrane protein structure determination by electron crystallography. Adv Protein Chem Struct Biol. 2010;81:33–60. doi: 10.1016/B978-0-12-381357-2.00002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubarretxena-Belandia I, Stokes DL. Membrane protein structure determination by electron crystallography. Curr Opin Struct Biol. 2012;22:520–528. doi: 10.1016/j.sbi.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valpuesta JM, Henderson R, Frey TG. Electron cryo-microscopic analysis of crystalline cytochrome oxidase. J Mol Biol. 1990;214:237–251. doi: 10.1016/0022-2836(90)90158-I. [DOI] [PubMed] [Google Scholar]
- Vink M, Derr K, Love J, Stokes DL, Ubarretxena-Belandia I. A high-throughput strategy to screen 2D crystallization trials of membrane proteins. Journal of Structural Biology. 2007;160:295–304. doi: 10.1016/j.jsb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Clark KM, Grayhack EJ, Dumont ME. Characteristics Affecting Expression and Solubilization of Yeast Membrane Proteins. J Mol Biol. 2008;365:621–636. doi: 10.1016/j.jmb.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener MC. A Census of Ordered Lipids and Detergents in X-ray Crystal Structures of Integral Membrane Proteins. Weinheim: Wiley-VCH Verlag GmbH & Co; 2006. [Google Scholar]
- Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S. Molecular Imprinting as a Signal-Activation Mechanism of the Viral RNA Sensor RIG-I. Mol Cell. 2014;55:511–523. doi: 10.1016/j.molcel.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Johnson MC, Schnell JR, Kanaoka Y, Haase W, Irikura D, Lam BK, Schmidt-Krey I. Two-dimensional crystallization conditions of human leukotriene C4 synthase requiring adjustment of a particularly large combination of specific parameters. J Struct Biol. 2010;169:450–454. doi: 10.1016/j.jsb.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Taraska J, Merz AJ, Gonen T. The prototypical H+/galactose symporter GalP assembles into functional trimers. J Mol Biol. 2010;396:593–601. doi: 10.1016/j.jmb.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Jr, Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: Sesame. J Struct Funct Genomics. 2003;4:11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]