Abstract
The question on the patterns and limits of reduction of plastid genomes in nonphotosynthetic plants and the reasons of their conservation is one of the intriguing topics in plant genome evolution. Here, we report sequencing and analysis of plastid genome in nonphotosynthetic orchids Epipogium aphyllum and Epipogium roseum, which, with sizes of 31 and 19 kbp, respectively, represent the smallest plastid genomes characterized by now. Besides drastic reduction, which is expected, we found several unusual features of these “minimal” plastomes: Multiple rearrangements, highly biased nucleotide composition, and unprecedentedly high substitution rate. Only 27 and 29 genes remained intact in the plastomes of E. aphyllum and E. roseum—those encoding ribosomal components, transfer RNAs, and three additional housekeeping genes (infA, clpP, and accD). We found no signs of relaxed selection acting on these genes. We hypothesize that the main reason for retention of plastid genomes in Epipogium is the necessity to translate messenger RNAs (mRNAs) of accD and/or clpP proteins which are essential for cell metabolism. However, these genes are absent in plastomes of several plant species; their absence is compensated by the presence of a functional copy arisen by gene transfer from plastid to the nuclear genome. This suggests that there is no single set of plastid-encoded essential genes, but rather different sets for different species and that the retention of a gene in the plastome depends on the interaction between the nucleus and plastids.
Keywords: plastid genome, nonphotosynthetic plants, genome reduction, gene loss, orchids
Introduction
One of the defining characteristics of plants is their capacity to photosynthesize. However, several species have lost this ability, a phenomenon that has occurred repeatedly in the course of evolution. Such plants have adapted so that they can obtain energy from organic compounds derived from other organisms—either from other plants (parasitism; Westwood et al. 2010) or from mycorrhizal fungi colonizing their roots (mycoheterotrophy; Hynson et al. 2013). Heterotrophy is not confined to any specific lineage of plants and occurs also in bryophytes and gymnosperms, although it is more common in flowering plants, where about 1% of species, representing different lineages of dicots and monocots, are heterotrophic. Plant heterotrophy is especially interesting in the context of evolutionary biology as it represents an example of convergent evolution, a term that refers to the emergence of the same trait in distant lineages.
Plant heterotrophy is commonly associated with distinct anatomical, physiological, and genomic features. One of these is a reduction in the size of the plastid genome, or plastome, which occurs when genes encoding proteins involved in photosynthesis become unnecessary and are lost or pseudogenized (Wolfe et al. 1992). The plastomes of most photosynthetic plants are known to be highly similar in gene content and organization, while information regarding nonphotosynthetic plant plastomes is only beginning to accumulate. Earlier reports suggested conservation of plastome gene order and presence of a large set of conserved genes (Wolfe et al. 1992; Wickett et al. 2008); however, recently, highly reduced (Delannoy et al. 2011; Wicke et al. 2013) and highly rearranged (Logacheva et al. 2014) plastomes have been identified. These findings suggest that plastomes of nonphotosynthetic plants may be much more diverse than previously thought. Consistent with this idea, it was recently reported that a nonphotosynthetic algae, Polytomella, completely lacks a plastid genome, even though it has a plastid. When genome and transcriptome sequencing were performed, the genome was found to not contain the genes required for plastid division, DNA replication, and repair, while the genes encoding plastid-targeted proteins involved in plastid metabolism were present and expressed (Smith and Lee 2014). As an example from higher plants, possible loss of the plastid genome in the parasitic plant Rafflesia was reported (Molina et al. 2014), although this may be explained by a dramatic reduction in plastid DNA copy number and gene content. Taken together, these examples highlight a fact that the patterns of nonphotosynthetic plant plastome evolution are still poorly understood and additional plastome sequences representing substantially different groups of plants are needed to resolve many evolutionary questions. Importantly, among heterotrophic plants, the plastomes of mycoheterotrophs are even less well characterized than those of parasitic plants.
As a source of previously unknown plastid genome variants, the monocot family Orchidaceae is particularly attractive as it is highly diverse and includes at least 30 independent transitions to mycoheterotrophy, including some that are very ancient (Freudenstein and Barrett 2010). In this study, we characterized the plastomes of two mycoheterotrophic orchid species, Epipogium aphyllum and Epipogium roseum, which are both fully mycoheterotrophic and associated with basidiomycete fungi, but exhibit two ecologically divergent mycoheterotrophic strategies. Epipogium aphyllum is mycorrhizal with Inocybe spp., which are themselves mycorrhizal on surrounding trees, the ultimate carbon source of the plant (Roy et al. 2009; Liebel and Gebauer 2011). In contrast, E. roseum is mycorrhizal with saprotrophic Coprinaceae, which recover carbon from soil litter (Yamato et al. 2005; Selosse et al. 2010). Epipogium species are often called “ghost orchids” because of their rarity and yellow-whitish, almost transparent color (fig. 1) and the genus is quite small, consisting only of mycoheterotrophic species. Its relationship with other orchids remains uncertain, but it is presently placed into a separate subtribe, Epipogiinae, which has been thought to be a part of an exclusively mycoheterotrophic tribe Gastrodieae (Dressler 1993), although it has also been claimed that Epipogium is closer to the photosynthetic genus Nervilia (Molvray et al. 2000). Because plastid genes are commonly used and important phylogenetic markers, there have been several attempts to amplify plastid genes from Epipogium, all of which have failed (Cameron 2004). This led to a hypothesis that Epipogium has a highly divergent and reduced plastome, or that it may even have lost its plastome. To address this question, we performed low-coverage genome sequencing (genome skimming) of two Epipogium species, E. aphyllum and E. roseum.
Fig. 1.—
General view and plastid genome map of Epipogium aphyllum (a) and Epipogium roseum (b). Genes shown inside the circle are transcribed clockwise and those outside the circle are transcribed counterclockwise. Genes are color coded according to their functions. Photo credits: E. A. Zvyagina (E. aphyllum) and M. S. Nuraliev (E. roseum).
Materials and Methods
DNA Extraction and Sequencing
Two accessions of E. aphyllum and five accessions of E. roseum were sampled (collection information is provided in supplementary table S1, Supplementary Material online). Total DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method (Doyle 1987). Libraries were prepared using a TruSeq DNA sample prep kit v.2 (Illumina). After post-PCR purification on agarose gel, libraries were quantified using both a Qubit fluorimeter and qPCR before paired-end sequencing using either a MiSeq or a HiSeq 2000 sequencer (Illumina). Library lengths and sequencing parameters are listed in supplementary table S2, Supplementary Material online. For the White Sea accession of E. aphyllum, we generated a long insert (mate pair) library using Nextera Mate Pair sample preparation kit (Illumina) in addition to a standard shotgun library.
Transcriptome libraries were constructed from rRNA-depleted (Ribo-Zero Plant Leaf rRNA Removal Kit; Epicentre) total RNA using a TruSeq mRNA stranded sample preparation kit (Illumina) and sequenced using a MiSeq instrument, which yielded read lengths of 259 + 259. The samples used for transcriptome sequencing were E. aphyllum White Sea and E. roseum Vietnam 2.
Read Preprocessing
Prior to assembly, pair-end reads were trimmed in order to remove adapters and low-quality ends using Trimmomatic 0.32 (Bolger et al. 2014). Reads were trimmed with minimal phred quality 3 from the 3′ end and with a sliding window size 5 and minimal average quality 10. Reads shorter than 50 bp were discarded. Overlapping MiSeq reads were concatenated using the fastq-join from the ea-utils toolset (http://code.google.com/p/ea-utils, last accessed April 6, 2015). The read sets were then edited to remove low-frequency k-mers using Kmernator 1.2 (https://github.com/JGI-Bioinformatics/Kmernator, last accessed April 6, 2015). This operation allowed the removal of most of the reads of nuclear origin, which constituted about 95% of all reads, and the trimming of reads containing errors. After removal of low-frequency k-mers, the plastome assembly was typically more rapid, with lower memory consumption and fewer errors. We removed k-mers of length 31 that were present fewer than 3 times. As an additional step of mate-pair read preprocessing, we also removed improperly oriented (i.e., in a forward–reverse direction) pairs using NextClip 0.8 (Leggett 2014).
De Novo Assembly
Paired-end reads generated with the HiSeq2000 were assembled using Velvet 1.2.10 (Zerbino and Birney 2008), CLC Genomics Workbench 6.0 (www.clcbio.com, last accessed April 6, 2015), and Spades 2.5.1 (Nurk et al. 2013). With the Velvet analysis, we typically considered all k-mers from 41 to 81 with step 5 and each k-mer genomes were assembled with all values of expected coverage from 10 to 2,000 multiplying at each step by 1.5. Coverage cutoff was set at each step to one-tenth of the expected coverage. With each set of Velvet parameters, assemblies were made both with and without scaffolding. Spades was run with default parameters except additional –careful parameter. CLC Genomics Workbench was used with all default parameters and both with and without scaffolding.
To assemble the plastomes sequenced using the MiSeq, which generates longer reads than HiSeq 2000, CLC Genomics Workbench 6.0 and Newbler 2.6 (http://454.com/products/analysis-software, last accessed April 6, 2015) were used. CLC was run with default parameters, while for Newbler assemblies were made with minimum lengths of the overlapping region 100, 200, 300, 400, identity of overlapping fragments 95%, 98%, 100%, and also with default values. For the E. aphyllum sample White Sea, which was sequenced using both the HiSeq 2000 and MiSeq, independent assemblies were generated from the read sets.
The resulting assemblies were searched for contigs and scaffolds of plastid origin. This was accomplished by aligning plastid gene sequences of the orchids Oncidium Gower Ramsey, Phalaenopsis aphrodite, Rhizanthella gardneri, and Neottia nidus-avis to contig and scaffold sets using BLAST 2.2.26 (Altschul et al. 1990), with a BLASTN e-value 10−10 and word size 9 nt. A comparison of the plastid contig sets allowed the selection of the best contig set for further analysis. Contigs were joined using Mapsambler 1.3.17 (Peterlongo and Chikhi 2012), which extends the ends of contigs in a graph-like mode, allowing the user to investigate possible extensions. After producing putative plastome assemblies, reads were mapped (read sets prior to reduction by Kmernator) on them in order to verify their quality, using CLC Assembly Cell 4.2 (www.clcbio.com) requiring 80% of the read to align with at least 98% identity, in a mode of mapping to circular sequences (–lengthfraction 0.8 –similarity 0.98 -z). Reads were mapped to the plastome represented in two orientations: The main orientation and another orientation with 10 kbp moved from the start to the end of the sequence. The second mapping allowed us to verify the circularization of plastid genome. As an additional confirmation of the E. aphyllum plastome organization, reads of the mate-pair library with an insert size of 10 kb were mapped using the CLC assembly cell with the same parameters as described above (supplementary fig. S1, Supplementary Material online).
Plastome Annotation
The assembled plastome sequences were annotated using the online tool DOGMA (Wyman et al. 2004). Taking into account the high degree of divergence between the plastomes of Epipogium and photosynthetic species, we used a sensitive set of parameters: e-value of a match to reference genes 10−5 and match identity 25% (minimal allowed value). Gene predictions suggested by DOGMA were manually checked. The validity of the DOGMA predictions was further assessed by aligning Oncidium Gower Ramsey, P. aphrodite, R. gardneri, and N. nidus-avis genes using BLASTN and TBLASTN. To generate more precise transfer RNA (tRNA) gene predictions, we scanned the whole plastomes using tRNAscan-SE (Lowe and Eddy 1997) with the mito/chloroplast prediction model at http://selab.janelia.org/tRNAscan-SE (last accessed April 6, 2015). Finally, we made a multiple comparison of Epipogium plastomes using mVista (Frazer et al. 2004), a web server for comparative genome analysis, to identify highly conserved but unannotated regions, presumably corresponding to unrecognized genes.
Analysis of Plastomes
To determine gene sequence identity, we performed multiple alignments of concatenated sets of common genes or proteins using MUSCLE 3.8.31 (Edgar 2004) (for RNA-coding genes and protein sequences) and TranslatorX+MUSCLE (for nucleotide sequences of protein-coding genes), and BioEdit 7.2.5 (Hall 1999) was used to build identity tables from the alignments. Insertions and deletions were counted using an in-house script.
A search of repeats in the plastomes was performed using Vmatch 2.2.1 (http://www.vmatch.de, last accessed April 6, 2015). Prior to the search, we removed one copy of inverted repeat (IR) from all plastomes under consideration. The requirements for repeat detection were as follows: Repeats should be no less then 20 bp in length, identity between the copies of the repeat should be no less than 90%, edit distance between them should be no more than 10 (the edit distance is the sum of numbers of mismatches and gaps). An overlap between two units of a repeat pair was not allowed and searches were made for both direct repeat and IR. There were no restrictions on maximum distance between repeat units.
The tree topology was taken from Górniak et al. (2010), where it was inferred from nuclear Xdh (xanthine dehydrogenase) gene sequences. The topology of the Epipogium subtree was inferred from sequences of nuclear 18S rRNA gene sequences found among de novo assembled contigs using BLAST. Sequences of 18S rRNA gene of Epipogium samples and N. nidus-avis, used as outgroup, were aligned using MUSCLE 3.8.31 (Edgar 2004) with default parameters. A tree was built using RAxML 8.2 (Stamatakis 2014) with a GTR+GAMMA model, with verification using 1,000 fast bootstrap replicates. The resulting topology was nested into the general orchid tree build by a maximum parsimony method by Górniak et al. (2010). Because Epipogium sequences were not used in that study, we extrapolated its position from that of its putative relatives, species from tribes Nervilieae, Gastrodieae, and Triphorae (Molvray et al. 2000; Rothacker 2007). All of them are basal braches of epidendroid orchids (Górniak et al. 2010). To add branch lengths, we used 14 protein-coding genes that are common to all orchids with a characterized plastome (rpl2, rpl14, rpl16, rpl36, rps2, rps3, rps4, rps7, rps8, rps11, rps14, rps19, accD, clpP). Nucleotide sequences were aligned using the respective amino acid sequences, using TranslatorX 1.1 (Abascal et al. 2010), which utilizes MUSCLE. PartitionFinder 1.1.1 (Lanfear et al. 2014) was then used to estimate an optimal separation of gene sequences into sets with different GTR + GAMMA model parameters. PartitionFinder combined the 42 input sets (14 genes with 3 codon positions in each) into 15 sets based on a corrected Akaike information criterion. RAxML then added branch lengths to the known topology (“-f e” mode) using the concatenated gene sequences and partition.
All dN/dS values were calculated using PAML 4.7 (Yang 2007). To test if there is difference between dN/dS values on branches of the Epipogium subtree and on branches of other orchids, we conducted two independent calculations using PAML. In the first case, it was assumed that all the branches have the same dN/dS (model = 0), while in the second case two different dN/dS ratios were allowed for Epipogium and non-Epipogium branches (model = 2). The P value of a particular hypothesis was calculated using a likelihood ratio test. We used a branch model with an estimation of codon usage using an F3x4 model. The initial dN/dS value was set to 0.5 and the initial ratio of transitions to transversions frequency to 2.0. The analysis was conducted with a concatenated set of common orchid protein-coding genes, aligned using TranslatorX+Muscle. The tree topology was inferred as described above. Branch lengths for the tree were computed using PAML (fix_blength = 0). In addition, we made a calculation in a mode that allows each branch to have its own dN/dS value (model = 1)
For pairwise dN/dS evaluations, we aligned orchid genes separately using TranslatorX+Muscle and then performed the PAML analysis in a pairwise mode (runmode = −2). The initial dN/dS and initial transition to transversion ratios were set to 0.5 and 2.0, respectively, with a codon frequency model F3x4.
Codon usage, amino acid usage, and Guanine-Cytosine (GC) content were estimated using CodonW 1.4.2 (Peden 1999). These analyses were made for orchids Oncidium Gower Ramsey, R. gardneri, and E. roseum sample Vietnam 2. The statistical significances of differences in codon and amino acid usage and GC-content were estimated using a pairwise proportion Z-test, with Bonferroni correction in cases of multiple comparisons.
To compare gene sets from different species, we used annotations deposited in GenBank or kindly provided to us by Susann Wicke from the University of Vienna (for annotations of Orobanchaceae). Unfortunately, some annotations have ambiguous gene descriptions and mistakes, so the results of our calculation of gene numbers may deviate slightly from those reported in corresponding publications.
Transcriptome Analysis
Reads from RNA sequencing were trimmed using a quality score of 3 with adapter removal using Trimmomatic. To manually confirm the presence of gene transcription, we mapped reads using CLC Assembly Cell. We used various requirements for mapping strictness, requiring a portion of a read that must map to vary from 0.3 to 1.0 and mapping identity from 80% to 100%. Stricter parameters help avoid nonspecific mapping, while less strict parameters allow reads with RNA editing and sequencing errors to be more readily mapped. To check for the presence of splicing, we artificially joined exons and mapped reads to them in a strict mode, demanding whole reads to map with 98% identity.
Results
Plastome Size, Structure, and Gene Content
We characterized the plastomes of five accessions of E. roseum and two accessions of E. aphyllum and determined that they are highly reduced and rearranged in both species compared with other plants, including those that are nonphotosynthetic. The plastome size of E. aphyllum is ∼30.5 kb and, in contrast to the other plastomes, which are either quadripartite, with two single-copy (SC) regions (one large one and one small), and an IR or lack of repeat region, it has an IR region, albeit one SC region (fig. 1). The E. aphyllum plastome is predicted to contain 27 unique genes, including 17 protein-coding genes (ribosomal proteins, a subunit of a chloroplast protease [clpP], acetyl-CoA carboxylase [accD], a translation initiation factor [infA]), 4 ribosomal RNA genes as well as 6 transfer RNAs. Two genes, rps11 and rps18, have a highly divergent 5′ end and are shorter than the orthologs from other species. Epipogium aphyllum retains four intron-containing genes: rpl2, clpP, rpl16, and rps12. clpP has only one intron, while in most species the orthologous gene has two introns, although the length of coding region is similar. In rpl2, the intron position is conserved and the gene is highly similar to its orthologs across its whole length, including the noncoding region (81% identity in the exons and 73% in the introns). Additionally, the rpl2 gene has an atypical start codon; one that is presumably converted to the canonical ATG codon by RNA editing, a feature commonly found in all monocots, whether photosynthetic or not. rpl16 has a very short first exon (9 nt), which complicates its identification, and a region with high similarity to the second exon is located near the IR–SC junction. We identified the predicted first exon of rpl16 as being located within the IR and separated from the second exon by a 914-bp region, ∼200 bp of which has moderate similarity (68%) with the rpl16 intron of other orchids. In addition, a gene encoding trnF-GAA is found within this region and, as far as we are aware, this represents the first report of transfer RNA gene within another gene. rps12 is a trans-splicing gene that consists of three exons, located in two different regions of plastome: The first exon is in the SC region and second and third are in the IR. In E. aphyllum, both parts of rps12 have high similarity with the corresponding sequences of other species, except for the last exon, which is either absent or highly divergent. We determined that the reduction of plastome size in E. roseum is more substantial due to the extreme contraction of the IR region—only a small region carrying the trnF gene is duplicated. The gene content of the E. roseum plastome is similar to that of E. aphyllum, except that E. roseum has two additional genes: A transfer RNA (trnQ-UUG) and a protein-coding gene rpl20. In contrast to E. aphyllum, the clpP gene of E. roseum has three exons, and we were able to identify only two exons in the E. roseum rps12 gene. Notably, in both species the fraction of coding DNA is very high (67–74%).
The plastomes of both Epipogium species show evidence of substantial rearrangement. For example, when compared with Oncidium, a photosynthetic orchid that has a typical plastome gene order, eight collinear blocks are present in E. aphyllum and three blocks are conserved between E. aphyllum and E. roseum (supplementary figs. S2 and S3, Supplementary Material online). Notably, despite numerous rearrangements, the genes that are known to constitute operons are conserved in terms of their order (e.g., rRNA genes, S10 operon, clpP-5′-rps12). Epipogium aphyllum and E. roseum have an almost identical gene order, except for the structure of the IR, which is highly reduced in E. roseum. Such a reduction could have arisen by two events: Inversion of the rps7–rrn5 part of IRb and almost complete loss of IRa (from rrn5 to rps3), or deletion of the rps7–rrn5 part of IRb and deletion of trnI–CAU–rps3.
Intraspecific and Interspecific Sequence Divergence
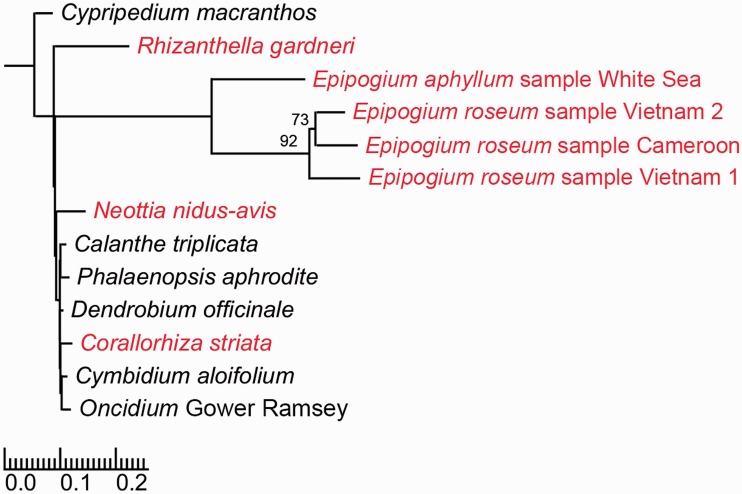
Sequencing of multiple individuals from the same species allowed us to assess intraspecific plastome polymorphism. In the two European E. aphyllum samples, sequence identity is very high (∼99%), with the differences corresponding to both single nucleotide substitutions and indels in both the noncoding and coding regions, and gene order and content are identical. In E. roseum, we revealed much higher diversity and the five samples analyzed from three equally distant groups. The first group includes two samples from Vietnam and a sample from Vanuatu, while the other two groups are represented by one sample each from Cameroon and Vietnam. The overall sequence similarity between these groups is 79–82% versus 98–99% within the Vietnam–Vanuatu group (supplementary table S3, Supplementary Material online). Noncoding sequences are the most polymorphic, with a prevalence of indels over single nucleotide substitutions. In the protein-coding sequences, the similarity is 0.994–0.999 within group and 0.836–0.864 between groups. rRNA genes comprise the most conserved part of the genome and their similarity within a group is 0.998 and between groups from 0.912 to 0.924. The sequence identity between E. aphyllum and E. roseum is ∼0.73–0.74 for protein-coding genes and 0.84–0.85 for rRNA genes. Among photosynthetic species, even those representing distant lineages such as Oncidium and Phalaenopsis, sequence similarity is much higher: 0.94–0.96 for protein-coding and 0.99 for rRNA genes (supplementary table S4, Supplementary Material online). An increase in substitution rate is also apparent in Neottia and Rhizanthella, although each has lost the capacity for photosynthesis independently (fig. 2). An increase in the substitution rate in plastomes of parasitic plants is a known phenomenon (Bromham et al. 2013), although to our knowledge such high divergence at an intraspecific level has not previously been reported.
Fig. 2.—
Phylogenetic tree of orchids with known plastome sequences (general tree topology is following Gorniak et al. 2010, branch lengths are inferred from the analysis of 14 shared plastid protein-coding genes). Plants that have lost the capacity for photosynthesis are labeled by red. The scale bar designates the number of substitution per nucleotide. Numbers above branches indicate bootstrap support values for the Epipogium subtree (inferred from the analysis of 18S rRNA gene sequences).
Indels between different E. roseum groups are abundant: We identified 200–250 with average length of 7.5–7.9 bp (30–43 indels between the genomes of one group). Length mutations predominantly occur in GC-poor regions (supplementary table S5, Supplementary Material online). Differences between E. roseum accessions also influence IR structure and gene content. In the sample from Cameroon, the two parts of IR are not identical, with only 93% similarity, but are identical in all other E. roseum samples. As mentioned above, the IR in Epipogium harbors the trnF gene. In most accessions of E. roseum (as well as in E. aphyllum), this is trnF-GAA, but in the Cameroon sample a mutation affecting the anticodon is found, converting trnF-GAA into trnF-AAA. trnF-GAA is typical for plant plastomes, whereas trnF-AAA has not been reported before. Its functionality in E. roseum Cameroon still requires validation (see Discussion), but regardless of whether or not this is a pseudogene or a new tRNA, we observed a difference in gene content in accessions of the same species.
GC Content
Epipogium plastomes have a very low GC content, with 30–31% in E. roseum and 32.8–32.9% in E. aphyllum (table 1). The distribution of nucleotides is very uneven across the genome, and the IR region in E. aphyllum and the homologous region in E. roseum are more GC rich (37%), while the SC region in E. aphyllum and homologous region in E. roseum have only 20% GC. The higher than average GC content in E. aphyllum is due to the fact that it has a long IR; specifically, the more GC rich part of the plastome is duplicated. In addition, the GC content differs greatly between the coding and noncoding parts of the plastome. Noncoding regions are the most GC poor: 8–14% in SC/SC-like intergenic spacers and 17–21% in IR/IR-like spacers (SC-like and IR-like refer to regions of the E. roseum plastome that are homologous to the IR and SC of E. aphyllum, respectively). In contrast, the most GC rich part of the plastome corresponds to rRNA genes (supplementary fig. S4, Supplementary Material online). In the protein-coding genes, the GC content in different positions of the codons differs, with the first positions being the most GC rich and third positions the most GC poor. The same is a general characteristic of both photosynthetic and nonphotosynthetic orchids; however, in Epipogium, the GC content in all three positions is significantly lower than in other orchids (P < 10−4). In contrast to other orchids, the difference between the GC content of the first and second positions in Epipogium is smaller (supplementary fig. S4, Supplementary Material online) and such convergence of GC content values is also observed in holoparasites from the Orobanchaceae (Wicke et al. 2013).
Table 1.
Summary of Epipogium Plastid Genome Features
| Sample | Plastome Length (bp) | GC Content (%) | Total Number of Genesa | Protein Codinga | rRNAa | tRNAa | Fraction of Coding DNA (%)b |
|---|---|---|---|---|---|---|---|
| Epipogium aphyllum sample White Sea | 30,650 | 32.8 | 27 | 17 | 4 | 6 | 67 |
| E. aphyllum sample France | 30,594 | 32.9 | 27 | 17 | 4 | 6 | 67 |
| Epipogium roseum sample Cameroon | 18,339 | 31.0 | 29 | 18 | 4 | 7 | 74 |
| E. roseum sample Vietnam 1 | 18,938 | 30.1 | 29 | 18 | 4 | 7 | 71 |
| E. roseum sample Vietnam 2 | 19,047 | 30.6 | 29 | 18 | 4 | 7 | 73 |
| E. roseum sample Vietnam 3 | 19,015 | 30.0 | 29 | 18 | 4 | 7 | 71 |
| E. roseum sample Vanuatu | 18,966 | 30.1 | 29 | 18 | 4 | 7 | 71 |
aNumber of unique genes (not including copies in IR).
bIncluding RNA genes.
Repeat Content
The fraction of repeats in the plastome of photosynthetic orchids is about 11–12%, and in Epipogium it ranges from 9.4 to 15.4% (supplementary table S6, Supplementary Material online). In contrast to other orchids, in Epipogium, repeats are extremely AT rich (92.5–94.2% AT) and most represent AT homopolymers and other low-complexity sequences. The distribution of repeats across the genome is uneven and they are much more abundant in noncoding than in coding regions, and in SC regions rather than in IR regions. The IR-like region of the E. roseum plastome also has a decreased density of repeats compared with a region that corresponds to the SC (supplementary fig. S5, Supplementary Material online).
Synonymous and Nonsynonymous Substitution Rates
To evaluate the selective pressure acting on the Epipogium plastid genes, we first analyzed a concatenated set of genes from Epipogium and other orchids. This analysis indicated that the selective force acting on Epipogium genes does not, on average, differ significantly from that acting on the genes of other orchids (P = 0.44). Averaged by the branches of the Epipogium clade, the dN/dS is 0.21, while on the branches of other orchids it is 0.20. Despite the fact that in Epipogium dS is substantially greater, the dN value increases proportionally and so the dN/dS ratio remains the same (supplementary fig. S6, Supplementary Material online). Pairwise comparisons of genes of Epipogium and a photosynthetic orchid Oncidium Gower Ramsey confirmed that the dN/dS value is low (supplementary fig. S7, Supplementary Material online). This supports the idea that selection pressure on Epipogium plastid gene sequences is not relaxed. The ratio between the numbers of nonsynonymous to synonymous substitutions within one species, E. roseum (pN/pS), is higher than dN/dS, further confirming negative selection (Eyrewalker 2006) (supplementary table S7, Supplementary Material online).
Evolution of tRNA Genes: High Sequence Divergence and Compensatory Mutations
Epipogium plastomes carry six (E. aphyllum) or seven (E. roseum) tRNA genes, most of which are conserved in other nonphotosynthetic plants, with the exceptions of trnF-GAA, which is absent from Phelipanche ramose (Wicke et al. 2013), and trnC-GCA which is absent from Epifagus virginiana (Wolfe et al. 1992). The gene trnQ-UUG is absent from E. aphyllum, although it is present in all nonphotosynthetic plants studied so far. The sequence similarity between tRNA genes of Epipogium and the photosynthetic orchid P. aphrodite (81–93%) is much less than usually observed for tRNAs. Epipogium trnY-GUA genes differ from those of Phalaenopsis not only with respect to substitutions but also by a 1-nt insertion. Length mutations in tRNAs are extremely rare; however, in silico tRNA folding analyses showed that most tRNAs have typical secondary structure and most positions that participate in hairpin formation are either conserved or substitutions occur in both nucleotides, such that complementarity is maintained (compensated substitutions; supplementary table S8, Supplementary Material online). The most striking observation related to the tRNA genes of Epipogium is the structure of trnF in the E. roseum sample from Cameroon. Although E. aphyllum and other E. roseum samples have a trnF-GAA that is typical of plant plastomes, in the Cameroon sample a substitution in the anticodon converts GAA into AAA. This sample has two slightly different trnF sequences (sequence similarity 93.5%), both of which have two noncompensated substitutions in the acceptor stem and low cove scores (supplementary table S8, Supplementary Material online; 40 and 23.8). (The lowest score required by tRNAscan-SE to accept a sequence as an organellar tRNA is 15).
Evolution of Protein-Coding Genes: Codon and Amino Acid Bias
Epipogium plastomes have higher AT content than it is usually observed in plant plastomes. The most AT-rich component is noncoding DNA, but the AT bias is also seen in protein-coding sequences and substantially changes codon usage. With respect to synonymous codons, those that are AT-rich ones have a strong predominance over GC-rich codons. Although a greater frequency of A/U-ending codons is common in most plastomes, in Epipogium it is significantly higher than in photosynthetic orchids. For example, for phenylalanine, which is encoded by UUU and UUC, the fraction of UUU is 65% in Oncidium and 91% in Epipogium (supplementary table S9, Supplementary Material online). Consistent with this, the effective number of codons (ENC) in Epipogium plastid genes is 36–40, while in photosynthetic orchids the equivalent range is 47–48. In other nonphotosynthetic orchids, ENC values remain within, or very close to, this range (supplementary table S10, Supplementary Material online). AT bias, which is a prominent feature of Epipogium plastomes, is apparent not only at the level of synonymous codon usage, but also at the amino acid usage level. A comparison of amino acid frequency in genes that are shared between Epipogium and other orchids showed that amino acids encoded by GC-poor codons, such as Phe and Ile, are significantly more frequent, and the frequency of amino acids encoded by GC-rich codons (Ala, Arg) is lower (fig. 3).
Fig. 3.—
Amino acid usage in three orchids. Amino acids are listed in the order of increase of average codon GC content (e.g., isoleucine has the codons ATA, ATT, and ATC with an average GC content 1/9, and glycine has the codons GGG, GGC, GGA, and GGT with an average GC content 10/12). Single asterisk denotes cases when amino acid usage in E. roseum differs significantly (P < 0.05) from both the other orchids. Two asterisks indicate that the difference is significant only from the green orchid Oncidium Gower Ramsey.
Expression of Plastid Genes
In order to test whether plastid genes in Epipogium are expressed, spliced, and edited, we mapped transcriptome reads onto the annotated plastome sequences. In both species, all the genes had mapped reads, except for rps18. In addition, all four intron-containing genes, reads that overlap predicted exon junctions were identified (supplementary fig. S8, Supplementary Material online). RNA-seq confirmed the presence of a single intron in E. aphyllum clpP, which differs from the two introns present in E. roseum and most other plants. However, the frequency of spliced transcripts was different. In clpP and rps12, the coverage of a region of the exon junction is approximately the same as within exons, while in rpl2 it is much lower, especially in E. roseum. This indicates a lower splicing rate. In addition to splicing, editing (C to U) of the second position of the rpl2 start codon was identified, although at a low frequency (83 out of 157 reads in E. aphyllum and 23 reads out of 176 in E. roseum).
Discussion
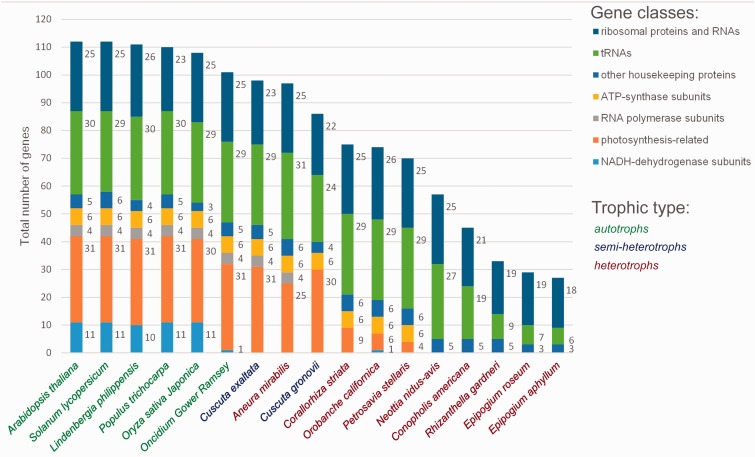
In this study, we characterized the plastomes of E. aphyllum and E. roseum, which we showed to be highly rearranged and reduced (31 and 19 kb, respectively). The latter is the smallest plastome reported to date and even the highly reduced residual plastomes of parasitic protists (such as Plasmodium and Helicosporidium) are larger, with size about 35 kb (Wilson et al. 1996; de Koning and Keeling 2006). Also, peridinin-containing dinoflagellates have plastomes in the form of several mini-circles 2–3 kb in length (Zhang et al. 1999) with a total length of 27–46 kb (Barbrook et al. 2014). The sizes of plant plastomes, including nonphotosynthetic species, range from 45.7 (Conopholis americana, a parasite) to 217 kb. We show here that the reduction can go further and affect ∼85% of the typical plastome size (∼150 kb) and 75% of the total gene complement. Besides heterotrophic plant species, a reduction in plastome size has also been reported in photosynthetic plants under several experimental conditions (Day and Ellis 1985; Cahoon, Cunningham, Stern 2003; Cahoon, Cunningham, Bollenbach, et al. 2003), but the patterns of reduction differ in these two cases. In experimental systems, different cell lines carry different deletions and the lines that started heterotrophic evolution at the same time have dissimilar degrees of reduction. In contrast, in heterotrophic plants, there is a clear parallel in the patterns of gene loss in lineages that transition to heterotrophy independently. This has led to a postulation of the existence of a conserved minimal gene set (Delannoy et al. 2011; Logacheva et al. 2011), to which all the reduced plastomes should converge. This core set was thought to include rRNA genes, several tRNA genes, most ribosomal protein genes, clpP, accD, ycf1, and ycf2. Later a model describing the pattern of gene loss during plastome degradation was proposed (Barrett and Davis 2012; Barrett et al. 2014). In postulates that changes in a plastome gene set of plants that lose their photosynthetic ability follow a specific path. First, the plastome loses genes encoding subunits of reduced nicotinamide adenine dinucleotide (NADH) dehydrogenase. This can be explained by the fact that this complex plays a supplementary role in photosynthesis and plants with knocked-out ndh genes are viable (Burrows et al. 1998). The second group of genes to be lost are those responsible for photosynthesis and the third in the sequence is the plastid-encoded RNA polymerase gene. This RNA polymerase, in contrast to the nuclear-encoded polymerase, is responsible mainly for the transcription of genes lost in the first two steps, and so it becomes unnecessary. Genes encoding transfer RNA and ribosome components and several other housekeeping genes—matK, clpP, infA, accD, ycf1, and ycf2—are the last ones. In general, the gene content of the Epipogium plastomes agrees with this model, being close to the end of this path (fig. 4).
Fig. 4.—
The patterns of plastid gene loss in different groups of photosynthetic and nonphotosynthetic plants. Species are ordered by a number of unique genes (not taking into account duplicated genes).
Despite high degree of reduction, several lines of evidence support that Epipogium plastome is not just a remnant of formerly functional plastome but a functional entity. First, there is a high percentage of coding DNA; more than in other nonphotosynthetic plants (Wicke et al. 2013) and even more than in photosynthetic orchids that have lost functional ndh genes but retain their pseudogenes in the plastome (Chang 2005; Wu et al. 2010). Second, there is a conservation of the nonrandom gene set, an absence of a dN/dS increase in proteincoding genes, and an abundance of compensatory substitutions in tRNA genes. Third, we found expression, editing, and splicing of plastid genes in both Epipogium species. Regulation of these processes relies completely on nuclear gene products (in photosynthetic plants, some elements of the transcription and splicing machinery, such as RNA polymerase and the splicing factor matK, are plastid encoded but this is not the case for Epipogium) and requires complex and co-ordinated nucleus-plastid interactions. This machinery is unlikely to be conserved in the absence of function and indeed it has been lost in the plastome-less alga Polytomella (Smith and Lee 2014). The only presumable pseudogene in Epipogium plastomes is trnF-AAA, found in one of E. roseum accession. trnF-AAA has not been reported to be present in plant plastomes; however, it is an isoacceptor of trnF-GAA that is commonly found in plant plastomes, including those that are nonphotosynthetic. Mutations in anticodons (anticodon shifts) occurred many times in the evolution of several genomes, with isoacceptor shifts being more frequent than alloacceptor shifts (Rogers and Griffiths-Jones 2014). The change of trnF-GAA to trnF-AAA may have been favorable, because ∼90% of all Phe codons in Epipogium are encoded by UUU. However, trnF-AAA does not retain a secondary structure that is typical of transfer RNAs as it has two noncompensated mutations in the acceptor stem. It has been shown in multiple experiments with Escherichia coli and other model systems that proper pairing in the acceptor stem is crucial for tRNA recognition by aminoacyl tRNA synthetase (Jahn et al. 1991). Thus, the presence of two noncompensated mutations in the acceptor stem strongly suggests nonfunctionality. However, a similar situation has been observed in the mitochondrial genomes of several invertebrates, including Lithobius, where most tRNAs have mismatches in the acceptor stem that are rectified by RNA editing (Lavrov et al. 2000). The same has been shown for plant mitochondrial transfer RNA trnF-GAA (Maréchal-Drouard et al. 1993). Similar mechanism may occur in E. roseum, restoring proper pairing in trnF-AAA.
Our analysis indicates that Epipogium plastomes underwent changes not only in gene content but also in structure. Although most plastomes have a quadripartite structure, with a long identical IR separated by two SC regions, E. aphyllum lacks the SSC and E. roseum has a very short IR. The loss of the SSC is expected as it usually contains ndh genes that are pseudogenized in many plants, including orchids (Chang 2005), as well as several photosynthesis-related genes. Comparative sequence analysis of E. aphyllum and E. roseum clearly indicates that the structure found for E. roseum is more derived. The E. roseum plastome region that corresponds to the IR has a higher GC content and is less divergent than that corresponding to the SC. In one E. roseum accession (Cameroon), we identified a nonidentical IR, while in most plants studied to date, parts of the IR are identical, even in plastomes with a very small IR. This identity is thought to be maintained by gene conversion (Palmer 1985). The occurrence of a nonidentical IR suggests that this mechanism may have been lost or suppressed in E. roseum.
Sequencing multiple accession of Epipogium allowed us to assess the level of intraspecific plastome polymorphism—the first intraspecific comparison of complete plastid genome sequences from nonphotosynthetic plants. We found an unexpectedly high divergence between E. roseum individuals, including those sampled not very far geographically from another (Vietnam 1 and 2 are situated about 100 km from each other). Although plastid DNA, especially in noncoding regions, displays some variation in most plant species, it is more than 10 times less than we found in E. roseum (Xu et al. 2012). Because little is known about the population genetics of E. roseum, which covers a wide range of longitudes and climates, we cannot exclude the possibility that these samples represent cryptic species. However, a recent survey of polymorphism in the atpB-rbcL region in another nonphotosynthetic orchid, N. nidus-avis, revealed that it is highly divergent between individuals from the same population (Cafasso and Chinali 2012). A comparison of several plastome regions from different populations of R. gardneri also showed high sequence divergence (Delannoy et al. 2011) suggesting that this is common for nonphotosynthetic plants.
A high substitution rate is not usual for plant plastomes. Among all DNA-containing organelles, those with the highest rate of substitution accumulation are animal mitochondria (Lynch and Blanchard 1998). Several hypotheses have been proposed to explain this: The absence of recombination and low effective number (i.e., despite a high number of mitochondria in somatic cells, few are present in the egg cell and are transmitted to the progeny) causing mutational drift (Neiman and Taylor 2009). These explanations may be applicable to the Epipogium plastome as well. Increased substitution rates in all three genomes were found in parasitic plants and it has been suggested that this may reflect a shorter generation time or decreased efficiency of DNA repair (Bromham et al. 2013). High substitution rates were also observed in the 18 S nuclear rRNA gene of mycoheterotrophic species (Lemaire et al. 2011), suggesting that rate acceleration is a characteristic of the genomes of all heterotrophic plants, and not only those that are parasitic. However, no evidence of accelerated sequence evolution was found in the mycoheterotrophic monocot Petrosavia stellaris (Logacheva et al. 2014). More data, including mitochondrial genome sequences, are necessary in order to determine whether the extremely high substitution rates of Epipogium plastomes are unique or reflect general trends of genome evolution in heterotrophic plants. Interestingly, a high substitution rate and high AT content are also typical in symbiotic prokaryotes with very small genomes (McCutcheon and Moran 2011).
Summarizing our findings, characterization of Epipogium plastomes reveals highly dynamic evolutionary patterns that are however not “chaotic” as in heterotrophic cell cultures and that are directed by high substitution rate and negative selection.
Several hypotheses have been proposed to explain the conservation of plastid genomes in nonphotosynthetic plants (reviewed in Barbrook et al. 2006). The most plausible is the essential tRNA hypothesis, which postulates that trnE-UUC is the main reason for plastome conservation since it is essential for heme biosynthesis. Our analysis of the Epipogium plastomes does not contradict this hypothesis as in both species trnE is intact; however, it does not explain the retention and expression of other plastid genes. Other than photosynthesis, plastids are also involved in several processes that are crucially important even for nonphotosynthetic plants. These processes include the synthesis of fatty acids and carotenoids, starch storage and gravitropism (Chen et al. 1999; Neuhaus and Emes 2000). In model plant species, complex interactions that involve feedback loops exist between the translation of plastid genes and other processes (Tiller and Bock 2014). These interactions mainly co-ordinate photosynthesis-related processes; however, it was shown that plastid translation is also required for Arabidopsis thaliana embryo development (Romani et al. 2012). Several lines of evidence indicate that this phenomenon is not related to photosynthesis, but rather to the necessity of the accD gene in fatty acid biosynthesis (Bryant et al. 2011). Epipogium plastomes encode only two genes, which are involved in functions other than translation and, notably, accD is one of these. Another gene, clpP, is essential for tobacco shoot development (Kuroda and Maliga 2003; but see Cahoon, Cunningham, Stern 2003; Cahoon, Cunningham, Bollenbach, et al. 2003). We suggest that associations between translation of these plastid genes and development, similar to those reported for A. thaliana and tobacco, exist in Epipogium and that this underlies the necessity of plastome retention.
Recently, the apparent absence of a plastid genome was reported in Rafflesia lagascae, a parasitic dicot (Molina et al. 2014). This seems to contradict the essential tRNA hypothesis as well as our hypothesis, which can be termed the “essential plastid translation hypothesis.” However, it is well known that different plant lineages have different necessity in plastid-encoded gene products. rpl22 and rps16 are essential for tobacco (Fleischmann et al. 2011), while in legumes they are lost from the plastome (Doyle et al. 1995). The accD gene of grasses and Campanulaceae has been lost from the plastome and its product has been replaced by a plastid-targeted, but nuclear-encoded gene (Rousseau-Gueutin et al. 2013). Consistent with this, the breakdown of translation in maize plastids does not lead to embryo lethality (Asakura and Barkan 2006). The same is true for clpP—as mentioned above, it is essential for tobacco, but not for maize (Cahoon, Cunningham, Stern 2003; Cahoon, Cunningham, Bollenbach, et al. 2003). Even more striking, in Brassica napus, a close relative of A. thaliana, plastid translation is not necessary for embryo development (Zubko and Day 1998) because accD gene encoded in the B. napus plastome is replaced by, or at least complemented with, the plastid-targeted nuclear gene product (Schulte et al. 1997). This emphasizes that even within closely related plant groups the requirements for plastid-encoded genes can differ. If the product of a plastid gene is essential for a plant, it can be lost from the plastome when a functional copy exists in the nuclear genome. Such copies are the results of gene transfer. Gene transfer from an organelle to the nucleus is a multistep process that includes: 1) integration of the sequence into the nuclear genome; 2) its activation (acquisition of translation and transcription); 3) gain of a transit peptide; and 4) loss from the organellar genome (Selosse et al. 2001). The first step, integration, is quite common in plants, as has been shown by both comparative genome analysis and direct experimental methods (Huang et al. 2003; Shahmuradov et al. 2003). The probability of integration correlates with the number of plastids (Smith et al. 2011) and thus can be species- or lineage specific. Although a functional gene is present in the plastome, all transformations involving its nuclear counterpart occur at random and are not under selection. Thus, we should not expect that their consequences are streamlined and lead to the same results in different species. There is therefore probably not a single set of “essential genes” for all nonphotosynthetic plants, but rather different sets of essential genes for different plants, depending on the interactions between the plastid and the nucleus that have been established over the course of evolution. We believe that exploration of nuclear genes and their structure and expression in both nonphotosynthetic plants and their photosynthetic relatives (including our forthcoming study on Epipogium transcriptomes) will provide new insights into these questions.
Supplementary Material
Supplementary figures S1–S8 and table S1–S10 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Hervé Christome (Biome company), Tatiana V. Neretina (Lomonosov Moscow State University), and Nikolai S. Mugue (Institute of Developmental Biology RAS) for their help in collection of E. aphyllum; to Marc Pignal and Guillaume Leotard for collection of E. roseum; to Ziheng Yang (University College London) for assistance with PAML; to Georgiy A. Bazykin and Alexey S. Kondrashov (Lomonosov Moscow State University) for helpful discussion and comments regarding manuscript; and to PlantScribe (www.plantscribe.com) for manuscript editing. The study was supported by Russian foundation for basic research (project №11-04-02031) and Ministry of education and science (project №11.G34.31.0008).
Literature Cited
- Abascal F, Zardoya R, Telford MJ. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010;38:W7–W13. doi: 10.1093/nar/gkq291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asakura Y, Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11:101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Voolstra CR, Howe CJ. The chloroplast genome of a Symbiodinium sp. clade C3 isolate. Protist. 2014;165:1–13. doi: 10.1016/j.protis.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Barrett CF, et al. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 2014;31:3095–3112. doi: 10.1093/molbev/msu252. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Davis JI. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 2012;99(9):1513–1523. doi: 10.3732/ajb.1200256. [DOI] [PubMed] [Google Scholar]
- Bolger A, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromham L, Cowman PF, Lanfear R. Parasitic plants have increased rates of molecular evolution across all three genomes. BMC Evol Biol. 2013;13:126. doi: 10.1186/1471-2148-13-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D. Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol. 2011;155:1678–1689. doi: 10.1104/pp.110.168120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows PA, Sazanov LA, Svab Z, Maliga P, Nixon PJ. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998;17(4):868–876. doi: 10.1093/emboj/17.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafasso D, Chinali G. Multiple and different genomic rearrangements of the rbc L gene are present in the parasitic orchid Neottia nidus-avis. Genome. 2012;55:629–637. doi: 10.1139/g2012-057. [DOI] [PubMed] [Google Scholar]
- Cahoon AB, Cunningham KA, Stern DB. The plastid clpP gene may not be essential for plant cell viability. Plant Cell Physiol. 2003;44(1):93–95. doi: 10.1093/pcp/pcg003. [DOI] [PubMed] [Google Scholar]
- Cahoon AB, Cunningham KA, Bollenbach TJ, Stern DB. Maize BMS cultured cell lines survive with massive plastid gene loss. Curr Genet. 2003;44:104–113. doi: 10.1007/s00294-003-0408-1. [DOI] [PubMed] [Google Scholar]
- Cameron KM. Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Mol Phylogenet Evol. 2004;31:1157–1180. doi: 10.1016/j.ympev.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Chang CC. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2005;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
- Chen R, Rosen E, Masson PH. Gravitropism in higher plants. Plant Physiol. 1999;120:343–350. doi: 10.1104/pp.120.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 2008;24:861–862. doi: 10.1093/bioinformatics/btm598. [DOI] [PubMed] [Google Scholar]
- Day A, Ellis TN. Deleted forms of plastid DNA in albino plants from cereal anther culture. Curr Genet. 1985;9:671–678. [Google Scholar]
- de Koning AP, Keeling PJ. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006;4:12. doi: 10.1186/1741-7007-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I. Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol. 2011;28:2077–2086. doi: 10.1093/molbev/msr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst Bot. 1995;20:272–294. [Google Scholar]
- Dressler RL. Cambridge: Cambridge University Press; 1993. Phylogeny and classification of the orchid family. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyrewalker A. The genomic rate of adaptive evolution. Trends Ecol Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Fleischmann TT, et al. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell Online. 2011;23:3137–3155. doi: 10.1105/tpc.111.088906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenstein JV, Barrett CF. Mycoheterotrophy and diversity in Orchidaceae. In: Seberg O, Petersen G, Barfod AS, Davis JI, editors. Diversity, phylogeny, and evolution in the monocotyledons. Aarhus (Denmark): Aarhus University Press; 2010. pp. 25–37. [Google Scholar]
- Górniak M, Paun O, Chase MW. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: congruence with organellar and nuclear ribosomal DNA results. Mol Phylogenet Evol. 2010;56(2):784–795. doi: 10.1016/j.ympev.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Huang CY, Ayliffe MA, Timmis JN. Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature. 2003;422:72–76. doi: 10.1038/nature01435. [DOI] [PubMed] [Google Scholar]
- Hynson NA, Madsen TP, Selosse M-A, Adam IKU, Ogura-Tsujita Y, Roy M, Gebauer G. 2013. The physiological ecology of mycoheterotrophy. In: Mycoheterotrophy, the biology of plants living on fungi. Berlin (Gerrmany): Springer. p. 297–342.
- Jahn M, Rogers MJ, Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991;352:258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Maliga P. The plastid clpP1 protease gene is essential for plant development. Nature. 2003;425:86–89. doi: 10.1038/nature01909. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 2014;14:82. doi: 10.1186/1471-2148-14-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov DV, Brown WM, Boore JL. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc Natl Acad Sci U S A. 2000;97:13738–13742. doi: 10.1073/pnas.250402997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett RM, Clavijo BJ, Clissold L, Clark MD, Caccamo M. NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics. 2014;30(4):566–568. doi: 10.1093/bioinformatics/btt702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire B, Huysmans S, Smets E, Merckx V. Rate accelerations in nuclear 18 S rDNA of mycoheterotrophic and parasitic angiosperms. J Plant Res. 2011;124:561–576. doi: 10.1007/s10265-010-0395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebel HT, Gebauer G. Stable isotope signatures confirm carbon and nitrogen gain through ectomycorrhizas in the ghost orchid Epipogium aphyllum Swartz. Plant Biol. 2011;13:270–275. doi: 10.1111/j.1438-8677.2010.00369.x. [DOI] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Nuraliev MS, Samigullin TH, Penin AA. The plastid genome of mycoheterotrophic monocot Petrosavia stellaris exhibits both gene losses and multiple rearrangements. Genome Biol Evol. 2014;6:238–246. doi: 10.1093/gbe/evu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva MD, Schelkunov MI, Penin AA. Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol Evol. 2011;3:1296–1303. doi: 10.1093/gbe/evr102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Blanchard JL. Deleterious mutation accumulation in organelle genomes. Genetica. 1998;102–103:29–39. [PubMed] [Google Scholar]
- Maréchal-Drouard L, Ramamonjisoa D, Cosset A, Weil JH, Dietrich A. Editing corrects mispairing in the acceptor stem of bean and potato mitochondrial phenylalanine transfer RNAs. Nucleic Acids Res. 1993;21:4909–4914. doi: 10.1093/nar/21.21.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- Molina J, et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol Biol Evol. 2014;31:793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molvray M, Kores PJ, Chase MW. Polyphyly of mycoheterotrophic orchids and functional influences on floral and molecular characters. In: Wilson KL, Morrison DA, editors. Monocots: systematics and evolution. Melbourne (Australia): CSIRO Publishing; 2000. pp. 441–448. [Google Scholar]
- Neiman M, Taylor DR. The causes of mutation accumulation in mitochondrial genomes. Proc R Soc B Biol Sci. 2009;276:1201–1209. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ. Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:111–140. doi: 10.1146/annurev.arplant.51.1.111. [DOI] [PubMed] [Google Scholar]
- Nurk S, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Peden JF. 1999. Analysis of codon usage [PhD thesis]. [Nottingham (United Kingdom)]: University of Nottingham. [Google Scholar]
- Peterlongo P, Chikhi R. Mapsembler, targeted and micro assembly of large NGS datasets on a desktop computer. BMC Bioinformatics. 2012;13:48. doi: 10.1186/1471-2105-13-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers HH, Griffiths-Jones S. tRNA anticodon shifts in eukaryotic genomes. RNA. 2014;20:269–281. doi: 10.1261/rna.041681.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani I, et al. Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J. 2012;72:922–934. doi: 10.1111/tpj.12000. [DOI] [PubMed] [Google Scholar]
- Rothacker EP. 2007. The primitive Epidendroideae (Orchidaceae): phylogeny, character evolution and the systematics of Psilochilus (Triphoreae). Dissertation. Athens: Ohio State University.
- Rousseau-Gueutin M, et al. Potential functional replacement of the plastidic acetyl-CoA carboxylase subunit (accD) gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 2013;161:1918–1929. doi: 10.1104/pp.113.214528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M, et al. Ectomycorrhizal Inocybe species associate with the mycoheterotrophic orchid Epipogium aphyllum but not its asexual propagules. Ann Bot. 2009;104:595–610. doi: 10.1093/aob/mcn269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte W, Töpfer R, Stracke R, Schell J, Martini N. Multi-functional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc Natl Acad Sci U S A. 1997;94:3465–3470. doi: 10.1073/pnas.94.7.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selosse MA, Albert B, Godelle B. Reducing the genome size of organelles favours gene transfer to the nucleus. Trends Ecol Evol. 2001;16:135–141. doi: 10.1016/s0169-5347(00)02084-x. [DOI] [PubMed] [Google Scholar]
- Selosse MA, Martos F, Perry BA, Padamsee M, Roy M, Pailler T. 2010. Saprotrophic fungal symbionts in tropical achlorophyllous orchids: Finding treasures among the ‘molecular scraps’? Plant Signaling and Behaviour 5: 1–5. [DOI] [PMC free article] [PubMed]
- Shahmuradov IA, Akbarova YY, Solovyev VV, Aliyev JA. Abundance of plastid DNA insertions in nuclear genomes of rice and Arabidopsis. Plant Mol Biol. 2003;52:923–934. doi: 10.1023/a:1025472709537. [DOI] [PubMed] [Google Scholar]
- Smith DR, Crosby K, Lee RW. Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol Evol. 2011;3:365–371. doi: 10.1093/gbe/evr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Lee RW. A plastid without a genome: evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014;164:1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R. The translational apparatus of plastids and its role in plant development. Mol Plant. 2014;7(7):1105–1120. doi: 10.1093/mp/ssu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15:227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Wicke S, et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25:3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett NJ, et al. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol. 2008;25:393–401. doi: 10.1093/molbev/msm267. [DOI] [PubMed] [Google Scholar]
- Wilson RJ, et al. Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J Mol Biol. 1996;261:155–172. doi: 10.1006/jmbi.1996.0449. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FH, et al. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010;10:68. doi: 10.1186/1471-2229-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- Xu Q, et al. Analysis of complete nucleotide sequences of 12 Gossypium chloroplast genomes: origin and evolution of allotetraploids. PLoS One. 2012;7(8):e37128. doi: 10.1371/journal.pone.0037128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamato M, Yagame T, Suzuki A, Iwase K. 2005. Isolation and identification of mycorrhizal fungi associating with an achlorophyllous plant, Epipogium roseum (Orchidaceae) Mycoscience 46:73–77.
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Green BR, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400:155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- Zubko MK, Day A. Stable albinism induced without mutagenesis: a model for ribosome-free plastid inheritance. Plant J Cell Mol Biol. 1998;15:265–271. doi: 10.1046/j.1365-313x.1998.00195.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.