Abstract
Viruses with single-stranded (ss) DNA genomes infect hosts in all three domains of life and include many medically, ecologically, and economically important pathogens. Recently, a new group of ssDNA viruses with chimeric genomes has been discovered through viral metagenomics. These chimeric viruses combine capsid protein genes and replicative protein genes that, respectively, appear to have been inherited from viruses with positive-strand RNA genomes, such as tombusviruses, and ssDNA genomes, such as circoviruses, nanoviruses or geminiviruses. Here, we describe the genome sequence of a new representative of this virus group and reveal an additional layer of chimerism among ssDNA viruses. We show that not only do these viruses encompass genes for capsid proteins and replicative proteins that have distinct evolutionary histories, but also the replicative genes themselves are chimeras of functional domains inherited from viruses of different families. Our results underscore the importance of horizontal gene transfer in the evolution of ssDNA viruses and the role of genetic recombination in the emergence of novel virus groups.
Keywords: ssDNA viruses, virus evolution, origin of viruses, genetic recombination, metagenomics
Introduction
Viruses with single-stranded (ss) DNA genomes infect organisms in all three domains of life and include many widespread, medically and economically important pathogens (Krupovic 2013). Until recently, the diversity of ssDNA viruses had been largely restricted to several families, such as Circoviridae, Nanoviridae, Geminiviridae, and Parvoviridae, which infect animals and plants, as well as Microviridae and Inoviridae, which infect bacteria. The ssDNA viruses have been thought to be much less abundant than RNA viruses in eukaryotes and double-stranded DNA viruses in prokaryotes. However, the appreciation of ssDNA viruses in the biosphere has been dramatically boosted by numerous recent metagenomic studies (Rosario and Breitbart 2011; Delwart and Li 2012; Rosario et al. 2012). Two key observations have been made: 1) ssDNA viruses are highly abundant in all studied environments, from the human gut to terrestrial hot springs (Rosario et al. 2009; Mochizuki et al. 2012; Roux et al. 2012; Whon et al. 2012; Dayaram, Goldstien, et al. 2013; Popgeorgiev et al. 2013; Yoshida et al. 2013; Zawar-Reza et al. 2014), and 2) they are highly diverse genetically (Roux et al. 2012; Dayaram, Potter, et al. 2013; Labonte and Suttle 2013). The global diversity of ssDNA viruses appears to be determined by two principal factors: Namely, extremely high nucleotide substitution rates that approach those of RNA viruses (Duffy et al. 2008; Duffy and Holmes 2008, 2009; Firth et al. 2009; Harkins et al. 2009, 2014; Grigoras et al. 2010; Sanjuan et al. 2010; De Bruyn et al. 2012) and pervasive recombination (Martin et al. 2011; Kraberger et al. 2013; Lefeuvre and Moriones 2015). Recombination route is especially important in the emergence of novel virus types, as exemplified by members of the family Bidnaviridae which have apparently evolved from genes of four groups of widely different viruses (Krupovic and Koonin 2014).
In 2012, in the course of metagenomic exploration of viral diversity in the Boiling Springs Lake (BSL), Diemer and Stedman have assembled a genome of a novel virus which they named RNA–DNA hybrid virus (BSL RDHV) (Diemer and Stedman 2012). Unexpectedly, the BSL RDHV genome was found to be a natural chimera: The gene for the rolling-circle replication initiation protein (RC-Rep) is inherited from a circovirus-like ancestor whereas the capsid protein (CP) gene is most closely related to that of RNA viruses of the Tombusviridae family (Diemer and Stedman 2012). ssDNA viruses appear to have access not only to the gene pool of other DNA viruses but also to that of viruses with RNA genomes, further expanding opportunities for recombination. Subsequently, several additional chimeric virus (CHIV) genomes have been assembled from metagenomic data (Roux et al. 2013; McDaniel et al. 2014; Kraberger et al. 2015). A different variety of a hybrid ssDNA virus has been serendipitously isolated as a contamination of nucleic acid-isolation spin columns, including a virus named NIH-CQV (Naccache et al. 2013; Xu et al. 2013). This virus encodes a CP that is affiliated with the CPs of parvoviruses and a replication protein that seems to belong to a distinct group distantly related to the replication proteins of circoviruses and nanoviruses. Thus, NIH-CQV appears to be a chimera consisting of genes derived from two families of ssDNA viruses.
These findings emphasize the mosaicism of ssDNA virus genomes and the key role of recombination, sometimes between different types of viruses, in their origin and evolution (Martin et al. 2011; Diemer and Stedman 2012; Krupovic 2013, 2012; Lefeuvre and Moriones 2015). Here, we report the genome sequence of a new CHIV, also derived from spin column-associated DNA (Xu et al. 2013), and examine the evolutionary implications of this finding.
Materials and Methods
Sample Preparation and Illumina High-Throughput Sequencing
A total of 92 sera samples from patients with non-A–E hepatitis, who were admitted to the Institute of Infectious Disease of Southwest Hospital, Third Military Medical University, China, between 1999 and 2007, were obtained and processed as previously described (Xu et al. 2013). Briefly, ten pools were made of sera from the 92 patients. After sterilization using Ultrafree-MC HV 0.45-µm filters (Millipore), the samples were digested with DNase and RNase to eliminate host nucleotide contamination, and the remaining nucleic acids were extracted using carrier RNA [synthetic poly(A)]. cDNA was synthesized from extracted viral nucleic acids and purified using the QIAquick PCR purification kit PCR (polymerase chain reaction purification kit (Qiagen). Samples were sheared by using Covaris S2 sonicator (Covaris, Woburn, MA) and the sheared DNAs were end-blunted using End-It DNA End Repair Kit (Epicentre) following manufacturer’s instruction. A 3′-end A-tailing was performed. Following ligation of paired-end (PE) adaptors (Illumina) to the repaired ends, the viral DNA was amplified using the PE PCR primers 1.0 and 2.0 (Illumina) for 17 cycles and the resulting products were resolved by agarose gel electrophoresis and fragments that ranged around 200–500 bp in length were excised and purified. The purified DNA was used directly for cluster generation and sequencing analysis using Illumina Hiseq 2000 Genome Analyzer following manufacturer’s protocol.
Overlapping PCR and Inverse PCR
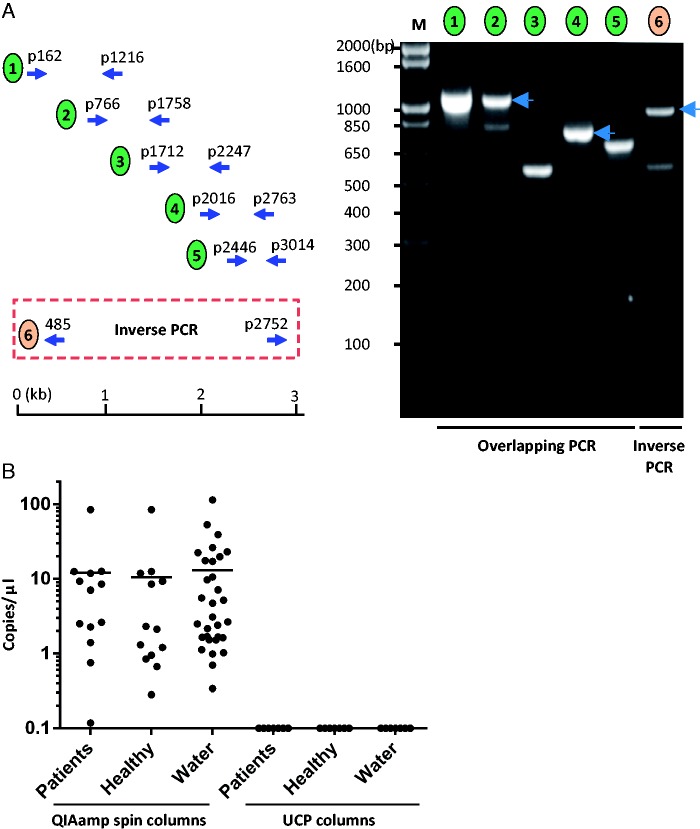
In order to verify the sequence of the viral genome assembled from the Illumina sequencing data, six sets of overlapping primer pairs were designed to amplify the viral fragments. To detect the circularized viral DNA, inverse PCR with a primer pair that oriented outwardly with respect to each other was used for amplification. PCR products were visualized on an agarose gel and all PCR products were Sanger sequenced. The complete CHIV14 genome sequence was deposited at GenBank under the accession number KM105952.
Evaluating Spin Columns for the Presence of CHIV14 by Quantitative Real-Time PCR
Nucleic acids were extracted from human serum samples or mock-extracted with water using a variety of spin columns (table 1), including QIAamp Viral RNA Mini Kit (Qiagen, kit catalog number 52906, lot number 436166748, and spin column lot number 139298432) and QIAamp ultraclean production (UCP) mini spin columns (Qiagen, reagents: catalog number 50112, lot number 142355460; spin columns: lot number 145033759). Nucleic acid extractions were performed following the manufacturers’ instructions or, in the case of QIAamp UCP, following a user-developed protocol (UDP). Briefly, in the UDP, 5.6 µg carrier RNA (Qiagen), 25 µl proteinase K, and 12.5 µl of APR buffer were added to 200 µl of the starting material (sample). Then, 200 µl of APL2 were added and incubated at 60 °C for 15 min. After a brief centrifugation, 800 µl of APB1 were added to the lysate and applied to the QIAamp UCP mini spin column in two rounds of centrifugation. The manufacturer’s protocol was followed thereafter to complete the DNA purification except for the final elution step where 60 µl of the AVE buffer were applied. In the case of QIAamp Viral RNA Mini Kit, the nucleic acids were also eluted with 60 μl of AVE buffer. As a negative control (mock sample), water (Ultra-pure, Quality Biological, Inc., Catalog number 351-029-131, lot number 719790) was used as a starting material for DNA extraction with QIAamp Viral RNA Mini Kit as well as other kits listed in table 1 using the protocols described above. Five microliters of the resulting DNA were used for analysis with the viral rep primers and rep probe (forward primer 5′-GTTGGCGAGTTATGGGTAAG-3′, reverse primer 5′-TGTACCAGAGGCAGTAACAG-3′, probe 5′FAM-CGAACAGGTACCAGGCTTTATTATGC-3′IABkFQ) purchased from Integrated DNA Technologies (Coralville, IA). All reactions were performed using the Chromo4 real-time detector (Bio-Rad). The reaction started with activation of the polymerase (PerfeCTa multiplex qPCR [quantitative real-time PCR] SuperMix, Quanta Biosciences) at 95 °C for 3 min, followed by 45 cycles of 15 s at 94 °C and 1 min at 60 °C. The quantitation of the amplicons was performed by interpolation with the standard curve to the synthesized rep gene with serial dilutions.
Table 1.
Kits and Spin Columns Used for Mock Extractions with Water and Water Elutions
| Kita | Spin Column | Catalog No. | Kit Lot | Column Lot | CHIV14 Presence |
|---|---|---|---|---|---|
| QIAamp Viral RNA Mini | QIAamp mini spin | 52906 | 436166748 | 139298432 | + |
| QIAamp Viral RNA Mini | QIAamp mini spin | 52904 | 140020818 | 145048587 | − |
| UCP PurePathogen Blood Kit | QIAamp UCP mini spin | 50112 | 142355460 | 145033759 | − |
| RNeasy Mini Kit (50) | RNeasy mini spin | 74104 | 142359481 | 142356163 | − |
| QIAamp MinElute Virus Spin Kit | QIAamp minElute | 57704 | 145020820 | 145018056 | − |
| QIAamp UltraSens Viral kit | QIAamp mini spin | 53704 | 42151888 | 127131210 | − |
| RNeasy Plus Mini Kit | RNeasy mini spin | 74134 | 145019932 | 14503787 | − |
| DNeasy Blood & Tissue Kit | DNeasy mini spin | 69506 | 427107496 | 133215292 | − |
| PureLink Viral RNA/DNA Mini Kit (Invitrogen) | Viral spin columns | 12280-050 | 1392776 | 1361245 | − |
aUnless stated otherwise, the kits were from Qiagen.
Computational Sequence Analysis
Domain recognition in the CHIV14 RC-Rep protein was performed using HHpred (Soding 2005) against the PFAM database. Structural modeling was performed using Modeller v9.9 (Marti-Renom et al. 2000), as described previously (Roux et al. 2013). X-ray structures of tomato bushy stunt virus (TBSV; PDB ID: 2TBV), melon necrotic spot virus (MNSV; PDB ID: 2ZAH), carnation mottle virus (CMV; PDB ID: 1OPO), and turnip crinkle virus (TCV; PDB ID: 3ZXA) were used as templates. Sequence of CHIV14 CP was aligned with the corresponding sequences of TBSV, MNSV, CMV, and TCV, and the resultant alignment was used to build a three-dimensional model of the putative CP of CHIV14. The initial model was optimized through multiple rounds of loop refinement with MODELLER. The stereochemical quality of the model was then assessed with ProSA-web (Wiederstein and Sippl 2007). ProSA-web quality (Z) score for the CHIV14 model was calculated to be −5.83, which is similar to the Z-scores determined for the template structures (TBSV, −5.18; MNSV, −6.26; CMV, −6.06; TCV, −3.39). The percent sequence identity between the CHIV CPs was mapped onto the structural model of the CHIV14 CP.
Sequences of the previously described CHIVs (Roux et al. 2013) were retrieved as GenBank-formatted files from Dryad Digital Repository, http://dx.doi.org/10.5061/dryad.19m2k (last accessed March 4, 2015). For phylogenetic analysis, protein sequences were aligned using PROMALS3D (Pei et al. 2008) and columns with low information content were removed from the alignment (alignments are available from the authors upon request). All alignments generated in the course of this study are available from the authors upon request. Maximum-likelihood phylogenetic analysis was carried out using PhyML 3.1 (Guindon et al. 2010), with the Jones–Taylor–Thornton model of amino acid substitutions, including a gamma law with four substitution rate categories.
Results and Discussion
CHIV14 Discovered by Deep Sequencing Is Traced Back to Spin-Column Contamination
De novo genome assembly and protein similarity search, which led to the discovery of a putative circular, ssDNA virus (CHIV14, 3141-nt) from deep sequencing data derived from ten serum pools of 92 patients with seronegative hepatitis, have been performed as described previously (Xu et al. 2013). The circular structure of the complete virus genome was confirmed by overlapping PCR and inverse PCR (fig. 1A). The origin of the virus was eventually traced to contaminated silica-binding spin columns used for nucleic acid extraction. Definitive confirmation of the origin of CHIV14 was obtained by in-depth analyses of water that was passed through contaminated spin columns (fig. 1B): The test for the presence of CHIV14 by PCR was positive for the non-A–E hepatitis sera, healthy sera control, and mock (water) extractions obtained using QIAamp Viral RNA Mini kit (Qiagen), but was negative for samples extracted using UCP spin columns (Qiagen). In addition, to support our assertion that the DNA originated from the QiaAmp mini spin columns, we have evaluated a variety of spin columns from different Qiagen and one Invitrogen kits (table 1). Water as well as mock extractions prepared following the corresponding manufacturers’ instructions were passed through the columns, collected, and evaluated by qPCR. CHIV14 was consistently found in only one kit, the QIAamp RNA mini kit described above. The elution buffers from each kit and water used for the elutions all tested negative for the presence of CHIV14 DNA. Based on the above results, we conclude that CHIV14 genome is a contamination specific to QIAamp mini spin columns.
Fig. 1.—
Confirmation of contaminated columns as origin of CHIV14 DNA by qPCR. (A) Verification of CHIV14 genome assembled from the Illumina deep-sequencing data by overlapping PCR and inverted PCR. Five sets of overlapping primer pairs and one set of inverted primer pair were designed and used to amplify overlapping DNA fragments. (Left) Schematic diagram of the positions of primer pairs for the overlapping PCR and inverted PCR. (Right) Amplification overlapping viral DNA fragments. The numbers above indicate the primer pair used for the PCR as illustrated on the left. The numbers on the left indicate the molecular weight in base pairs. (B) Scatterplot showing copy number of CHIV14 per microliter of DNA extraction. DNA from patients (n = 13), healthy controls (n = 13), and water (n = 31) was extracted using QIAamp mini spin columns (QIAamp Viral RNA Mini kit; Qiagen). In parallel, seven DNA extractions for each specimen type (patients, healthy individuals, and water) were performed using the UCP columns. Each dot represents one specimen. Bars show the average copy numbers of the viral genome.
All previously described CHIV genomes were recovered from aquatic environments and it was suggested that these viruses might infect unicellular algae (Roux et al. 2013). The discovery of CHIV14 in spin columns further supports this prediction and suggests that CHIV14 is associated with algae that constitute the silica matrix used in the spin columns, as previously concluded for NIH-CQV (Naccache et al. 2013), or introduced during the extensive water washing of the spin columns during manufacturing. Ambiguity regarding the actual host notwithstanding, analysis of the CHIV14 genome provided important insights into virus evolution, as described below.
CHIV14 Is a Chimera of Genes from RNA and DNA Viruses
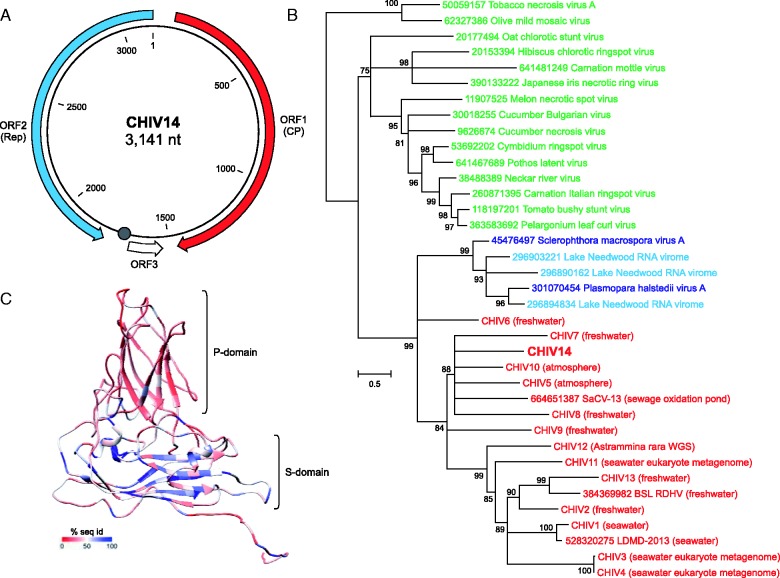
The CHIV14 genome contains three predicted open reading frames (ORF) larger than 45 codons (fig. 2A). When the sequence of the ORF1 product (445 amino acids) was used as a query in BLASTp search against National Center for Biotechnology Information (NCBI) nonredundant protein sequence database, the top hit was to the uncharacterized Sewage-associated circular virus-13 (SaCV-13; E = 1e-49), whereas the second best hit (E = 5e-40) was to the CP of the unclassified oomycete-infecting RNA virus Plasmopara halstedii virus A (SmV-A) (Heller-Dohmen et al. 2011). Further hits, with somewhat lower expectation values, were to the CP of BSL RDHV and to numerous plant-infecting RNA viruses of the family Tombusviridae. Thus, CHIV14 ORF1 encodes a tombusvirus-like CP, as also has been found for all BSL RDHV-like CHIVs (Diemer and Stedman 2012; Roux et al. 2013). We next compared the CHIV14 CP with the corresponding proteins encoded by the previously reported CHIVs which are available through the Dryad Digital Repository (Roux et al. 2013), and we found that CP of CHIV14 was most closely related to that of CHIV10 (E = 8e-65; fig. 2B). The analysis also revealed that the history of the CHIV group included a single event of CP gene transfer from an RNA virus. Consistent with previous findings (Roux et al. 2013), CHIV CPs formed a sister group to the clade consisting of SmV-A/PhV-A CPs and sequences recovered from the Lake Needwood RNA virome (Djikeng et al. 2009) (fig. 2B).
Fig. 2.—
Characterization of CHIV14. (A) Genome map of CHIV14. Predicted protein-coding genes are indicated with arrows, indicating the direction of transcription. A circle indicates the position of a potential origin of replication containing the nonanucleotide motif (AAGTATTAC) which is identical to the one found in BSL RDHV genome. (B) Maximum-likelihood phylogenetic analysis of the tombusvirus-like CPs. CHIVs are highlighted in red, tombusviruses in green and unclassified ssRNA viruses are either in blue when isolated, or in cyan when assembled from the Lake Needwood RNA virome. Tobacco necrosis virus A and Olive mild mosaic virus, both members of the genus Necrovirus within Tombusviridae, have CPs lacking the projection (P) domain and were used as an outgroup. Numbers at the branch points represent SH-like local support values. Nodes with support values less than 75% were collapsed. NCBI GI numbers are indicated for all reference sequences. The respective origins of the viromes from which the CHIV genomes were assembled are indicated next to the CHIV names. (C) Structural model of the CHIV14 CP. The P and shell (S) domains are indicated. The coloring represents sequence conservation among CHIV CPs. The color key is provided at the bottom of the panel.
Tombusvirus CPs contain two jelly-roll (antiparallel eight-stranded β-barrel) domains (fig. 2C). The shell (S) domain participates in the formation of the icosahedral capsid, whereas the projection (P) domain faces away from the capsid surface and might be involved in virus–host interactions. Multiple sequence alignment of CHIV CPs showed nonhomogeneous distribution of conservation, as has been previously observed with a smaller data set of CHIV proteins (Roux et al. 2013). To examine the potential functional implications of this observation, we constructed a three-dimensional model of the CHIV14 CP (see Materials and Methods) and mapped onto it the conservation/divergence pattern of the CHIV CPs obtained from the multiple sequence alignment. The sequence conservation is markedly higher in the S-domain compared with the P-domain (fig. 2C), consistent with the distinct functional roles of the two domains. Specifically, these observations indicate that the P-domain substantially diverged in the CHIV group following the CP gene acquisition from an RNA virus, consistent with its predicted role in virus–host interactions.
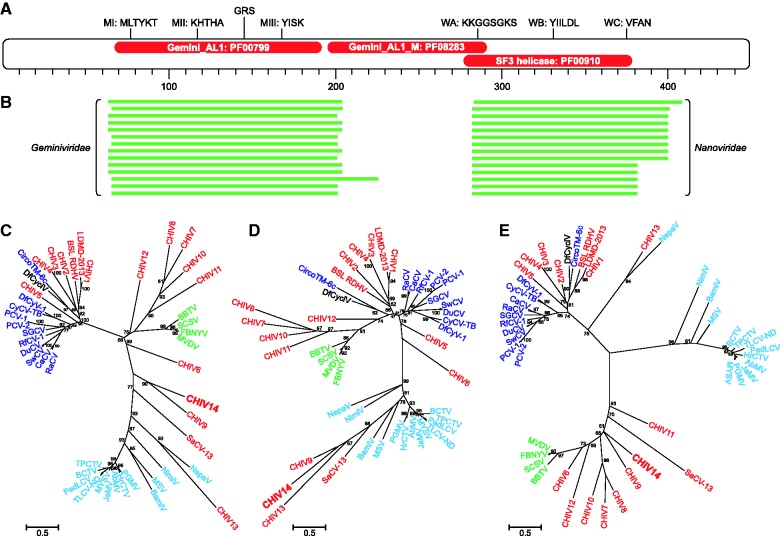
ORF2 is located on the complementary strand of the CHIV genomes and encodes a putative RC-Rep (449 amino acids), the most common replication protein in ssDNA viruses (Krupovic 2013). An HHpred analysis revealed three domains in the protein: The N-terminal Gemini_AL1 endonuclease domain (PF00799), the central geminivirus-specific Gemini_AL1_M domain (PF08283), and the C-terminal superfamily 3 (SF3) helicase domain (PF00910) (fig. 3A). The fusion between the rolling-circle endonuclease domain and the SF3 helicase domain is a signature of eukaryotic ssDNA viruses (Koonin 1993; Rosario et al. 2012; Krupovic 2013) and is also found in some bacterial and eukaryotic plasmids (Krupovic et al. 2009; Dayaram, Goldstien, et al. 2013; Krupovic 2013). All three diagnostic motifs (MI–III) of RC-Rep proteins (Ilyina and Koonin 1992) were identified in the N-terminal domain of the CHIV14 RC-Rep (fig. 3A). In addition, the N-terminal domain contains the geminivirus-specific motif, GRS, which is essential for geminivirus genome replication (Nash et al. 2011). The SF3 helicase domain is characterized by distinct versions of the three signature motifs of P-loop NTPases (A–C), all of which are conserved in the CHIV14 RC-Rep (fig. 3A).
Fig. 3.—
Analysis of the chimeric RC-Rep protein of CHIV14. (A) Domain organization of the CHIV14 RC-Rep. Red ellipses indicate the positions of the three identified domains: The N-terminal endonuclease domain Gemini_AL1 (PF00799, residues 65–191), central domain Gemini_AL1_M (PF08283; residues 196–291), and the C-terminal SF3 helicase domain (PF00910; residues 278–390). The diagnostic motifs of the RC-Rep proteins (MI-III) as well as Walker A, B, and C (WA–C) motifs of the SF3 helicase domain are shown at the top. The position of the geminivirus RC-Rep-specific motif GRS (geminivirus Rep sequence) defined by Nash et al. (2011) is also indicated. (B) Distinct sets of best BLASTp hits for two different regions of the CHIV14 RC-Rep. The N-terminal and C-terminal domains were most similar to the corresponding regions of RC-Reps from geminiviruses and nanoviruses, respectively. Maximum-likelihood phylogenetic analysis of the full-length RC-Rep protein (C), endonuclease domain (D), and SF3 helicase domain (E) of CHIV14. CHIVs are highlighted in red, whereas members of the families Circoviridae, Geminiviridae, and Nanoviridae are shaded blue, cyan, and green, respectively. Numbers at the branch points represent SH-like local support values. Abbreviations and NCBI GI: RaCV, Raven circovirus (115334608); CaCV, Canary circovirus (18875310); SwCV, Swan circovirus (156079716); DuCV, Duck circovirus (71658852); RfCV, Rhinolophus ferrumequinum circovirus 1 (389568560); SGCV, Silurus glanis circovirus (365269059); PCV-1, Porcine circovirus-1 (94451274); PCV-2, Porcine circovirus-2 (404553515); CyCV-TB, Cyclovirus bat/USA/2009 (318069480); DfCyV-1, Dragonfly cyclovirus 1 (324309814); DfCyclV, Dragonfly cyclicusvirus (406870761); CircoTM-6c, Circoviridae TM-6c (297598949); LDMD-2013, Circo-2 LDMD-2013 (528320274); SaCV-13, Sewage-associated circular virus-13 (664651387); BBTV, Banana bunchy top virus (81993219); SCSV, Subterranean clover stunt virus (82005379); FBNYV, Faba bean necrotic yellows virus (20143454); MVDV, Milk vetch dwarf virus (82005916); NepaV, Nepavirus (403044759); NimiV, Niminivirus (404352299); MSV, Maize streak virus (14794722); BamiV, Baminivirus (403044751); PGMV, Pepper golden mosaic virus (22128601); HrCTV, Horseradish curly top virus (1255063); AbMV, Abutilon mosaic virus (39980674); JaMV, Jatropha mosaic virus (612184447); MYSV, Macroptilium yellow spot virus (417355462); TLCV-ND, Tomato leaf curl New Delhi virus (562890733); PedLCV, Pedilanthus leaf curl virus (224581833); BCTV, Beet curly top virus (46254388); TPCTV, Tomato pseudo-curly top virus (20564197).
ORF3, predicted to encode a short protein of 48 amino acids, does not share sequence similarity with proteins in public databases, nor does it contain a homolog in available CHIV genomes. The sequence analysis described above indicates that CHIV14 belongs to the expanding group of putative BSL RDHV-like CHIVs, which evolved as the result of recombination between virus ancestors with RNA and DNA genomes. However, like in the case of all other CHIVs, the viral nature of CHIV14 has to be confirmed experimentally.
CHIV14 RC-Rep Is a Chimera of Domains from Different Groups of ssDNA Viruses
Recent analysis of RC-Rep diversity in CHIVs has revealed an unexpectedly frequent RC-Rep gene transfer in the CHIV lineage (Roux et al. 2013). In contrast to the monophyly of the CHIV CPs, the RC-Reps of these viruses segregated into three groups with respective closest relatives in different families of eukaryotic ssDNA viruses, namely Circoviridae, Nanoviridae, and Geminiviridae. The BLASTp analysis of the CHIV14 RC-Rep has shown that the N-terminal domain (residues 1-280) is most closely related to the corresponding domain of the proteins encoded by geminiviruses (fig. 3B), consistent with the HHpred analysis (fig. 3A). In contrast, when the SF3 helicase domain (residues 280-499) was used as a seed, significant hits to geminiviruses were not retrieved. Instead, the CHIV14 SF3 domain showed highly significant similarity to the corresponding domains of the RC-Rep proteins of nanoviruses and to some uncharacterized environmental viruses. This observation suggests that the RC-Rep protein of CHIV14 is likely a chimera, in which the N- and C-terminal domains have different evolutionary histories.
To further investigate the provenance of CHIV14 RC-Rep, we performed maximum-likelihood phylogenetic analysis of the full-length RC-Reps from different groups of ssDNA viruses and from the corresponding endonuclease and helicase domains separately (fig. 3C–E). In all cases, geminiviruses, nanoviruses, and circoviruses formed distinct clades, indicating that the two domains in these viruses have coevolved and there was no interfamilial exchange of either the full-length RC-Rep genes or gene fragments encoding separate domains. Similarly, nuclease and helicase domains of CHIV1–CHIV5, LDMD-2013, and BSL RDHV consistently grouped with circoviruses; those of CHIV7, CHIV8, CHIV10, CHIV11, and CHIV12 grouped with nanoviruses; and CHIV13 branched with geminiviruses (fig. 3C–E). In contrast, the endonuclease domains of CHIV14, CHIV9, and SaCV-13 formed a deeply branching clade with geminiviruses (fig. 3D), and their helicase domains grouped with those of nanoviruses (fig. 3E). Consequently, RC-Reps of the three CHIVs are chimeras that consist of endonuclease and helicase domains that do not share a common evolutionary history.
Conclusions
The importance of horizontal gene transfer in virus evolution cannot be overestimated. It has been extensively investigated in the case of viruses with dsDNA genomes, where gene fragments, individual genes, and even multigene operons are known to have been exchanged between different viruses, plasmids, and their hosts (Krupovic and Bamford 2007; Filée et al. 2008; Fischer et al. 2010; Krupovic et al. 2011; Yutin and Koonin 2012; Filée 2013; Forterre and Prangishvili 2013; Koonin and Dolja 2014; Yutin et al. 2014; Krupovic and Koonin 2015). However, in the case of viruses with small genomes, such as ssDNA viruses, horizontal gene transfer had not been characterized in comparable detail although given the small number of genes, it is expected to have a more profound effect on the genetic “identity” (and taxonomic affiliation) of the small viruses. Here, we show that BSL RDHV-like ssDNA viruses are chimeric at two levels. Not only do they combine genes from viruses with different types of genomic nucleic acids (RNA and DNA) but also the genes themselves in some of these hybrid viruses are chimeric, with different functional domains donated by viruses from different families. Although genomic recombination is frequent in many groups of eukaryotic ssDNA viruses (Martin et al. 2011; Lefeuvre and Moriones 2015), it is noteworthy that horizontal gene transfer and domain shuffling described here so far have not been observed in this class of viruses. Recombinant viruses are likely to suffer from disruption of favorable coevolved genetic interactions (Monjane et al. 2014) and are usually unfit to compete with the parental viruses, which ultimately leads to their elimination from the population. However, the example of CHIVs indicates that occasionally such recombinant viruses do succeed, resulting in the emergence of multiple, novel groups of viruses that conceivably could occupy new ecological niches.
Acknowledgment
This work was supported in part by the Intramural Research Program of the National Institute of Health, National Heart, Lung, and Blood Institute, and National Library of Medicine.
Literature Cited
- Dayaram A, Goldstien S, et al. Novel ssDNA virus recovered from estuarine Mollusc (Amphibola crenata) whose replication associated protein (Rep) shares similarities with Rep-like sequences of bacterial origin. J Gen Virol. 2013;94:1104–1110. doi: 10.1099/vir.0.050088-0. [DOI] [PubMed] [Google Scholar]
- Dayaram A, Potter KA, et al. High global diversity of cycloviruses amongst dragonflies. J Gen Virol. 2013;94:1827–1840. doi: 10.1099/vir.0.052654-0. [DOI] [PubMed] [Google Scholar]
- De Bruyn A, et al. East African cassava mosaic-like viruses from Africa to Indian ocean islands: molecular diversity, evolutionary history and geographical dissemination of a bipartite begomovirus. BMC Evol Biol. 2012;12:228. doi: 10.1186/1471-2148-12-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwart E, Li L. Rapidly expanding genetic diversity and host range of the Circoviridae viral family and other Rep encoding small circular ssDNA genomes. Virus Res. 2012;164:114–121. doi: 10.1016/j.virusres.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemer GS, Stedman KM. A novel virus genome discovered in an extreme environment suggests recombination between unrelated groups of RNA and DNA viruses. Biol Direct. 2012;7:13. doi: 10.1186/1745-6150-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One. 2009;4:e7264. doi: 10.1371/journal.pone.0007264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Holmes EC. Phylogenetic evidence for rapid rates of molecular evolution in the single-stranded DNA begomovirus tomato yellow leaf curl virus. J Virol. 2008;82:957–965. doi: 10.1128/JVI.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Holmes EC. Validation of high rates of nucleotide substitution in geminiviruses: phylogenetic evidence from East African cassava mosaic viruses. J Gen Virol. 2009;90:1539–1547. doi: 10.1099/vir.0.009266-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Filée J. Route of NCLDV evolution: the genomic accordion. Curr Opin Virol. 2013;3:595–599. doi: 10.1016/j.coviro.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Filée J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by nucleocytoplasmic large DNA viruses. BMC Evol Biol. 2008;8:320. doi: 10.1186/1471-2148-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth C, Charleston MA, Duffy S, Shapiro B, Holmes EC. Insights into the evolutionary history of an emerging livestock pathogen: porcine circovirus 2. J Virol. 2009;83:12813–12821. doi: 10.1128/JVI.01719-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci U S A. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre P, Prangishvili D. The major role of viruses in cellular evolution: facts and hypotheses. Curr Opin Virol. 2013;3:558–565. doi: 10.1016/j.coviro.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Grigoras I, et al. High variability and rapid evolution of a nanovirus. J Virol. 2010;84:9105–9117. doi: 10.1128/JVI.00607-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Harkins GW, et al. Dating the origins of the maize-adapted strain of maize streak virus, MSV-A. J Gen Virol. 2009;90:3066–3074. doi: 10.1099/vir.0.015537-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins GW, Martin DP, Christoffels A, Varsani A. Towards inferring the global movement of beak and feather disease virus. Virology. 2014 doi: 10.1016/j.virol.2013.11.033. 450-451:24-33. [DOI] [PubMed] [Google Scholar]
- Heller-Dohmen M, Gopfert JC, Pfannstiel J, Spring O. The nucleotide sequence and genome organization of Plasmopara halstedii virus. Virol J. 2011;8:123. doi: 10.1186/1743-422X-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Dolja VV. Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol Mol Biol Rev. 2014;78:278–303. doi: 10.1128/MMBR.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraberger S, et al. Evidence that dicot-infecting mastreviruses are particularly prone to inter-species recombination and have likely been circulating in Australia for longer than in Africa and the Middle East. Virology. 2013;444:282–291. doi: 10.1016/j.virol.2013.06.024. [DOI] [PubMed] [Google Scholar]
- Kraberger S, et al. Characterisation of a diverse range of circular Rep-encoding DNA viruses recovered from a sewage treatment oxidation pond. Infect Genet Evol. Forthcoming 2015;31:73–86. doi: 10.1016/j.meegid.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Krupovic M. Recombination between RNA viruses and plasmids might have played a central role in the origin and evolution of small DNA viruses. Bioessays. 2012;34:867–870. doi: 10.1002/bies.201200083. [DOI] [PubMed] [Google Scholar]
- Krupovic M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr Opin Virol. 2013;3:578–586. doi: 10.1016/j.coviro.2013.06.010. [DOI] [PubMed] [Google Scholar]
- Krupovic M, Bamford DH. Putative prophages related to lytic tailless marine dsDNA phage PM2 are widespread in the genomes of aquatic bacteria. BMC Genomics. 2007;8:236. doi: 10.1186/1471-2164-8-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Koonin EV. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci Rep. 2014;4:5347. doi: 10.1038/srep05347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Koonin EV. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat Rev Microbiol. Forthcoming 2015;13:105–115. doi: 10.1038/nrmicro3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Prangishvili D, Hendrix RW, Bamford DH. Genomics of bacterial and archaeal viruses: dynamics within the prokaryotic virosphere. Microbiol Mol Biol Rev. 2011;75:610–635. doi: 10.1128/MMBR.00011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupovic M, Ravantti JJ, Bamford DH. Geminiviruses: a tale of a plasmid becoming a virus. BMC Evol Biol. 2009;9:112. doi: 10.1186/1471-2148-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labonte JM, Suttle CA. Previously unknown and highly divergent ssDNA viruses populate the oceans. ISME J. 2013;7:2169–2177. doi: 10.1038/ismej.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P, Moriones E. Recombination as a motor of host switches and virus emergence: geminiviruses as case studies. Curr Opin Virol. 2015;10C:14–19. doi: 10.1016/j.coviro.2014.12.005. [DOI] [PubMed] [Google Scholar]
- Marti-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Martin DP, et al. Recombination in eukaryotic single stranded DNA viruses. Viruses. 2011;3:1699–1738. doi: 10.3390/v3091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel LD, Rosario K, Breitbart M, Paul JH. Comparative metagenomics: natural populations of induced prophages demonstrate highly unique, lower diversity viral sequences. Environ Microbiol. 2014;16:570–585. doi: 10.1111/1462-2920.12184. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, et al. Archaeal virus with exceptional virion architecture and the largest single-stranded DNA genome. Proc Natl Acad Sci U S A. 2012;109:13386–13391. doi: 10.1073/pnas.1203668109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monjane AL, et al. Extensive recombination-induced disruption of genetic interactions is highly deleterious but can be partially reversed by small numbers of secondary recombination events. J Virol. 2014;88:7843–7851. doi: 10.1128/JVI.00709-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naccache SN, et al. The perils of pathogen discovery: origin of a novel parvovirus-like hybrid genome traced to nucleic acid extraction spin columns. J Virol. 2013;87:11966–11977. doi: 10.1128/JVI.02323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash TE, et al. Functional analysis of a novel motif conserved across geminivirus Rep proteins. J Virol. 2011;85:1182–1192. doi: 10.1128/JVI.02143-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popgeorgiev N, Temmam S, Raoult D, Desnues C. Describing the silent human virome with an emphasis on giant viruses. Intervirology. 2013;56:395–412. doi: 10.1159/000354561. [DOI] [PubMed] [Google Scholar]
- Rosario K, Breitbart M. Exploring the viral world through metagenomics. Curr Opin Virol. 2011;1:289–297. doi: 10.1016/j.coviro.2011.06.004. [DOI] [PubMed] [Google Scholar]
- Rosario K, Duffy S, Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol. 2012;157:1851–1871. doi: 10.1007/s00705-012-1391-y. [DOI] [PubMed] [Google Scholar]
- Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- Roux S, et al. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat Commun. 2013;4:2700. doi: 10.1038/ncomms3700. [DOI] [PubMed] [Google Scholar]
- Roux S, Krupovic M, Poulet A, Debroas D, Enault F. Evolution and diversity of the Microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One. 2012;7:e40418. doi: 10.1371/journal.pone.0040418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuan R, Nebot MR, Chirico N, Mansky LM, Belshaw R. Viral mutation rates. J Virol. 2010;84:9733–9748. doi: 10.1128/JVI.00694-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- Whon TW, et al. Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J Virol. 2012;86:8221–8231. doi: 10.1128/JVI.00293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, et al. Hybrid DNA virus in Chinese patients with seronegative hepatitis discovered by deep sequencing. Proc Natl Acad Sci U S A. 2013;110:10264–10269. doi: 10.1073/pnas.1303744110. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Yoshida M, Takaki Y, Eitoku M, Nunoura T, Takai K. Metagenomic analysis of viral communities in (hado)pelagic sediments. PLoS One. 2013;8:e57271. doi: 10.1371/journal.pone.0057271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Koonin EV. Hidden evolutionary complexity of Nucleo-Cytoplasmic Large DNA viruses of eukaryotes. Virol J. 2012;9:161. doi: 10.1186/1743-422X-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf YI, Koonin EV. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology. 2014;466–467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawar-Reza P, et al. Diverse small circular single-stranded DNA viruses identified in a freshwater pond on the McMurdo Ice Shelf (Antarctica) Infect Genet Evol. 2014;26:132–138. doi: 10.1016/j.meegid.2014.05.018. [DOI] [PubMed] [Google Scholar]