Abstract
The ancestors to the Australian marsupials entered Australia around 60 (54–72) Ma from Antarctica, and radiated into the four living orders Peramelemorphia, Dasyuromorphia, Diprotodontia, and Notoryctemorphia. The relationship between the four Australian marsupial orders has been a long-standing question, because different phylogenetic studies have not been able to consistently reconstruct the same topology. Initial in silico analysis of the Tasmanian devil genome and experimental screening in the seven marsupial orders revealed 20 informative transposable element insertions for resolving the inter- and intraordinal relationships of Australian and South American orders. However, the retrotransposon insertions support three conflicting topologies regarding Peramelemorphia, Dasyuromorphia, and Notoryctemorphia, indicating that the split between the three orders may be best understood as a network. This finding is supported by a phylogenetic reanalysis of nuclear gene sequences, using a consensus network approach that allows depicting hidden phylogenetic conflict, otherwise lost when forcing the data into a bifurcating tree. The consensus network analysis agrees with the transposable element analysis in that all possible topologies regarding Peramelemorphia, Dasyuromorphia, and Notoryctemorphia in a rooted four-taxon topology are equally well supported. In addition, retrotransposon insertion data support the South American order Didelphimorphia being the sistergroup to all other living marsupial orders. The four Australian orders originated within 3 Myr at the Cretaceous–Paleogene boundary. The rapid divergences left conflicting phylogenetic information in the genome possibly generated by incomplete lineage sorting or introgressive hybridization, leaving the relationship among Australian marsupial orders unresolvable as a bifurcating process millions of years later.
Keywords: Notoryctes, Australian marsupials, marsupial phylogeny, retrotransposons, SINE, network analysis
Introduction
Considerable effort has been invested into resolving the relationships among the more than 300 living marsupial species (Szalay 1982; Kirsch et al. 1997; Nowak 2005; Meredith et al. 2008, 2011; Nilsson et al. 2010; Mitchell et al. 2014). The most dramatic revelation was when a small South American marsupial was taxonomically moved from within the order Didelphimorphia to its own monotypic order, Dromiciopsa (later: Microbiotheria; Szalay 1982). A single morphological character suggested that Microbiotheria might be closely related to Australian marsupials (Szalay 1982). Molecular analyses have since identified its position as sistergroup to all Australian marsupials (Nilsson et al. 2010; Meredith et al. 2011). However, the relationship among the four Australian marsupial orders (Peramelemorphia, Dasyuromorphia, Diprotodontia, and Notoryctemorphia) is still highly controversial and has yet to be settled (Nilsson et al. 2010; Meredith et al. 2011; Mitchell et al. 2014). In particular the phylogenetic position of Notoryctemorphia (marsupial moles) is debated because morphological and molecular approaches result in conflicting topologies or nonsignificant branch support (supplementary table S1, Supplementary Material online). Most molecular studies favor placing Peramelemorphia, Dasyuromorphia, and Notoryctemorphia in conflicting constellations to the exclusion of Diprotodontia. Notoryctemorphia consist of one living genus, Notoryctes, which contains two species, the northern (Notoryctes caurinus) and the southern marsupial mole (Notoryctes typhlops). Marsupial moles resemble the placental moles (e.g., golden moles, Chrysochloridae) due to their convergent adaptations to the subterrestrial, fossorial lifestyle (Kirsch et al. 1997; Nowak 2005; Archer et al. 2011). The phenotypic adaptation has led to highly derived characters that complicate morphological phylogenetic analyses of the position of Notoryctemorphia (Horovitz and Sánchez-Villagra 2003; Asher et al. 2004).
The analysis of presence and absence of retrotransposon insertions in mammalian genomes is a powerful tool to resolve conflicting phylogenetic hypotheses (Shedlock and Okada 2000; Boore 2006) and to recover evolutionary processes that would remain hidden by nucleotide sequence analyses (Terai et al. 2003). The availability of whole-genome sequence data has revolutionized the use of retrotransposon insertion approaches for phylogenetic questions by increasing the number of investigable loci (Warren et al. 2008; Nishihara et al. 2009). Retrotransposons propagate from a few active “source elements” in the genome following the “copy-and-paste” principle using RNA-intermediates (Kramerov and Vassetzky 2011). Once new retrotransposon copies are inserted in the genome they are usually unable to propagate further (Shedlock and Okada 2000). With the knowledge that insertions that occurred in the common ancestor will be transferred to the offspring in the same orthologous position, the fossilized retrotransposon insertions in the mammalian genome are ideal to disentangle relationships among species (Shedlock and Okada 2000).
Retrotransposon insertion analyses are less affected by the stochastic processes that can confound sequence-based studies (Shedlock and Okada 2000; Ray et al. 2006). Nucleotide composition biases, varying evolutionary rates, and parallel/convergent substitutions from limited character states (G, A, T, C) cause uncertainties in sequence-based tree reconstruction (Boore 2006). Retrotransposons have the advantage of a clear knowledge of the ancestral state, which is absence (Ray et al. 2006). The probability of an independent retrotransposon insertion occurring at the same target sequence in two or more taxa is extremely low. Similarly, the likelihood of precise deletions of retrotransposons occurring is negligible (van de Lagemaat et al. 2005). Retrotransposon insertion analyses have resolved relationships within several marsupial and placental mammalian groups (Shimamura et al. 1997; Churakov et al. 2009; Nilsson et al. 2010). The availability of a dasyuromorphian marsupial genome (Tasmanian devil) made it possible to use retrotransposon insertion analysis for investigating the controversial phylogenetic relationships among Australian marsupial orders. An experimental screen of 158 in silico identified introns with suitable retrotransposon insertions revealed 20 phylogenetically informative insertions for resolving intra- and interordinal relationships in both South American and Australian marsupial orders (supplementary table S2, Supplementary Material online).
The South American Didelphimorphia is the Sistergroup to all Marsupials
The earliest fossil crown group marsupials bear morphological similarities to the South American order Didelphimorphia (opossums), which has been regarded as the sistergroup to all other living marsupial orders (Horovitz and Sánchez-Villagra 2003; Horovitz et al. 2009). However, some sequence-based phylogenetic studies place the South American order Paucituberculata (shrew opossums) as the sistergroup to living marsupials (Meredith et al. 2011; Mitchell et al. 2014). The apparent conflict between sequence data and the fossil record, that favor two different marsupial orders at the basal position of all living marsupials, has so far not been further investigated (Meredith et al. 2011; Mitchell et al. 2014).
To obtain statistically significant support for a branch, three retrotransposon insertions are required (Waddell et al. 2001). We identified three retrotransposon insertions (MIR3, Mar1_MD, MIRc) present in all living marsupial orders supporting the monophyly of marsupials (fig. 1), but none for conflicting topologies, yielding a [3 0 0] pattern of support and a probability for this branch of P = 0.037 (Waddell et al. 2001).
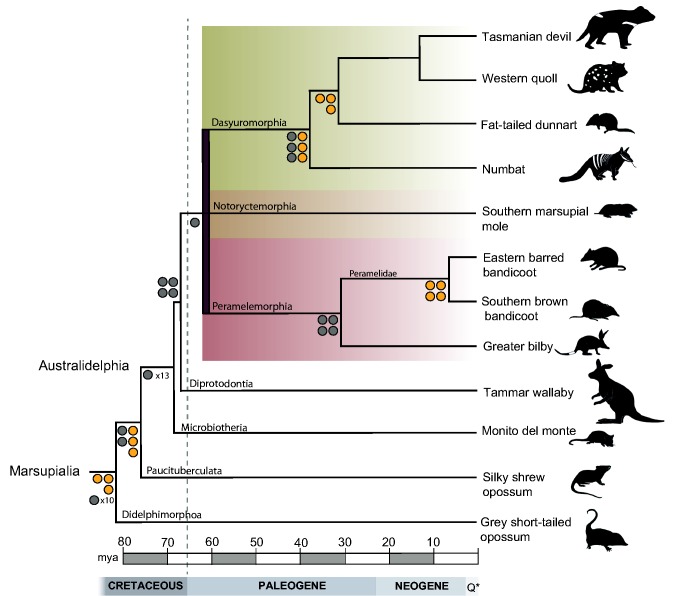
Fig. 1.—
Phylogenetic tree of the seven marsupial orders based on 16 of the 20 phylogenetically informative retrotransposon insertions and sequence data. Markers for different nodes in the marsupial tree are shown as orange circles. Branches showing retrotransposon support based on Nilsson et al. (2010) are indicated as gray circles. The tree includes representatives of all seven living marsupial orders, and has been scaled to divergence time (Mitchell et al. 2014). The phylogenetic tree is based on a Bayesian analysis of 32,253 nt from 28 nuclear gene fragments (supplementary fig. S1, Supplementary Material online). The branches leading to Peramelemorphia, Dasyuromorphia, and Notoryctemorphia have been collapsed. Additional markers supporting the relationship between Peramelemorphia (red shaded area), Dasyuromorphia (green shaded area), and Notoryctemorphia (brown shaded area) are shown separately in figure 2. Australidelphia refers to the grouping of the South American Microbiotheria and the four Australian orders (Szalay 1982). Q*, quaternary period.
In the course of our screening we found three retrotranposons (WSINE1, WALLSI1, RTESINE2) favoring Didelphimorphia as sistergroup to all other living marsupials (fig. 1). The two SINE (Short Interspersed Element) types WALLSI1 and RTESINE2 do not exist in the didelphimorphian genome (Gentles et al. 2007), further strengthening the support for placing Didelphimorphia as the earliest divergence. Previous analysis of retrotransposon insertions identified two SINE insertions ([2 0 0] P = 0.111) supporting Didelphimorphia as sistergroup to all other living marsupials (Nilsson et al. 2010), resulting in a total of five independent retrotransposon insertions ([5 0 0] P = 0.001) without any conflicting markers. However, given the discrepancies between the retrotransposon insertions and previous sequence analyses it is still possible that analyses of future genome data, in particular that of Paucituberculata, might result in conflicting data.
Intraordinal Relationships among and between Australian Marsupials
Within Marsupialia, the order Dasyuromorphia, the carnivorous marsupials, have attracted considerable attention in morphological and molecular phylogenetic studies, because they represent the dominant terrestrial mammalian carnivores of Australia and New Guinea. The order is subdivided into three families (Archer 1984). They include the extinct Tasmanian wolf (Thylacinus), the endangered numbat, and the species-rich family Dasyuridae. The latter includes 69 species such as quolls, dibblers, dunnarts, and the Tasmanian devil (Nowak 2005). The earliest dasyuromorphian fossils from the middle Miocene (23.5–11 Ma) represent mainland Australian Thylacinus species (Wroe et al. 1999). The ant-eating numbat (Myrmecobiidae) is only known since the Holocene (since 10,000 years ago), and only few species belonging to Dasyuridae have been found at Riversleigh, an Oligocene/Miocene fossil site, 25–15 Ma old (Archer 1984; Long et al 2002). The three Dasyuridae subfamilies are known from Pliocene deposits, 5.3–2.6 Ma (Wroe 2003). Given the poor fossil record from Dasyuromorphia, phylogenetic conclusions, divergence times, and evolutionary correlations to environmental changes during the Miocene (23.0–5.3 Ma) depend largely on molecular data (Krajewski, Blacket, et al. 2000; Krajewski, Wroe, et al. 2000). Three novel retrotransposon insertions further strengthen the divergence between Dasyuridae and Myrmecobiidae ([3 0 0] P = 0.037) and the monophyly of Dasyuromorphia ([3 0 0] P = 0.037; fig. 1).
The order Peramelemorphia consists of 23 species that are divided into two families, Peramelidae (bandicoots) and Thylacomyidae (bilbies); (Nowak 2005). Thylacomyidae contain just one extant species, the Greater Bilby (Macrotis lagotis), as the Lesser Bilby (Macrotis leucura) became extinct during the 20th century (Nowak 2005; Gurovich et al. 2014). Peramelemorphian fossils are known from fossil sites of Oligocene/Miocene age (e.g., Riversleigh), with the earliest crown-group fossils being Middle Miocene (Gurovich et al. 2014, Travouillon et al. 2014). Previous molecular phylogenetic analysis and divergence time estimation placed the age for the basal split within Peramelemorphia at 27 Ma (Westerman et al. 2012). We identified four retrotransposon insertions ([4 0 0] P = 0.012) that support the monophyly of Peramelinae.
The relationships within Peramelemorphia and Dasyuromorphia have been extensively investigated using nuclear genes, complete mitochondrial genomes as well as retrotransposons (Krajewski, Blacket, et al. 2000; Krajewski, Wroe, et al. 2000; Meredith et al. 2011; Westerman et al. 2012; Zemann et al. 2013; Gallus et al. 2015; Mitchell et al. 2014). The results of our retrotransposon insertion analysis are statistically significant and in agreement with previous studies.
Conflicting Signals for the Position of Notoryctemorphia among Australian Marsupial Orders
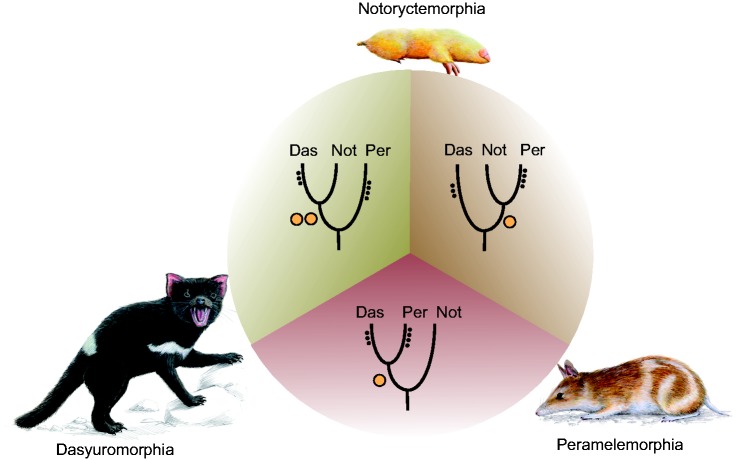
We identified four phylogenetically informative, but conflicting, retrotransposon insertions for the position of Notoryctemorphia, supporting three different topologies ([2 1 1] P = 0.284; fig. 2). Two WSINE1 insertions support a sistergroup relationship between Dasyuromorphia and Notoryctemorphia that is most often recovered in sequence-based studies (supplementary table S1, Supplementary Material online). However, one independent insertion supports a relationship between Notoryctemorphia and Peramelemorphia, previously suggested based on syndactyly, a morphological character possibly in common to both orders (Szalay 1982). Syndactyly, the joining of the third and fourth digit on the hind foot occurs in two unrelated Australian orders, Diprotodontia and Peramelemorphia, but its presence in Notoryctemorphia is debated (Weisbecker and Nilsson 2008). Finally, one retrotransposon marker was identified for the third possible topology in a rooted four-taxon tree. This marker supports a sistergroup relationship between Peramelemorphia and Dasyuromorphia (fig. 2), a relationship in accordance with nuclear sequence data (Meredith et al. 2011). A recent study combining a novel mitogenomic and a nuclear gene data set (Meredith et al. 2011) recovered the same topology, but with less support (Mitchell et al. 2014).
Fig. 2.—
The relationship between Dasyuromorphia, Peramelemorphia, and Notoryctemorphia. The retrotransposon screen revealed four informative retrotransposon markers that weakly support three different topologies of the three orders. Retrotransposon support for each topology is indicated by orange colored circles representing the identified number of retrotransposon markers. The small black circles represent markers for intraordinal relationships inside the respective orders (fig. 1). Das, Dasyuromorphia (Tasmanian devil); Not, Notoryctemorphia (marsupial mole); Per, Peramelemorphia (Eastern barred bandicoot/Southern brown bandicoot). Animal figures were provided by Jon Baldur Hlíðberg (www.fauna.is).
The strongest support in our analysis came from two retrotransposon insertions that agree with the topology from most sequence-based studies, showing Dasyuromorphia as sistergroup to Notoryctemorphia (supplementary table S1, Supplementary Material online). The pattern of retrotransposon insertions, however, indicates that there is conflict in the genome and mirrors the inconsistent results based on previous sequence data analyses (supplementary table S1, Supplementary Material online). The four Australian marsupial orders diverged in a rapid succession close to the Cretaceous–Paleogene (KPg) boundary around 67–64 Ma (Mitchell et al. 2014). During the relatively short time spanning the divergences, only a limited number of mutations could accumulate. In addition, new mutations occur continuously in the species genome, creating phylogenetic noise (Leaché et al. 2014). Incomplete lineage sorting (ILS), a phenomenon where allelic variation does not have time to sort during speciation can lead to problems during phylogenetic reconstruction (Hudson 1983; Tajima 1983; Pinho and Hey 2010). Another issue is that introgressive hybridization or gene flow might have taken place between the diverging marsupial populations (Hudson 1983; Tajima 1983; Pinho and Hey 2010; Phillips et al. 2013). It is difficult to assess if, and how much ILS and gene flow have contributed to the observed phylogenetic conflict (Pinho and Hey 2010). As neither genome data for Peramelemorphia nor Notoryctemorphia is currently available it is not possible to do an in silico whole-genome screen to find phylogenetically informative retrotransposon insertions. Thus, we caution that when complete genome data are available for Peramelemorphia or Notoryctemorphia allowing for a more exhaustive marker screen, it could reach other conclusions, as we cannot completely rule out the possibility of exact deletion or precise insertions of elements in our markers (van de Lagemaat et al. 2005).
Previous studies showed that some nodes in the placental mammalian tree have experienced ILS and possible ancestral gene flow and are impossible to resolve using retrotransposons or sequence-based phylogenetic analyses. Such patterns are observed when the radiation between species is within 2–4 Myr, a time frame in which lineage sorting of alleles may not be completed or hybridization between species is still possible (Hallström and Janke 2010). The deepest splits in the placental mammalian tree have been particularly problematic to resolve (Churakov et al. 2009; Nishihara et al. 2009; Hallström and Janke 2010). Retrotransposon insertion analyses have revealed conflicting signals for relationships among Cetartiodactyla, Carnivora, Perissodactyla, and Chiroptera (Nishihara et al. 2006; Hallström et al. 2011), which were not resolvable as a bifurcating tree by phylogenomic analysis (Hallström et al. 2011). It is highly unlikely that the observed signals of ILS in placental mammals are caused by erosion of the retrotransposons, as this would require erosion in all species and orders at the same rate. Or vice versa, meaning that all species within the placental mammalian orders acquired elements independently at the exact same position.
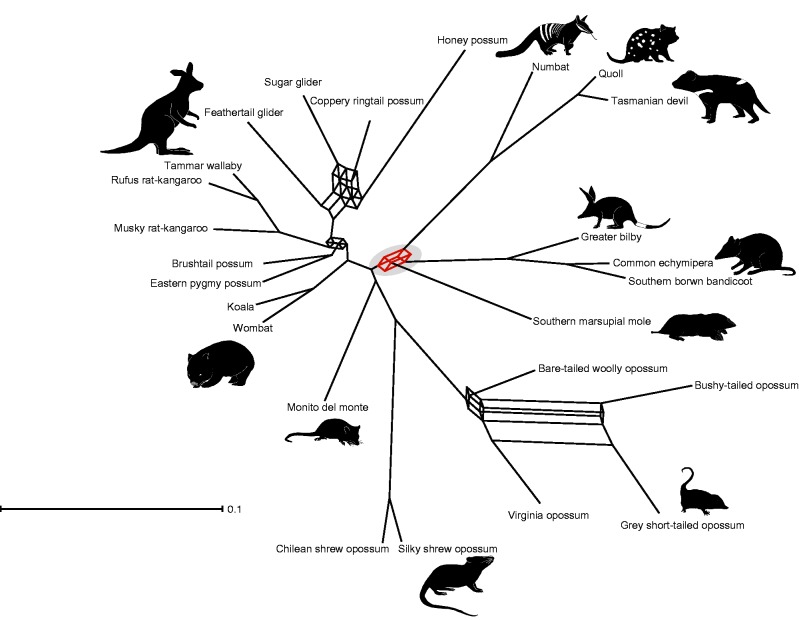
The results from our retrotransposon screen suggest that aside from a possibility of lack of data, it might be not possible to find a single sistergroup to Notoryctemorphia due to the short divergence times between the Australian marsupial orders. Thus we conclude, that some ordinal relationships might be better depicted as an evolutionary network rather than a bifurcating tree. Networks allow the depiction of conflicting signals from phylogenetic trees that are otherwise ignored when striving for a strict bifurcating species tree (Bapteste et al. 2013). Therefore, we reinvestigated a published data set of marsupial nuclear genes (Meredith et al. 2011) for conflicting signals (fig. 3). The reanalysis shows that there is a strong signal for grouping Dasyuromorphia, Peramelemorphia, and Notoryctemorphia to the exclusion of Diprotodontia, which has been suggested by sequence-based analyses as well as one retrotransposon marker (Nilsson et al. 2010). However, the consensus network analysis suggests that there is equally strong phylogenetic signal for each of the three possible topologies among Dasyuromorphia, Peramelemorphia, and Notoryctemorphia (fig. 3). High levels of conflicting signals in the sequence data are also found within two orders, the Australian Diprotodontia and the South American Didelphimorphia.
Fig. 3.—
Consensus network based on 20 individual Bayesian gene trees (> 600 nt) from 25 marsupial species representing all seven orders. The consensus network (threshold 0.07) shows the possible phylogenetic splits. Red lines and the gray shaded circle highlight the trifurcation between Dasyuromorphia, Notoryctemorphia, and Peramelemorphia.
The consensus-network analysis coupled with the findings from the retrotransposon insertions suggest that the evolution of the Australian orders was complex and cannot be depicted as a bifurcating tree. The root of the marsupial tree might resemble the basal splits in the placental mammalian phylogenetic tree, which seem to have undergone extensive ILS as suggested by the analysis of several complete genomes (Hallström and Janke 2008, 2010; Churakov et al. 2009; Nishihara et al. 2009). It is tempting to interpret the occurrence of syndactyly in different orders, as a consequence of the very rapid divergence. Syndactyly could possibly show that there was gene flow between the diverging populations as it is only found in two unrelated orders. In the absence of genome data, we identified retrotransposon markers that weakly support three different topologies for the relationship between Dasyuromorphia, Peramelemorphia, and Notoryctemorphia. The phylogenetic finding neatly summarizes the efforts of previous sequence-based studies that could not agree on the relationship of the Australian orders. The divergence between the four Australian orders took place in a window of a few million years at the KPg boundary (Mitchell et al. 2014) hindering the accumulation of sufficient phylogenetic information to yield a clear species tree around 60 Myr later.
Materials and Methods
Phylogenetically Informative Retrotransposon Insertion Analysis
We continued the work on an existing data set comprising 158 marsupial intron markers (Gallus et al. 2015) that were identified by extracting single-copy introns from the Tasmanian devil genome (WTSI_Devil_ref v7.0; Murchison et al. 2012) and aligning them with homologous loci in the South Amerian opossum (Didelphimorphia; Mikkelsen et al. 2007) and Tammar wallaby (Diprotodontia; Renfree et al. 2011). We selected SINEs that were, based on previous studies, expected to be active at the time when the four Australian orders (Diprotodontia, Notoryctemorphia, Peramelemorphia, and Dasyuromorphia) diverged (Nilsson et al. 2010; Gallus et al. 2015). In order to avoid a bias in the transposable element selection process, 38 introns without retrotransposable elements were selected by random, to increase the chance of finding independent insertions in species not covered by genome data.
Primers were located in conserved exonic regions flanking the introns with the selected SINE insertions (supplementary table S3, Supplementary Material online). All introns were amplified and sequenced in representatives from the respective marsupial orders (supplementary table S4, Supplementary Material online). The Amplicon Taq containing VWR Master Mix (VWR International GmbH, Darmstadt, Germany) was used in a touch-down polymerase chain reaction (PCR) approach to amplify selected loci. The resulting PCR fragment sizes were inspected on agarose gels for size differences. Where size-shifts between species were observed the locus was Sanger-sequenced in the relevant species (ABI 3730 DNA Analyzer with BigDye terminator sequencing kit 3.1, Applied Biosystems). If direct sequencing of the PCR product was not possible, the PCR-products were cloned into pDrive (Qiagen) or TOPO-TA (Life Technologies) vectors according to the manufacturers recommendations before Sanger sequencing. Sequences were masked with the online tools CENSOR (Kohany et al. 2006) or Repeatmasker (Smit et al. 2013). Sequences for each locus were aligned in Geneious V6 (BioMatters) using the embedded alignment tool, and manually inspected, and corrected. All alignments were visually screened for retrotransposon type, orientation of retrotransposon, target site duplications, and exon/intron homology to ensure the homology of each SINE insertion. The statistical evaluation of SINE insertions was performed according to Waddell et al. (2001) where three insertions per branch are required to obtain significant support [P = 0.037]. The approach calculates branch support under a standard Wright–Fisher coalescent model with panmictic mating, nonoverlapping generations and a constant population size (Waddell et al. 2001).
Consensus Network Analyses
Marsupial nuclear gene sequences were downloaded from GenBank (NCBI), the ensembl browser (EMBL-EBI and Wellcome-Trust Sanger Institute), and extracted from the Tasmanian devil, opossum, and wallaby genome using BLAT (Kent 2002; supplementary table S5, Supplementary Material online). In total, sequences of 25 marsupial species (supplementary table S4, Supplementary Material online) were aligned for 26 genes (28 gene fragments, supplementary table S6, Supplementary Material online), using the MUSCLE (Edgar 2004) algorithm in Geneious (v6; BioMatters). The Recombination Activation Gene 1 was split in two smaller fragments, as was Apolipoprotein B due to large amounts of missing sequence information in some species.
All sequence alignments were trimmed to minimize the total number of gaps and ambiguous nucleotides (nt) in the beginning and end of each alignment. Alignments were visually inspected and corrected if necessary. Best fitting DNA substitution models were calculated for each alignment using the jModelTest (Darriba et al. 2012) with Ґ models with four rate categories and a proportion of invariable sites (supplementary table S7, Supplementary Material online). We used the next complicated substitution model (in most cases general time reversible model), if the proposed model from jModelTest was not implemented in the MrBayes.
Bayesian gene trees were created using MrBayes V3.2.2 for each of the 28 gene fragments (Ronquist and Huelsenbeck 2003). MrBayes was run for 4,000,000 generations using random starting trees with default priors. The burn in was set at 250 (25% of 1,000 generations). Consensus networks were constructed in SplitsTree4 (version 4.13.1; Huson and Bryant 2006), using a threshold value of 7% for two different data sets, one consisting of all 28 gene trees, and one data set excluding all trees (n = 8) based on alignments shorter than 600 nt (fig. 3, supplementary fig. S1, Supplementary Material online). At this threshold level, multifurcations due to unresolved branches by single genes are not shown, making the network easier to interpret. Additionally, the single genes were concatenated to a data set consisting of 32,253 nt. The data set was partitioned to allow different substitution models for each fragment. MrBayes was run using the same parameters on the concatenated data set as for the single gene data set (supplementary fig. S2, Supplementary Material online), showing no differences to the consensus networks for the main marsupial groups.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors are grateful to Dr Björn Hallström (Science for Life, Sweden) for the screening of the Tasmanian devil genome. We thank the three reviewers for their helpful comments that improved the manuscript. This work was supported by Deutsche Forschungsgemeinschaft grant NI-1284/1-1 to M.A.N. This study was conducted at the Biodiversity and Climate Research Centre Frankfurt (BiK-F), and funded by the research-funding program “The Biodiversity and Climate Research Centre Frankfurt (BiK-F) is funded by LOEWE—Landes Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz” of Hesse’s Ministry of Higher Education, Research, and the Arts.
Literature Cited
- Archer M. The Australian marsupial radiation. In: Archer M, Clayton G, editors. Vertebrate zoogeography and evolution in Australasia. Perth: Hesperian Press; 1984. pp. 633–708. [Google Scholar]
- Archer M, et al. Australia’s first fossil marsupial mole (Notoryctemorphia) resolves controversies about their evolution and palaeoenvironmental origins. Proc R Soc B Biol Sci. 2011;278:1498–1506. doi: 10.1098/rspb.2010.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher RJ, Horovitz I, Sánchez-Villagra MR. First combined cladistics analysis of marsupial mammal interrelationships. Mol Phylogenet Evol. 2004;33:240–250. doi: 10.1016/j.ympev.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Bapteste E, et al. Networks: expanding evolutionary thinking. Trends Genet. 2013;29:439–441. doi: 10.1016/j.tig.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Boore JL. The use of genome-level characters for phylogenetic reconstruction. Trends Ecol Evol. 2006;21:439–446. doi: 10.1016/j.tree.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Churakov G, et al. Mosiac retroposon insertion patterns in placental mammals. Genome Res. 2009;19:868–875. doi: 10.1101/gr.090647.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallus S, et al. Evolutionary histories of transposable elements in the genome of the largest living marsupial carnivore, the Tasmanian devil. Mol Biol Evol. 2015 doi: 10.1093/molbev/msv017. Advance Access published January 28, 2015, doi: 10.1093/molbev/msv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles AJ, et al. Evolutionary dynamics of transposable elements in the short-tailed opossum Monodelphis domestica. Genome Res. 2007;17:992–1004. doi: 10.1101/gr.6070707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurovich Y, Travouillon KJ, Beck RMD, Muirhead J, Archer M. Biogeographical implications of a new mouse-sized fossil bandicoot (Marsupialia: Peramelemorphia) occupying a dasyurid-like ecological niche across Australia. J Syst Paleontol. 2014;12:265–290. [Google Scholar]
- Hallström BM, Janke A. Resolution among major placental mammal interordinal relationships with genome data imply that speciation influenced their earliest radiations. BMC Evol Biol. 2008;8:162. doi: 10.1186/1471-2148-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström BM, Janke A. Mammalian evolution may not be strictly bifurcating. Mol Biol Evol. 2010;27:2804–2816. doi: 10.1093/molbev/msq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallström BM, Schneider A, Zoller S, Janke A. A genomic approach to examine the complex evolution of laurasiatherian mammals. PLoS One. 2011;6:e28199. doi: 10.1371/journal.pone.0028199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz I, et al. Cranial anatomy of the earliest marsupials and the origin of opossums. PLoS One. 2009;4:e8278. doi: 10.1371/journal.pone.0008278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horovitz I, Sánchez-Villagra MR. A morphological analysis of marsupial mammal higher-level phylogenetic relationships. Cladistics. 2003;19:181–212. [Google Scholar]
- Hudson RR. Testing the constant-rate neutral allele model with protein sequence data. Evolution. 1983;37:203–217. doi: 10.1111/j.1558-5646.1983.tb05528.x. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch JAW, Springer MS, Lapointe FJ. DNA-hybridization studies of marsupials and their implications for metatherian classification. Aust J Zool. 1997;45:211–280. [Google Scholar]
- Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;25:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski C, Blacket MJ, Westerman M. DNA sequence analysis of familial relationships among dasyuromorphian marsupials. J Mamm Evol. 2000;7:95–108. [Google Scholar]
- Krajewski C, Wroe S, Westerman M. Molecular evidence for the pattern and timing of cladogenesis in dasyurid marsupials. Zool J Linn Soc. 2000;130:375–404. [Google Scholar]
- Kramerov DA, Vassetzky NS. Origin and evolution of SINEs in eukaryotic genomes. Heredity. 2011;107:487–495. doi: 10.1038/hdy.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaché AD, Harris RB, Rannala B, Yang Z. The influence of gene flow on species tree estimation: a simulation study. Syst Biol. 2014;63:17–30. doi: 10.1093/sysbio/syt049. [DOI] [PubMed] [Google Scholar]
- Long J, Archer M, Flannery T, Hand S. Prehistoric mammals of Australia and New Guinea: one hundred million years of evolution. Sydney: University of New South Wales Press; 2002. [Google Scholar]
- Meredith RW, et al. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science. 2011;334:521–524. doi: 10.1126/science.1211028. [DOI] [PubMed] [Google Scholar]
- Meredith RW, Westerman M, Case JA, Springer MS. A phylogeny and timescale for marsupial evolution based on sequences for five nuclear genes. J Mamm Evol. 2008;15:1–36. [Google Scholar]
- Mikkelsen TS, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, et al. Molecular phylogeny, biogeography, and habitat preference evolution of marsupials. Mol Biol Evol. 2014;31:2322–2330. doi: 10.1093/molbev/msu176. [DOI] [PubMed] [Google Scholar]
- Murchison EP, et al. Genome sequencing and analysis of the Tasmanian devil and its transmissible cancer. Cell. 2012;148:780–791. doi: 10.1016/j.cell.2011.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson MA, et al. Tracking marsupial evolution using archaic genomic retroposon insertions. PLoS Biol. 2010;8:e1000436. doi: 10.1371/journal.pbio.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Hasegawa M, Okada N. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci U S A. 2006;103:9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Maruyama S, Okada N. Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc Natl Acad Sci U S A. 2009;106:5235–5240. doi: 10.1073/pnas.0809297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak RM. Walker’s marsupials of the world. Baltimore: The Johns Hopkins University Press; 2005. [Google Scholar]
- Phillips MJ, Haouchar D, Pratt RC, Gibb GC, Bunce M. Inferring kangaroo phylogeny from incongruent nuclear and mitochondrial genes. PLoS One. 2013;8:e57745 doi: 10.1371/journal.pone.0057745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho C, Hey J. Divergence with gene flow: models and data. Annu Rev Ecol Evol Syst. 2010;41:215–230. [Google Scholar]
- Ray DA, Xing J, Salem A-H, Batzer MA. SINEs of a nearly perfect character. Syst Biol. 2006;55:928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- Renfree MB, et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12:R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shedlock AM, Okada N. SINE insertions: powerful tools for molecular systematics. Bioessays. 2000;22:148–160. doi: 10.1002/(SICI)1521-1878(200002)22:2<148::AID-BIES6>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Shimamura M, et al. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013. RepeatMasker Open-4.0. (2013–2015) Available from http://www.repeatmasker.org.
- Szalay FS. A new appraisal of marsupial phylogeny and classification. In: Archer M, editor. Carnivorous marsupials. Mossman. Australia: Royal Zoological Society of New South Wales; 1982. pp. 621–640. [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai Y, Takahashi K, Nishida M, Sato T, Okada N. Using SINEs to probe ancient explosive speciation: "hidden" radiation of African cichlids? Mol Biol Evol. 2003;20:924–930. doi: 10.1093/molbev/msg104. [DOI] [PubMed] [Google Scholar]
- Travouillon KJ, Hand SJ, Archer M, Black KH. Earliest modern bandicoot and bilby (Marsupialia, Peramelidae and Thylacomyidae) from the Miocene oft he Riversleigh World Heritage Area, northwestern Queensland, Australia. J Vertebr Paleontol. 2014;34:375–382. [Google Scholar]
- van de Lagemaat LN, Gagnier L, Medstrand P, Mager DL. Genomic deletions and precise removal of transposable elements mediated by short identical DNA segments in primates. Genome Res. 2005;15:1243–1249. doi: 10.1101/gr.3910705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell PJ, Kishino H, Ota R. A phylogenetic foundation for comparative mammalian genomics. Genome Inform Ser Workshop Genome Inform. 2001;12:141–154. [PubMed] [Google Scholar]
- Warren WC, et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbecker V, Nilsson M. Integration, heterochrony, and adaptation in pedal digits of syndactylous marsupials. BMC Evol Biol. 2008;8:160. doi: 10.1186/1471-2148-8-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman M, et al. Phylogenetic relationships of living and recently extinct bandicoots based on nuclear and mitochondrial DNA sequences. Mol Phylogenet Evol. 2012;62(1):97–108. doi: 10.1016/j.ympev.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Wroe S. Australian marsupial carnivores: recent advances in palaeontology. In: Jones M, Dickerman C, Archer M, editors. Predators with pouches: the biology of marsupial carnivores. Collingwood: Commonwealth Scientific and Industrial Research Organisation (CSIRO); 2003. pp. 102–123. [Google Scholar]
- Wroe S, Myers TJ, Wells RT, Gillespie A. Estimating the weight of the Pleistocene marsupial lion, Thylacoleo carnifex (Thylacoleonidae:Marsupialia): implications for the ecomorphology of a marsupial super-predator and hypotheses of impoverishment of Australian marsupial carnivore faunas. Aust J Zool. 1999;47:489–498. [Google Scholar]
- Zemann A, et al. Ancestry of the Australian termitivorous numbat. Mol Biol Evol. 2013;30:1041–1045. doi: 10.1093/molbev/mst032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.