Abstract
Understanding how gene regulation evolves is a key area in the current evolutionary field. Gene regulation occurs at various levels. Previous work on the evolution of gene regulation has largely focused on gene transcription. In this study, we used a recently developed ribosomal footprint profiling method to investigate how gene regulation evolves at both the transcription (mRNA abundance) and translation (ribosomal density) levels. By constructing a hybrid between Saccharomyces cerevisiae (Scer) and Saccharomyces bayanus (Sbay), which diverged ∼20 Ma, and quantifying transcriptome and translatome in both parental strains and their hybrid, we showed that translation is much more conserved than transcription, mostly due to the buffering effect of translational regulation for the transcriptional divergence. More conservation in translation than transcription is also confirmed by the inheritance mode of transcription and translation between two species. Furthermore, cis and trans effects are widely involved in changes at both transcription and translation levels. Finally, our results showed that genes with certain functions and sequence features might employ specific modes for evolution at these two critical levels of gene regulation. Our results demonstrated that it is essential to investigate the evolution of gene regulation at various levels from different genetic backgrounds to obtain a complete picture of its evolutionary modes in nature.
Keywords: evolution of gene regulation, RNA-Seq, ribosomal profiling
Introduction
Gene regulation translates static genomic information into dynamic organism phenotypes. It is increasingly clear that regulatory changes play important roles in determining phenotypic diversity between and within species. Therefore, a thorough understanding of how gene regulation evolves will have profound impacts on both basic and biomedical research (King and Wilson 1975; Wray 2007). Technological strides have enabled great progress, such as the recently published ENCyclopedia Of DNA Elements (ENCODE) consortium project (Consortium et al. 2012), in understanding gene regulation and its evolution (Rockman and Kruglyak 2006; Wray 2007; Zheng et al. 2011; Gordon and Ruvinsky 2012; Wittkopp and Kalay 2012). It is well known that the expression of a gene is regulated at multiple levels, including chromatin structure (Wyrick et al. 1999), gene transcription (O'Malley et al. 1977), posttranscriptional regulation (Filipowicz et al. 2008), translation, and protein modification (Westermann and Weber 2003), all of which determine the fate of its transcripts. Understanding the evolution of regulation at these various levels will enable a complete picture underlying the evolution of phenotypic adaptation in nature.
Gene transcriptional change might be a rapid and efficient approach for environmental adaptation during evolution. However, posttranscriptional regulation, which is more directly relevant to the amount of functional proteins in cells (Ingolia et al. 2009), also likely contributes to phenotypic adaptation. Gene translation regulation can be affected by multiple factors such as 5′- or 3′- untranslated regions (UTRs) composition, mRNA structure, RNA binding protein (RBP), and ribosomal proteins. Changes at these regulatory components could lead to translational divergence, and thus possible phenotypic consequences. However, the evolution of gene translation regulation has been understudied, likely due to the technical limitation with a lack of efficient and high-throughput approaches for translation quantification. Ribosomal footprint profiling (RFP) was recently invented to use the next-generation sequencing to monitor in vivo translation (Ingolia et al. 2009). By profiling the ribosome protected fragments on mRNAs after nuclease digestion, the method can enable determining the positions of ribosomes, thus providing snapshots of the ribosome locations during translation across the whole genome. The approach is now widely used to address various issues related with translation (Ingolia 2014) and has been recently employed to investigate the evolution of translation regulation (Artieri and Fraser 2014; Coate et al. 2014; McManus et al. 2014). The overall goal of this study is to use this approach and yeasts as model systems to investigate the evolution of gene translation regulation.
The genetic causes of gene transcription divergence can be generally classified into two categories: 1) cis element regulation (e.g., changes in promoters and enhancers), which affects the expression and mRNA stability of nearby genes on the same chromosome (Wittkopp et al. 2008; Tirosh et al. 2009); 2) trans element regulation, which results from functional divergence of transcription factors and chromatin modifiers, or from other components that transduce external signal into gene expression changes. The cis and trans effects for transcription divergence can be uncovered by conducting expression quantitative trait loci (eQTL) mapping (Gilad et al. 2008) or the hybrid experiments (Tirosh et al. 2009). In the hybrid cell, both alleles are under the same cellular environment, thus changes in gene transcription between alleles can only be attributed to the cis effect. In contrast, allelic gene transcription changes between the parental strains can be caused by both trans and cis effects. The advantage of the hybrid approach in comparison to the eQTL method is that it has a higher reliability in distinguishing the cis- and trans-acting elements (Schaefke et al. 2013) and can also be used to investigate mechanisms underlying regulatory divergence other than the ones at the transcription level. The approach has been widely applied in inter and intraspecific hybrids of yeast (Tirosh et al. 2009; Emerson et al. 2010; Khan et al. 2012; Schaefke et al. 2013), fly (Wittkopp et al. 2004, 2008), maize (Springer and Stupar 2007), Arabidopsis (Shi et al. 2012), and mouse (Goncalves et al. 2012) to study divergence in various aspects of gene regulation, such as gene transcription (Tirosh et al. 2009; Bullard et al. 2010; Emerson et al. 2010; Tirosh and Barkai 2011; Shi et al. 2012), replication timing (Muller and Nieduszynski 2012), protein abundance (Khan et al. 2012), nucleosome positioning (Tirosh et al. 2010), and mRNA degradation (Dori-Bachash et al. 2011).
In this study, we used the RNA-Seq and RFP approaches to investigate how gene transcription and translation regulation evolved between two remotely related yeast species. To achieve this, we constructed a hybrid between Saccharomyces cerevisiae (Scer) and Saccharomyces bayanus (Sbay) that diverged around 20 Ma (Lin et al. 2006), two most remotely related species hybrid constructed so far in addressing the evolution of gene translation. Transcriptome (mRNA) and translatome (RFP) were then sequenced for both parental strains and their hybrid. We compared orthologous gene transcription and translation between two species and demonstrated that translation is much more conserved than transcription during evolution. Furthermore, the cis and trans effects are widely involved in changes at both transcription and translation levels. Our major conclusions are consistent with previous work comparing transcriptome (Bullard et al. 2010) and proteome (Khan et al. 2012), and two very recent publications using a similar RFP approach with hybrids between two closely related yeast species, S. cerevisiae and Saccharomyces paradoxus, that diverged ∼5 Ma (Artieri and Fraser 2014; McManus et al. 2014). We also reported some unique discoveries using the hybrid constructed between two much more remotely related species in this study, indicating that survey of various levels of gene regulation from different genetic backgrounds can enable a more comprehensive understanding of the gene regulation evolutionary modes in nature.
Materials and Methods
Yeast Strains and Growth Condition
Scer strains BY4741 (MATa, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and BY4742 (MATα, hoD::NatMX, his3Δ1, lys2Δ0, ura3Δ0, leu2Δ0), and Sbay strain JRY8154 (MATα, hoD::NatMX, his3Δ1, lys2Δ5, trpΔ1, ura3Δ1) were used in this study. The BY4741 and JRY8154 were hybridized to obtain the diploid strain. The parental strains were cocultured to eliminate possible experimental variations. To avoid the potential hybridization of the parental strains in the coculture condition, we replaced BY4741 with the isogenetic opposite mating type strain BY4742. All strains were cultured in YPD at 25 °C, 200 rpm.
RNA-Seq and RFP
Both parental strains (cocultured) and diploid strain (hybrid) were cultured in YPD medium starting at OD600 = 0.1 and harvested when OD600 reached 0.6–0.8. Cells were treated immediately with cycloheximide (CHX, to a final concentration of 100 µg/ml) for 2 min to inhibit cytosolic translation elongation. The treated cells were then separated into two parts. One part was used for mRNA sequencing: the total RNA was extracted using hot phenol method (Ingolia 2010), mRNA was then purified using oligo-dT DynaBeads, and the cDNA and the sequencing library was constructed using the method described by Zhong et al. (2011). The other part was prepared for RFP sequencing by the method described by Ingolia (2010).
To conduct RFP sequencing, cell pellet with 2.5 ml polysome lysis buffer (20 mM Tris–HCl [pH 8.0], 140 mM KCl, 1.5 mM MgCl2, 100 µg/ml CHX, 1% [v/v] Triton X-100) was grounded in a mortar with liquid nitrogen. The cell powder was thawed at 30 °C for 2 min and centrifuged at 3,000 × g in 4 °C for 5 min to get rid of the cell debris. The supernatant was then centrifuged at 20,000 × g in 4 °C for 10 min. The cell extract was diluted to A260 = 200 with the lysis buffer. To digest the polysome to monosome, 7.5 µl RNase I (Ambion, AM2294) was added to 250 µl cell extract for 1 h at 25 °C and then the reaction was stopped by adding 5 µl SUPERase.In (Ambion, AM2694). The sucrose density gradients (centrifuged at 150,000 × g in 4 °C for 3 h) were used to collect the 80S monosome fraction from the cell extract. The purified footprint fragments by a standard SDS/phenol method were dephosphorylated at its 3′ end by the T4 polynucleotide kinase. The polyacrylamide denaturing gel purification was conducted to purify the mRNA footprint fragments. The region near the 28 nt marker was excised. RNA was recovered from the gel slice and quantified by BioAnalyzer (Agilent).
After adding a poly-A tail to each RNA fragment by Escherichia coli poly-A polymerase (NEB, M0276S), reverse transcription for the polyadenylated RNA samples was performed with primers that are linked to unique barcodes. The reverse transcription products were then purified on 10% denaturing polyacrylamide gel, and the region between 90 and 130 nt was excised and cDNA was recovered. The CircLigase (Epicentre Biotechnologies CL4111K) was added to the gel extraction products and the mix was incubated for 1 h at 60 °C and then inactivated by heating at 80 °C for 10 min. The circularized DNA was amplified by Phusion polymerase for 12 cycles, and the PCR products were purified on 8% nondenaturing polyacrylamide gel. The PCR products at ∼120 bp were finally recovered from the excised gel slice and quantified by BioAnalyzer (Agilent). Sequencing was performed on Illumina Hiseq2000 platform (50 bp, single-end). We conducted two biological replicates for each sample.
Reads Mapping and Transcription/Translation Quantification
Raw reads for both mRNA and RFP were cleaned by removing the barcodes and low-quality reads using custom perl scripts. For the RFP data, reads were further cleaned by removing the 3′ end polyA and then only those with length between 16 and 35 bp were retained. The wide range of read length was applied to include the ribosomal footprints that might be over or underdigested. Narrowing the read size (27–30 bp) reduced the total read number for each gene but essentially did not change the level of translation for each gene (supplementary table S4, Supplementary Material online). We removed reads mapped to rRNA genes using bowtie (Langmead 2010). The remaining reads were then aligned to the combined Scer and Sbay genomes with no more than two mismatches allowed. Allele-specific reads were assigned to either species when satisfying the following criteria: 1) the read was uniquely mapped to one species, or had at least two mismatches difference between the best alignment and the second best alignment if it mapped to multiple locations in one species; or 2) the read was mapped to both genomes, but the alignment to one species was significantly better than the other (with at least two mismatches difference). Reads for each gene were counted using htseq-count (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html, last accessed April 7, 2015), and genes with more than ten mapped reads in both mRNA and RFP were retained for further analysis.
To test the statistical significance of allelic differential transcription/translation (Sbay/Scer) in the parental strains, estimation of variability throughout the dynamic range of the sequence data and a suitable error model for statistical analysis are essential. We used a method by Anders and Huber (2010) that was developed to analyze differential expression using the sequence count data. The method is based on a negative binomial model and estimates the variance and mean of data by local regression (Anders and Huber 2010). We used the DESeq package which implements the method using R. To determine a significant transcription/translation difference (Sbay/Scer ≠ 1), we applied the criteria of false discovery rate (FDR) ≤ 0.05 and fold change ≥1.5 throughout the article.
Identifying the Buffering/Amplifying Effects in Translation Efficiency
Translation efficiency (TE) for each gene in the parental strains was calculated from RFP/mRNA. To test the significance of TE difference between two parental strains, we employed a likelihood ratio test (LRT), as described by Khan et al. (2013). For each gene, the method uses the framework of nested linear models to compare the fit of the null model (TE = c + ε, where c is a constant for each gene and ε measures the random errors) that correspond to no difference between species, to the alternative model (TE = c + ε + β*species, where β measures the regression coefficient for species) that correspond to significant differences between species. Sequence data for two biological replicates in each species were used for the regression analysis. The P values from LRT for all genes in the analysis were adjusted for the multiple test correction by the Benjamini–Hochberg method (Benjamini and Hochberg 1995) to control for the FDR.
We defined the buffering or amplifying effects in TE as described by McManus et al. (2014). Briefly, to consider the buffering or amplifying TE effect on the transcription divergence, genes that showed significant differences in both transcription and TE were investigated. The amplifying effect was defined if the translation changes were at least 1.5-fold higher than the transcription changes. In contrast, the buffering effect was assigned if translation changes were at least 1.5-fold lower than the transcription changes. Genes with diverged transcription but no TE divergence were classified as no amplifying or buffering effect.
Inheritance of Gene Transcription/Translation
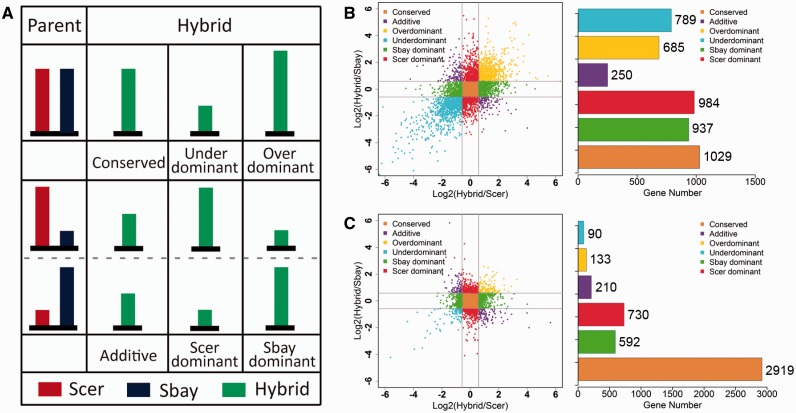
The inheritance modes of gene transcription and translation were shown in figure 3A, as defined by McManus et al. (2010). To be specific, the overall transcription or translation for both alleles in the hybrid strain (sum of the counts in both Scer and Sbay, hereafter named as Hall) were compared with that of either Scer (PScer) or Sbay (PSbay) in the parental strains using the DESeq package, as described earlier. We defined genes as being similarly transcribed (or translated) if the fold change <1.5. Under this definition, genes in the hybrid showing similar transcription (or translation) with both parental Scer and Sbay were considered as having the conserved inheritance mode. The other five nonconserved modes were defined as the following: 1) underdominant: the Hall was 1.5-fold lower than both PScer and PSbay; 2) overdominant: the Hall was 1.5-fold higher than both PScer and PSbay; 3) additive: the Hall was an intermediate between PScer and PSbay; 4) Scer dominant: Hall was similar to PScer but different from PSbay; 5) Sbay dominant: Hall was similar to PSbay but different from PScer.
Fig. 3.—
The inheritance modes of gene transcription and translation. (A) Diagram depicting the six inheritance modes in gene regulation. The overall mRNA (or RFP) level of two alleles for each gene in the hybrid was compared with the gene in either parental strain. Genes showing similar mRNA (or RFP) levels between the hybrid and both parental strains were classified as the “conserved” category, whereas the genes with lower or higher mRNA/RFP levels than both parents were designated as “underdominant” or “overdominant”, respectively. The “additive” mode was defined for a gene if its mRNA (or RFP) level in the hybrid was an intermediate between the two parents. The gene with a similar mRNA (or RFP) level between the hybrid and only one of the two parental strains was classified into “Scer dominant” or “Sbay dominant”. The inheritance modes of gene transcription (B) and translation (C) are shown. Transcription or translation divergence between the hybrid and the parental strains in each category are shown in a scatter plot and the number of genes in each category is displayed to the right. The gray lines in B and C represent the log2-transformed minimum cutoff of the fold change (1.5-fold difference) used to define the regulation divergence.
Identifying the Cis/Trans Effects in Transcription and TE
The schematic diagram for the cis and trans effects for gene transcription (or TE) was shown in figure 1B. Generally, the hybrid strain eliminates the trans effect, thus the changes between alleles are only subject to the cis effect regulation. In the parental strains, however, as both trans and cis effects are present, the fraction of gene transcription (or TE) divergence not explained by the cis changes can be attributed to the differences caused by the trans effect (Tirosh et al. 2009; Emerson et al. 2010; Shi et al. 2012; Schaefke et al. 2013; Artieri and Fraser 2014; McManus et al. 2014). Following this logic, we estimated the cis regulatory effects for those genes showing significantly different transcription (or TE) between Scer and Sbay in the hybrid ([Sbay/Scer]hybrid ≠ 1). The statistical methods determining the cis effects in transcription and TE (in the hybrid) were the same to those described earlier that measured the allelic differential transcription and TE between the parental strains.
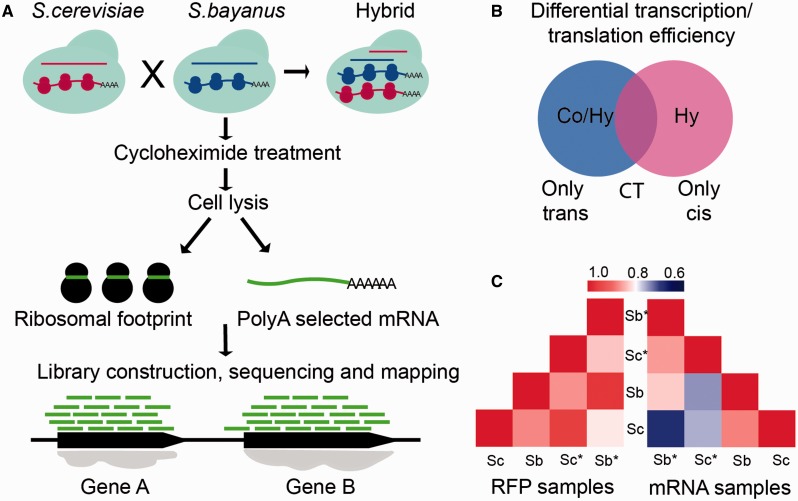
Fig. 1.—
The workflow for RNA-Seq and RFP. (A) The mixed parental strains or their hybrid were cultured in the YPD medium to the log phase. Cells were collected and treated by CHX for 2 min, and then divided into two aliquots, one for RNA-Seq and the other for RFP. (B) Diagram depicting the cis and trans effects for gene regulation. In the hybrid, genes face the same trans environment, so the divergence in gene regulation between alleles is only subject to the cis effect. Whereas in the parental strains, the divergence is subject to either trans or cis effect, or both. (C) Pearson correlation coefficient (r) among mRNA and RFP data between biological replicates and alleles (Sc: Saccharomyces cerevisiae, Sb: Saccharomyces bayanus). The asterisk indicates the hybrid strain.
To infer the trans effect for transcription (or TE), we identified genes that have significantly different ratio of (Sbay/Scer) for transcription (or TE) between the parental and hybrid strains. We applied the Fisher’s exact test (FET) using the sequence counts to identify genes with significant trans effect in transcription. The trans effect in TE for each gene was inferred using the above LRT method, with the null model of no-trans effect, Y = c + ε, where Y is the ratio of TE (Sbay/Scer), c is a constant for the cis effect in TE (c = 1 if there is no cis effect), and ε measures the random errors. The alternative model with a significant trans effect is Y = c + ε + β*treatment, where β measures the regression coefficient for treatment (coculture or hybrid). The parameters determining the significance of the cis or trans effect were subject to the criteria mentioned earlier. The log2-transformed cis and trans regulatory divergence for transcription (or TE) were then represented by |log2(Sbay/Scer)hybrid| and |log2(Sbay/Scer)parental − log2(Sbay/Scer)hybrid|, respectively.
Gene ontology (GO) Enrichment Analysis and Statistics
GO enrichment analysis was conducted using an online service of DAVID (Huang et al. 2009). The corrected P value ≤5% was used to define significant functional enrichment. All statistics in this study was performed using the R version 2.15.2 (R Core Team 2012). FDRs were calculated using the Benjamini–Hochberg method implemented in the p.adjust() command. The FET and the linear regression were conducted using the fisher.test() and lm() functions.
Data Access
The high-throughput sequencing data in this study has been deposited in NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra, last accessed April 7, 2015) under the accession number: SRR1175760, and SRR1175763 to SRR1175769. The genome sequence and annotation for Scer used in this study was downloaded from the Saccharomyces Genome Database (http://www.yeastgenome.org/, last accessed April 7, 2015) with the latest version (R64-1-1), and the sequence for Sbay was downloaded from Yeast Gene Order Browser (YGOB; http://ygob.ucd.ie/, last accessed April 7, 2015).
Results
We performed cellular mRNA and RFP sequencing to investigate the transcription and translation divergence between cocultured parental strains of Scer and Sbay, as well as their hybrid (fig. 1A and B). The sequencing statistics are shown in supplementary table S1, Supplementary Material online. Orthologous genes between Scer and Sbay were predicted using a method based on a Markov Cluster algorithm (Li et al. 2003), and then only one to one ortholog groups were kept for further analysis. Altogether 4,774 ortholog pairs were included (supplementary table S2, Supplementary Material online). Of these genes, 4,674 genes were both transcribed and translated (defined as having ≥10 reads per transcript) and thus used in the analysis. Our results indicated that each sample had high correlation between biological replicates (Pearson correlation coefficient, r ∼ 0.98 for mRNA and r ∼ 0.99 for RFP), which is more significant than the correlation between orthologous genes between two species (r ∼ 0.88 for mRNA, r ∼ 0.87 for RFP) or between corresponding alleles within the hybrid (r ∼ 0.86 for mRNA, r ∼ 0.83 for RFP; fig. 1C).
We also compared our transcriptome and translatome data for the parental S. cerevisiae with the S. cerevisiae data from two recently published work with a similar experimental design using hybrids between two closely related yeast species, S. cerevisiae and S. paradoxus (Artieri and Fraser 2014; McManus et al. 2014). Using the same analysis pipeline, we noted that our sequence reads for S. cerevisiae were highly correlated with these publications for both mRNA (r ∼ 0.80–0.87) and RFP (r ∼ 0.83–0.89), whereas the r between these two previous studies for mRNA is from 0.86 to 0.87 and RFP from 0.89 to 0.90 (supplementary table S3, Supplementary Material online), implying that our sequencing reads are comparable with these previous work, and thus providing an important quality check before further analyses.
Gene Translation Is More Conserved than Transcription during Evolution
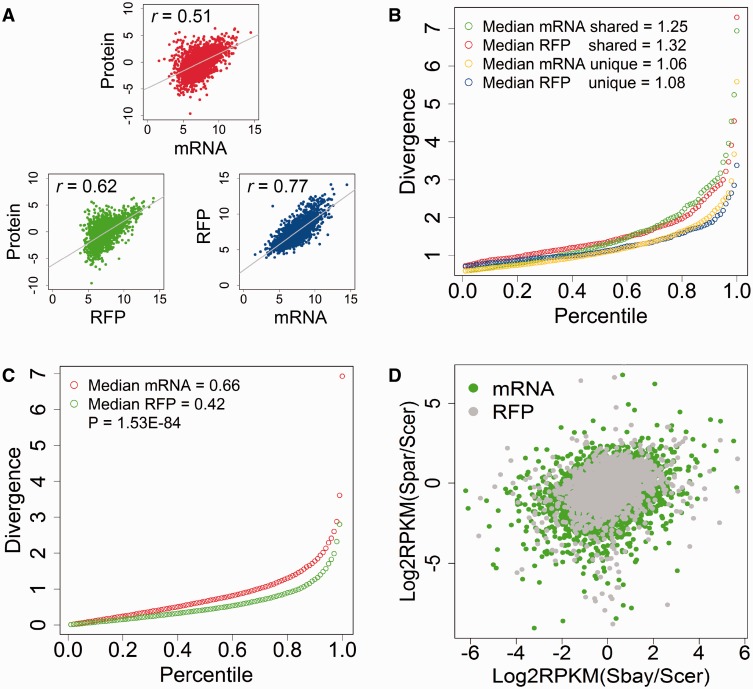
To illustrate the relationship between transcriptome and translatome with respect to the protein product, we first explored the correlation of our transcriptome and translatome data with the whole-cell proteome data in S. cerevisiae (Picotti et al. 2013). As shown in figure 2A, the Pearson correlation coefficient between translatome and proteome (r ∼ 0.62) is much higher than that between transcriptome and proteome (r ∼ 0.51). Of note, the correlation between transcriptome and translatome (r ∼ 0.77) is still much higher than that between translatome and proteome, confirming that there are a large amount of unknown regulatory steps shaping protein biosynthesis. In the following, we just used RFP data to represent translation.
Fig. 2.—
Comparison of transcriptome, translatome, and proteome in yeast species. (A) Pearson correlation coefficient (r) between transcriptome (mRNA), translatome (RFP), and proteome (Protein) data in Saccharomyces cerevisiae. Reads per kilo per million (RPKM) for mRNA and RFP were calculated and compared with the normalized proteome data from Picotti et al. (2013). The line in each plot indicates the linear fitting with P values all essentially 0. (B) Comparison of transcription and translation for genes that had significant changes between the parental strains in mRNA (2,509 genes) and RFP (901 genes). The “shared” means that those genes had significant changes in both mRNA and RFP (559 genes), whereas the “unique” means that those genes had changes in either mRNA or RFP, but not both. Divergence was defined as |log2(fold change)| between the parental strains. The figure shows the quantile-normalized divergence for mRNA and RFP, and the number indicated the median divergence for the gene group. The shared genes have significantly higher divergence than the unique genes (WRT: P = 1.24E−10 for mRNA and P = 1.59E−11 for RFP). (C) The quantile-normalized divergence of transcription and translation for all genes. The transcription (median mRNA divergence = 0.66) has a significantly higher divergence than the translation (median RFP divergence = 0.42, WRT: P = 1.53E−84). (D) Comparison of the transcription and translation divergence between three yeast species, S. cerevisiae, S. bayanus, and S. paradoxus. The RPKM values of mRNA and RFP for each species were calculated and averaged between replicates. The log2-transformed RPKM ratios of transcription and translation were plotted. The plot for mRNA is more dispersed than RFP for the same species pairs. Significant more transcription than translation divergence was observed between S. cerevisiae and S. paradoxus (WRT: P < 2.22E−16, data were obtained from McManus et al. [2014]).
To investigate what genes show differential transcription and translation, we compared the mRNA and RFP abundance between species using a method based on a negative binomial model that was developed by Anders and Huber (2010). We applied the criteria of a FDR ≤ 0.05 and a fold change ≥1.5 to define genes with differential transcription or translation in the respective dataset. Although significant genes (FDR ≤ 0.05) with small fold changes might still be biologically meaningful, we set the criteria of fold change ≥1.5 to reduce the impact from noise in the sequencing data. In total, 2,509 out of 4,674 studied genes (∼54%) showed significant transcriptional changes between two parental species. Using the same criteria, only 901 genes showed significant translational changes. Among all studied genes, 559 genes showed differences at both transcription and translation levels, and 425 (76%) of them showed difference in the same direction. Furthermore, genes that changed at both transcription and translation levels had significantly higher regulation divergence [represented as |log2(fold change)|] than those genes with changes in only transcription or translation (fig. 2B). To reduce the impact of noise in the sequencing data, we also increased the minimum cutoff of the number of mapped reads to 100 to define that a gene is transcribed/translated. With more stringent criteria, the numbers of differentially transcribed and translated genes were both reduced, but the general conclusion that more genes show transcription than translation divergence between two species was still similar (supplementary fig. S1, Supplementary Material online).
At the genome level, RFP also had a lower divergence (median divergence = 0.42) than that of mRNA (median divergence = 0.66, Wilcoxon rank-sum test [WRT]: P = 1.53E−84, fig. 2C), indicating that the evolution of gene translation might be slower than gene transcription. To test whether this is a common phenomenon for other organisms, we used the same approach to reanalyze the recently published mRNA and RFP data from S. cerevisiae and S. paradoxus (McManus et al. 2014). A similar pattern was observed for these two species that had a significant lower RFP divergence than transcription divergence (fig. 2D).
Dominant Buffering Effect in Translation Efficiency
TE was calculated by normalizing the RFP reads with the mRNA reads of the same genes. In almost all previous RFP research, TE was used as a proxy to represent the rate of translation. It is worthy to note that this term is indeed the “ribosomal density,” which could be different from TE if the rate of ribosome movement varies in different type of genes. However, in a recent study (Ingolia et al. 2011), the authors monitored the kinetics of protein synthesis globally by tracing runoff elongation using the ribosome profiling approach, and the results showed that the average ribosome movement speed is around 5.6 amino acids per second, very close to the single gene measurement by a classical biochemical method (6 amino acids per second [Bostrom et al. 1986]). More importantly, the results indicated that translation rate is remarkably consistent among different classes of mRNAs, regardless of gene length, transcription level, and codon usage bias, indicating that the overall rate of translation elongation is generally constant. To be consistent with these previous literatures, we kept using TE for RFP/mRNA for each gene throughout the following discussion.
Changes in gene TE and transcription could occur at the same (amplifying effect) or opposite (buffering effect) direction, as defined by McManus et al. (2014). We found that more than seven times genes have the buffering effect in translation than those with the amplifying effect (N = 1,522 vs. N = 207), consistent with the observation that gene translation is more conserved than transcription. Functional enrichment analysis revealed that genes with the buffering effect in translation were highly associated with oxidation reduction (GO:0055114, P = 3.63E−5) and sterol metabolism (GO:0008202, P = 3.71E−4). The sterol biosynthesis pathway, which is responsible for synthesizing the lipid components of yeast cell plasma membrane, was significantly downregulated in gene transcription (9 out of 14 genes) in Scer, which is consistent with the previous observations (Fraser et al. 2010; Chang et al. 2013). However, our results indicated that gene transcription divergence for this pathway was buffered during translation, so the genes in this pathway might not show the same scale of difference at the protein level. Genes with the amplifying effect during translation were more related to amino acid biosynthetic processes, like cysteine biosynthetic process (GO:0019344, P = 5.73E−4), sulfur amino acid biosynthetic process (GO:0000097, P = 5.70E−4), and methionine biosynthetic process (GO:0009086, P = 2.38E−2), consistent with the fact that these strains have different auxotrophies in the amino acid biosynthesis. Genes with no buffering or amplifying effect were strongly associated with mitochondrial translation (GO:0032543, P = 7.84E−7) and other mitochondrion-related functions.
Inheritance Patterns of Gene Transcription and Translation
The inheritance of gene transcription and translation was measured by comparing the dominance of gene transcription and translation between the parental strains and the hybrid as described previously (McManus et al. 2010). As shown in the Materials and Methods and figure 3A, genes with <1.5-fold differences were considered as the conserved genes, whereas other genes were classified into five categories based on their relative levels among different genetic backgrounds. For gene transcription inheritance, ∼22.0% of the investigated genes were classified as the conserved genes (1,029/4,674). The other 78% genes were assigned to the following categories: additive (250, ∼5.3%), overdominant (685, ∼14.7%), underdominant (789, ∼16.9%), Scer dominant (984, ∼21.1%), and Sbay dominant (937, ∼20.0%, fig. 3B).
The gene transcription inheritance could be modified by posttranscriptional regulation. In our analysis, 20–81% genes in the five nonconserved inheritance categories of transcription were buffered by the translational process, whereas only 4–6% of them were amplified during translation. The inheritance of translation was more stable than transcription (fig. 3C). Indeed, ∼62.5% (2,919/4,674) genes had the conserved inheritance pattern in translation. Genes with the additive, overdominant, or underdominant inheritance modes in translation only accounted for 4.5% (210 genes), 2.8% (133 genes), and 1.9 % (90 genes), respectively, of the studied genes. The inheritance patterns showed big differences between transcription and translation, suggesting the involvement of posttranscriptional regulation in modifying gene expression inheritance. (fig. 3B and C).
Cis and Trans Effects Involved in the Evolution of Transcription Regulation
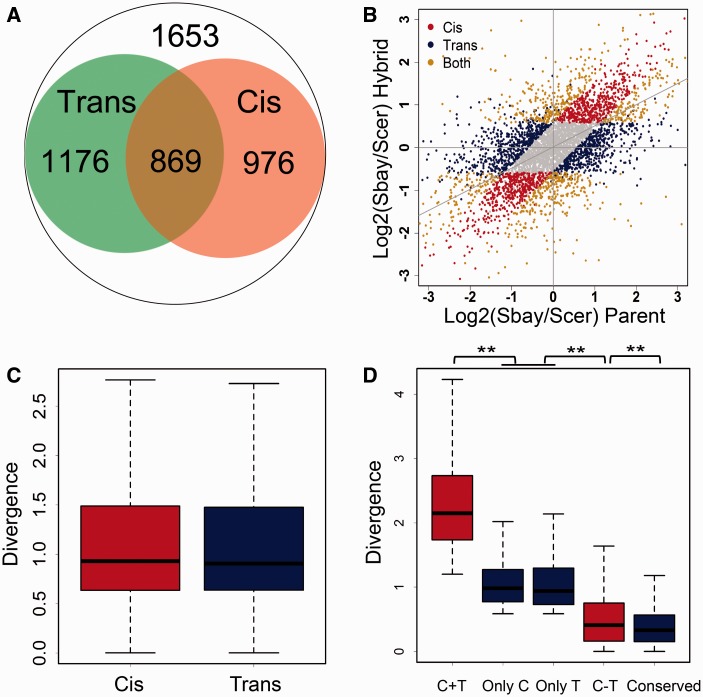
Changes in gene transcription can result from either cis or trans effect (Rockman and Kruglyak 2006). We measured the cis effect on gene transcription by directly comparing the gene transcription difference between Sbay and Scer in the hybrid cell, and the trans effect was measured by comparing the changes of transcription ratio (Sbay/Scer) in the parental strains to that in the hybrid, as defined in previous studies (Tirosh et al. 2009; Emerson et al. 2010; Shi et al. 2012; Schaefke et al. 2013; Artieri and Fraser 2014; McManus et al. 2014). Results showed that the trans effect impacted slightly more genes (10.8%) than the cis effect in contributing to the divergence in transcription between Scer and Sbay (2,045 genes with the trans effect vs. 1,845 with the cis effect, fig. 4A and B), whereas the two had similar contribution to the magnitude of gene transcription divergence (median divergencecis = 0.93, divergencetrans = 0.91, WRT: P = 0.34, fig. 4C). Among genes with an identifiable cis or trans effect, 869 genes showed both cis and trans regulation (defined as CT genes, fig. 4A). The cis and trans effects might act at the same direction (an enhancing CT effect) or opposite direction (a compensating CT effect) during transcription regulation. We found 288 (∼33%) genes had the enhancing CT effect, and the other 581 (∼67%) genes had the compensating CT effect. Expression of genes with the enhancing CT effect was significantly higher than those with only cis or trans effect (WRT: P = 2.25E−111), whereas the expression of genes with the compensating CT effect was significantly lower (WRT: P = 2.64E−109, fig. 4D). This result is consistent with the hypotheses that diversifying selection that promotes gene transcription divergence could lead to the enhancing cis and trans interaction, whereas purifying selection that would reduce gene expression divergence results in compensating cis and trans interplaying (Shi et al. 2012). A previous study performed an analysis of allele-specific expression using RNA-Seq on the hybrid between Scer and Sbay (but different strain from this study). Comparison between our results and the previous one revealed 1.5-fold differential expressed genes identified in this study using the same criteria (FDR ≤ 5% and fold change ≥1.5). Moreover, the trans factors cannot be inferred as the transcriptome was only profiled in the hybrid (Bullard et al. 2010).
Fig. 4.—
The cis and trans effects on the evolution of gene transcription. (A) The cis effect was defined by comparing the transcription difference between alleles in the hybrid strain (FDR ≤ 0.05 and fold change ≥1.5, 1,845 genes in total). The trans effect was defined by measuring the ratio change of allelic transcription between the parental strains and the hybrid (2,045 genes). The overlapping genes from the two categories are those that have both the cis and trans effects (869 genes). (B) Scatter plot for the cis, trans and CT genes in transcription regulation. The gray points show genes without allelic transcription divergence (FDR > 0.05 or fold change <1.5), and the gray diagonal line represents the linear fitting using all the data. (C) Boxplot for the transcription divergence between the parental strains for the cis and trans genes. The cis and trans effects have similar contributions to the magnitude of transcription divergence (median divergencecis = 0.93, divergencetrans = 0.91, P = 0.34). (D) Boxplot for the transcription divergence between the parental strains for the only cis, the only trans, the enhancing CT (C + T), the compensatory CT (C − T), and the conserved genes. The ** means P < 1.50E−6.
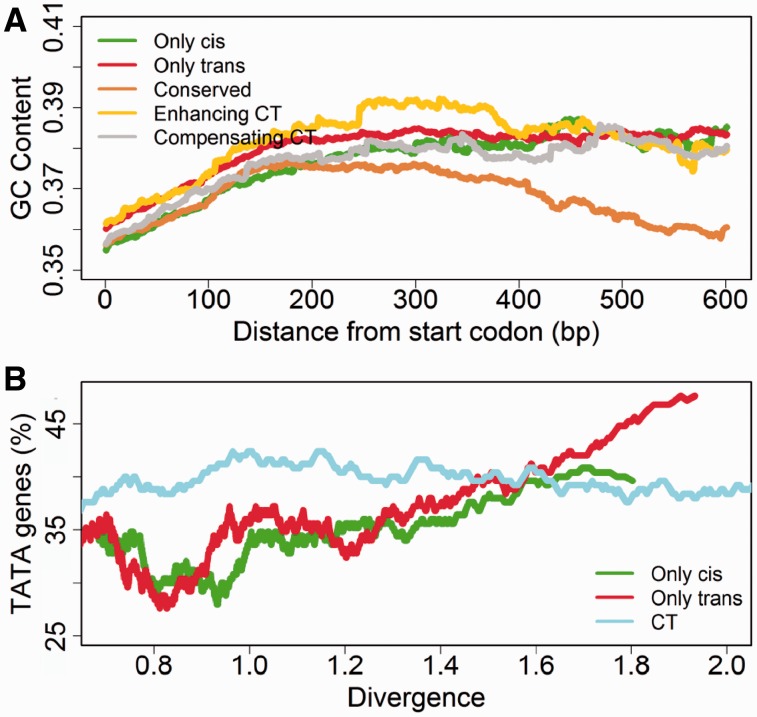
We further investigated several parameters that might be associated with the evolution of gene transcription. The 1 kb sequences upstream of genes from the only cis, only trans, CT (enhancing CT and compensating CT), and conserved categories were analyzed. Our results indicated that both cis and trans regulated genes had higher average GC content in the upstream sequences than the conserved genes (paired WRT: P = 3.40E−64). The enhancing CT genes had the highest GC content (P = 2.82E−40), which might be related with their complicated multiplex regulation (fig. 5A). Previous study using other closely related yeast species revealed that gene transcription plasticity was correlated with the presence of TATA box (Tirosh and Barkai 2008), and especially in trans-regulated genes (Tirosh et al. 2009). In this work, we discovered that the TATA box was enriched in both trans (FET: P = 4.15E−14) and cis genes (P = 2.88E−12). Interestingly, the percentage of CT genes with TATA box remained relatively constant, regardless of the level of gene transcription divergence between species, whereas that in the cis or trans only genes tend to be increasingly associated with the level of gene transcription divergence (fig. 5B). We further investigated the nucleosome structure near different categories of genes and found an enrichment of trans-related genes (only trans [P = 9.31E−3] and CT [P = 7.52E−4]) with the occupied proximal nucleosome (high occupancy close to the transcription start site [TSS] and low occupancy at the more distal region), and no enrichment for either cis and trans genes were found in the depleted proximal nucleosome (low occupancy close to the TSS and high occupancy at the more distal region), which are consistent with previous reports (Tirosh et al. 2010) (supplementary fig. S2, Supplementary Material online).
Fig. 5.—
The impact of GC content and TATA box on gene transcription divergence. (A) Comparison of the GC content in 1 kb sequences upstream of genes with the cis or trans effects. The GC content was calculated by sliding a 400 bp window with the step size of 1 bp on the upstream sequences. Both the cis and trans genes have higher GC contents in the upstream sequences than the conserved genes (P = 3.40E−64). The enhancing CT genes had the highest GC content (P = 2.82E−40). (B) The association between the TATA box and the transcription divergence between two species. Genes were sorted based on their transcription divergence between two parental strains and a sliding window of 250 genes with the step size of one gene was used to calculate the transcription divergence and the percentage of genes with the TATA boxes. The TATA box is enriched in both the trans (P = 4.15E−14) and cis genes (P = 2.88E−12). The percentage of CT genes with the TATA box remained constant, regardless of the level of gene transcription divergence between species, whereas that in the cis or trans only genes tend to be increasingly associated with the level of gene transcription divergence. Different window sizes and steps sizes in the analysis lead to similar conclusions in both A and B (data not shown).
GO analysis revealed different functional enrichment between genes with the cis and trans effects: the cis regulatory genes are enriched in the basic biological processes, such as protein transport (GO:0015031, P = 5.28E−4), ncRNA metabolic process (GO:0034660, P = 7.71E−3), and ribosome biogenesis (GO:0042254, P = 7.76E−3), whereas the trans genes are enriched in the stress response related processes, like dephosphorylation (GO:0016311, P = 1.40E−3), cell redox homeostasis (GO:0045454, P = 5.53E−3), cellular response to stress (GO:0033554, P = 2.24E−3), and tetrapyrrole biosynthetic process (GO:0033014, P = 5.51E−4). Additionally, the CT genes are more related with cell death (GO:0008219, P = 7.41E−7) and mitochondrial functions: oxidative phosphorylation (GO:0006119, P = 6.49E−4), ion transport (GO:0006811, P = 3.40E−4), and cellular respiration (GO:0045333, P = 2.53E−3).
Cis and Trans Effects Involved in the Evolution of TE
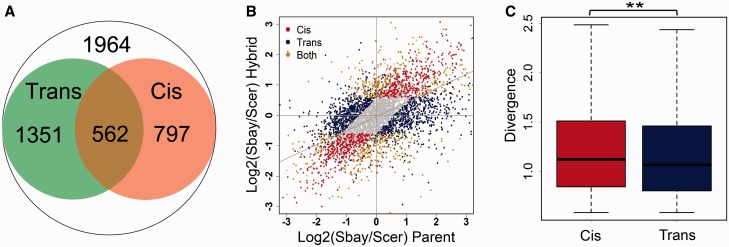
We conducted similar analysis to explore the cis and trans effects in the divergence of TE. There were 1,359 and 1,913 genes changed translational efficiency between two species due to the cis and trans effects, respectively. Among these genes, 797, 1,351, and 562 genes were caused by the cis only, trans only, and CT effects, respectively (fig. 6A and B). The enhancing CT genes accounted for ∼29% of all the CT genes and the other ∼71% CT genes had the compensating CT effect. The trans effect was dominant (∼41% more than the cis effect) in the number of regulated genes in TE divergence, but the cis effect contributed slightly larger than the trans effect to the magnitude of divergence in TE between two species (WRT: P = 6.13E−3, fig. 6C).
Fig. 6.—
The cis and trans effects on the evolution of TE. (A) TE was defined by dividing the RFP reads with the mRNA reads for each gene. The cis effect was obtained by comparing the TE differences between alleles in the hybrid strain (FDR ≤ 0.05 and fold change ≥1.5, 1,359 genes in total). The trans effect was defined by measuring the TE ratio changes of alleles between the parental strains and the hybrid (1,913 genes). The overlapping genes from the two categories have both the cis and trans effects in their TE regulation (562 genes). (B) Scatter plot for the cis, trans, and CT genes in TE regulation. The gray points show genes without allelic TE divergence (FDR > 0.05 or fold change <1.5), and the gray diagonal line represents the linear fitting using all the data. (C) Boxplot for the TE divergence between the parental strains for the cis and trans genes. The divergence of the cis effect genes was slightly higher than that of the trans genes (**, WRT: P < 0.01).
Translation is a tightly regulated step for protein biosynthesis and is known to be influenced by many features of mRNA transcripts, such as existence or the length of 5′-UTR, frequency of upstream stop codons, uORF activity, and codon usage bias(Ingolia et al. 2009; Brar et al. 2012; Arribere and Gilbert 2013; McManus et al. 2014). We investigated whether RNA binding motif (RBM) and mRNA secondary structure can also affect TE divergence by using the genome level RBM and mRNA structure data from Freeberg et al. (2013) and Kertesz et al. (2010), respectively. We discovered that genes with detected RBM (FDR ≤ 0.05, as defined in the Freeberg’s study) in 3′-UTR were significantly enriched in genes with divergence in TE (P = 3.49E−5), but not the conserved genes (P = 0.01). Furthermore, the trans effect was more associated with genes having RBM at both 5′-UTR (P = 1.34E−04) and 3′-UTR (P = 8.74E−05) than the cis effect (P = 0.01 for 5′-UTR, P = 7.59E−4 for 3′-UTR).
Our results also showed that the genes with detected mRNA secondary structure (FDR < 0.05, as defined in Kertesz et al. [2010]) were significantly enriched in genes with divergence in TE (FET: P = 2.45E−6), whereas no enrichment was observed in the conserved genes (FET: P = 0.11). The paired mRNA structure (represented by the magnitude of parallel analysis of RNA structure [PARS] score, which was based on the deep sequencing fragments of RNAs that were treated with structure-specific enzymes [Kertesz et al. 2010]) was slightly but significantly anticorrelated with the TE for genes with low TE values (defined as TE < 1, r = − 0.15, P = 1.95E−8) but not for those genes with high TE values (TE > 1, r = 0.04, P = 0.18, supplementary fig. S3, Supplementary Material online). In addition, although the genes with detected mRNA secondary structure are enriched in both cis and trans genes (FET: P values are 1.59E−4 and 3.88E−4, respectively), the trans genes had higher PARS scores than the cis genes (WRT: P = 6.42E−11). These results indicate that the RBM and mRNA secondary structure are important determinants for the evolution of gene translational efficiency.
Discussion
Different Conservation Levels for Transcription and Translation
In this study, we showed that gene translation is more conserved than gene transcription. Indeed, ∼78% interspecies transcription divergence cannot be observed at the translation level for two remotely related yeast species, S. cerevisiae and S. bayanus. Recently, two studies using S. cerevisiae and another closely related yeast species, S. paradoxus, reported similar conclusion (Artieri and Fraser 2014; McManus et al. 2014). The discordance between transcription and translation is most likely due to the buffering effect of the translational process. Buffering effect of translation on gene transcription divergence was also revealed in another study (Khan et al. 2013), which demonstrated more conserved evolution at the protein level than the gene transcript level, indicating that gene transcription divergence could overestimate the speciation difference in gene regulation.
The inheritance modes of gene transcription and translation also support the finding that gene translation is more conserved. Around 63% genes show conserved inheritance mode in translation whereas only ∼22% genes in transcription. Regulation at different levels seem to have a great impact on inheritance, for example, in the transcription inheritance, there are similar number of genes with the Scer dominant mode and the Sbay dominant mode, but in the translation inheritance, this difference is enlarged to 3-fold. However, the opposite pattern (more Sbay-dominant genes than Scer-dominant genes) was observed in the inheritance of protein abundance (Khan et al. 2012).
Factors Affecting Transcription and Translation Divergence
We measured gene transcription and translation in the haploid parental strains. In previous studies using the yeast hybrid approach to investigate the evolution of gene regulation, both haploid (Tirosh et al. 2009; Emerson et al. 2010; Schaefke et al. 2013; Artieri and Fraser 2014) and diploid (McManus et al. 2014) parental strains have been used. Certain genes have been demonstrated to be differentially regulated between isogenetic haploid and diploid strains, such as those involved in mating, meiosis, budding pattern, and ploidy. However, the differential regulation of these genes responsible for the cellular ploidy state might not affect the genome-wide conclusion in this study because a previous work by Galitski et al. (1999) compared gene expression between isogenic S. cerevisiae strains with different ploidy backgrounds and identified only 17 genes that are related to ploidy, suggesting that the effect of ploidy on gene expression is minimum. In a similar experimental design using the hybrid approach to identify the cis/trans effects on gene regulation divergence between S. cerevisiae and S. paradoxus (Tirosh et al. 2009), the authors conducted expression profiling for both haploid and diploid parental strains, and found that the gene expression is highly similar between the haploid and diploid strains (r = 0.94 and 0.93 for Scer and Spar, respectively). Therefore, we kept using the haploid as our parental strains in this study.
It was shown before that GC content differences between orthologous genes could inflate transcription divergence (Bullard et al. 2010). We confirmed this observation using our dataset (supplementary fig. S4, Supplementary Material online). We also found a significant correlation between the GC difference and the translation divergence (supplementary fig. S4, Supplementary Material online) for orthologous genes. These observations need to be taken with caution because the Illumina sequencing technology can be affected by the GC content in the reads. Furthermore, the correlations between the GC difference and the regulation divergence, even significant, are weak. We also investigated whether the differences of length between orthologous genes could bias our results. No significant correlations between gene length and transcription (P > 0.07) or translation (P > 0.06) were detected (supplementary fig. S5, Supplementary Material online).
The cis and trans regulatory factors differ in influencing the evolution of gene regulation and their inheritance patterns. Numerous studies using the hybrid approach with different species indicated that the cis effect might be the dominant regulatory factor in contributing to gene transcription divergence between species, whereas the trans effect is more responsible for the transcription divergence within species (Tirosh et al. 2009; Emerson et al. 2010; Li et al. 2012; Shi et al. 2012; Suvorov et al. 2013). However, some other studies also observed that the trans effect has a larger impact than the cis effect in contributing to the evolution of gene transcription or translation between different species (McManus et al. 2010; Artieri and Fraser 2014; McManus et al. 2014). Our results showed that the trans effect is more important than the cis effect in affecting both transcription and translation divergence between the two species we investigated. The mechanisms underlying these different observations warrant further investigation.
A previous study on the evolution of transcription and mRNA decay using a hybrid of S. cerevisiae and S. paradoxus revealed that the regulatory factors for gene transcription and mRNA decay were coupled (Dori-Bachash et al. 2011). Notably, the transcriptional and posttranscriptional gene regulation might be coupled due to the fact that some cis motifs located at the 5′-UTR/3′-UTR are important for both transcription and translation. Some trans factors, such as RBPs can also influence gene transcription and translation simultaneously (Morris et al. 2010). In our study, the cis and trans effects of transcription and TE are intuitively related because the inference of TE relies on the ratio of ribosome footprint to the mRNA level of each gene. Indeed, ∼50% and ∼10% of the cis genes for TE overlap with the cis and trans genes for transcription, respectively. On the other hand, ∼9% and ∼46% of the trans genes for TE overlap with the cis and trans genes for transcription, respectively. This result suggests a possible coupling relationship among the cis factors and among the trans factors during the gene transcription and translation processes.
Conclusion
We used two remotely related yeast species (S. cerevisiae and S. bayanus with a divergence ∼20 Ma) as models to investigate the evolution of gene regulation during transcription and translation. The evolutionary distance between these two species is much further than yeast species pairs previously used with the similar hybrid approach to investigate the evolution of gene translation. Our results agreed with multiple previous reports that translation is more conserved than transcription, leading to a more stable proteome than that appreciated by comparing gene transcription alone. We also discovered some unique results using our experimental design. For example, at the gene transcription level, the TATA box, which was reported to be only enriched in the trans genes, was discovered to be overrepresented in both cis and trans genes between the species pair used in this study. We also found that the trans effects are more responsible than the cis effects for the divergence at both levels of gene regulation. For genes whose transcription or translation are affected by both the trans and cis factors (the CT genes), these factors usually function in the opposite directions (the compensating CT effect), indicating the stability of regulation for these genes. Our results demonstrate that surveys of various levels of gene regulation from different genetic backgrounds can enable a more comprehensive understanding of the gene regulation evolutionary modes in nature.
Supplementary Material
Supplementary tables S1–S4 and figures S1–S5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors deeply appreciated the comments from several colleagues when the results were presented as a poster in SMBE, Chicago, 2013. They also thank Dr Jian Lu and Dr Xiaoqiu Liu for discussion during the implementation of the ribosomal profiling protocol, Paul Bill-Ross and Kaixiong Ye for reading the manuscript. This work was supported by startup funds from Cornell University, NSF MCB-1243588 and NIH 1R01AI085286 awarded to Z.G. X.S. is supported by a visiting student fellowship from China Scholarship Council.
Literature Cited
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribere JA, Gilbert WV. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res. 2013;23:977–987. doi: 10.1101/gr.150342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. Evolution at two levels of gene expression in yeast. Genome Res. 2014;24:411–421. doi: 10.1101/gr.165522.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple Testing. J R Stat Soc Series B Methodol. 1995;57:289–300. [Google Scholar]
- Bostrom K, et al. Pulse-chase studies of the synthesis and intracellular-transport of Apolipoprotein-B-100 in hep G2 cells. J Biol Chem. 1986;261:3800–3806. [PubMed] [Google Scholar]
- Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Mostovoy Y, Dudoit S, Brem RB. Polygenic and directional regulatory evolution across pathways in Saccharomyces. Proc Natl Acad Sci U S A. 2010;107:5058–5063. doi: 10.1073/pnas.0912959107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, et al. The molecular mechanism of a cis-regulatory adaptation in yeast. PLoS Genet. 2013;9:e1003813. doi: 10.1371/journal.pgen.1003813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate JE, Bar H, Doyle JJ. Extensive translational regulation of gene expression in an allopolyploid (Glycine dolichocarpa) Plant Cell. 2014;26:136–150. doi: 10.1105/tpc.113.119966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dori-Bachash M, Shema E, Tirosh I. Coupled evolution of transcription and mRNA degradation. PLoS Biol. 2011;9:e1001106. doi: 10.1371/journal.pbio.1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JJ, et al. Natural selection on cis and trans regulation in yeasts. Genome Res. 2010;20:826–836. doi: 10.1101/gr.101576.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fraser HB, Moses AM, Schadt EE. Evidence for widespread adaptive evolution of gene expression in budding yeast. Proc Natl Acad Sci U S A. 2010;107:2977–2982. doi: 10.1073/pnas.0912245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeberg MA, et al. Pervasive and dynamic protein binding sites of the mRNA transcriptome in Saccharomyces cerevisiae. Genome Biol. 2013;14:R13. doi: 10.1186/gb-2013-14-2-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galitski T, Saldanha AJ, Styles CA, Lander ES, Fink GR. Ploidy regulation of gene expression. Science. 1999;285:251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Pritchard JK. Revealing the architecture of gene regulation: the promise of eQTL studies. Trends Genet. 2008;24:408–415. doi: 10.1016/j.tig.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves A, et al. Extensive compensatory cis-trans regulation in the evolution of mouse gene expression. Genome Res. 2012;22:2376–2384. doi: 10.1101/gr.142281.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon KL, Ruvinsky I. Tempo and mode in evolution of transcriptional regulation. PLoS Genet. 2012;8:e1002432. doi: 10.1371/journal.pgen.1002432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. Ribosome profiling: new views of translation, from single codons to genome scale. Nat Rev Genet. 2014;15:205–213. doi: 10.1038/nrg3645. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, et al. Quantitative measurement of allele-specific protein expression in a diploid yeast hybrid by LC-MS. Mol Syst Biol. 2012;8:602. doi: 10.1038/msb.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, et al. Primate transcript and protein expression levels evolve under compensatory selection pressures. Science. 2013;342:1100–1104. doi: 10.1126/science.1242379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M-C, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics. 2010;32:11.17.11–11.17.14. doi: 10.1002/0471250953.bi1107s32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CM, Tzeng JN, Sung HM. Effects of cis and trans regulatory variations on the expression divergence of heat shock response genes between yeast strains. Gene. 2012;506:93–97. doi: 10.1016/j.gene.2012.06.034. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Byrnes JK, Hwang JK, Li WH. Codon-usage bias versus gene conversion in the evolution of yeast duplicate genes. Proc Natl Acad Sci U S A. 2006;103:14412–14416. doi: 10.1073/pnas.0606348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, et al. Regulatory divergence in Drosophila revealed by mRNA-seq. Genome Res. 2010;20:816–825. doi: 10.1101/gr.102491.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus J, May GE, Spealman P, Shteyman A. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 2014;24:422–430. doi: 10.1101/gr.164996.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. Systematic analysis of posttranscriptional gene expression. Wiley Interdiscip Rev: Syst Biol Med. 2010;2:162–180. doi: 10.1002/wsbm.54. [DOI] [PubMed] [Google Scholar]
- Muller CA, Nieduszynski CA. Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res. 2012;22:1953–1962. doi: 10.1101/gr.139477.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley BW, Towle HC, Schwartz RJ. Regulation of gene expression in eucaryotes. Annu Rev Genet. 1977;11:239–275. doi: 10.1146/annurev.ge.11.120177.001323. [DOI] [PubMed] [Google Scholar]
- Picotti P, et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;494:266–270. doi: 10.1038/nature11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman MV, Kruglyak L. Genetics of global gene expression. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- Schaefke B, et al. Inheritance of gene expression level and selective constraints on trans- and cis-regulatory changes in yeast. Mol Biol Evol. 2013;30:2121–2133. doi: 10.1093/molbev/mst114. [DOI] [PubMed] [Google Scholar]
- Shi X, et al. Cis-and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids. Nat Commun. 2012;3:950. doi: 10.1038/ncomms1954. [DOI] [PubMed] [Google Scholar]
- Springer NM, Stupar RM. Allelic variation and heterosis in maize: how do two halves make more than a whole? Genome Res. 2007;17:264–275. doi: 10.1101/gr.5347007. [DOI] [PubMed] [Google Scholar]
- Suvorov A, et al. Intra-specific regulatory variation in Drosophila pseudoobscura. PLoS One. 2013;8:e83547. doi: 10.1371/journal.pone.0083547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Inferring regulatory mechanisms from patterns of evolutionary divergence. Mol Syst Biol. 2011;7:530. doi: 10.1038/msb.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Reikhav S, Levy AA, Barkai N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–662. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N. Divergence of nucleosome positioning between two closely related yeast species: genetic basis and functional consequences. Mol Syst Biol. 2010;6:365. doi: 10.1038/msb.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Weber K. Post-translational modifications regulate microtubule function. Nat Rev Mol Cell Biol. 2003;4:938–947. doi: 10.1038/nrm1260. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Evolutionary changes in cis and trans gene regulation. Nature. 2004;430:85–88. doi: 10.1038/nature02698. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 2008;40:346–350. doi: 10.1038/ng.77. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet. 2012;13:59–69. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, et al. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Zheng W, Gianoulis TA, Karczewski KJ, Zhao HY, Snyder M. Regulatory variation within between species. Annu Rev Genomics Hum Genet. 2011;12:327–346. doi: 10.1146/annurev-genom-082908-150139. [DOI] [PubMed] [Google Scholar]
- Zhong S, et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc. 2011:940–949. doi: 10.1101/pdb.prot5652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.