Abstract
Unlike many other photosynthetic dinoflagellates, whose plastids contain a characteristic carotenoid peridinin, members of the genus Lepidodinium are the only known dinoflagellate species possessing green alga-derived plastids. However, the precise origin of Lepidodinium plastids has hitherto remained uncertain. In this study, we completely sequenced the plastid genome of Lepidodinium chlorophorum NIES-1868. Our phylogenetic analyses of 52 plastid-encoded proteins unite L. chlorophorum exclusively with a pedinophyte, Pedinomonas minor, indicating that the green-colored plastids in Lepidodinium spp. were derived from an endosymbiotic pedinophyte or a green alga closely related to pedinophytes. Our genome comparison incorporating the origin of the Lepidodinium plastids strongly suggests that the endosymbiont plastid genome acquired by the ancestral Lepidodinium species has lost genes encoding proteins involved in metabolism and biosynthesis, protein/metabolite transport, and plastid division during the endosymbiosis. We further discuss the commonalities and idiosyncrasies in genome evolution between the L. chlorophorum plastid and other plastids acquired through endosymbiosis of eukaryotic photoautotrophs.
Keywords: genome reduction, Lepidodinium, Pedinophyceae, secondary plastids, plastid replacement
Introduction
Plastids are photosynthetic organelles that are found in diverse lineages, scattered across the vast diversity of eukaryotes (Keeling 2004; Kim and Archibald 2009). A cyanobacterium-eukaryote endosymbiosis, called primary endosymbiosis, produced the first photosynthetic eukaryote (De Clerck et al. 2012), which subsequently diverged into three lineages with “primary plastids,” namely red algae, green algae/land plants, and glaucophytes (Rodriguez-Ezpeleta et al. 2005; Keeling 2010). Plastids further spread into diverse eukaryotes through multiple events involving the uptake and retention of either a green or red alga by a heterotrophic eukaryote, a phenomenon referred to as secondary endosymbiosis (Douglas 1998; Archibald 2012). Henceforth, we refer to the plastids that emerged from secondary endosymbioses as “secondary plastids.” Euglenophytes and chlorarachniophytes acquired their plastids from different endosymbiotic green algae, a prasinophyte and a member of core chlorophytes, respectively (Rogers et al. 2007; Turmel, Gagnon, et al. 2009). Thus, green alga-involved secondary endosymbiosis has occurred multiple times (at least twice) in eukaryotes. On the other hand, the vast majority of photosynthetic dinoflagellates, photosynthetic lineages basal to apicomplexan parasites (i.e., Chromera velia and Vitrella brassicaformis), haptophytes, cryptophytes, and heterokont algae, possesses red alga-derived plastids; however, it remains controversial whether their red alga-derived plastids result from a single or multiple endosymbiotic events (Archibald 2009; Keeling 2009).
Dinoflagellates are a part of a large microeukaryotic assemblage termed Alveolata, and about 50% of the known species are photosynthetic (Taylor 1987). The vast majority of the photosynthetic species possesses red alga-derived secondary plastids, which contain chlorophylls (Chls) a and c, plus a dinoflagellate-specific carotenoid, peridinin (Schnepf and Elbrachter 1999). However, some dinoflagellate species replaced the original (peridinin-containing) plastids with others bearing distinct carotenoid compositions. For instance, plastids in members of the genera Karenia, Karlodinium, and Takayama contain Chls a and c, plus 19′ hexanoyloxyfucoxanthin, instead of peridinin. The 19′ hexanoyloxyfucoxanthin-containing plastids most likely originated from an endosymbiotic haptophyte (Tengs et al. 2000; Keeling 2004). Likewise, Durinskia baltica and other closely related species possess diatom-derived plastids containing Chls a and c, plus fucoxanthin (Jeffrey et al. 1975; Keeling 2004).
The dinoflagellate genus Lepidodinium possesses plastids containing Chls a and b, which are believed to be remnants of an endosymbiotic green alga (Watanabe et al. 1990; Matsumoto et al. 2012). Prior to this study, there were two lines of studies on Lepidodinium chlorophorum: 1) A survey of green algal genes endosymbiotically transferred to the host genome, and 2) an investigation of the precise origin of the Lepidodinium plastids. First, surveys of the host nuclear transcripts in L. chlorophorum detected putative plastid genes of both green algal and peridinin-containing dinoflagellate origins. In organismal phylogenies, L. chlorophorum affiliates with lineages possessing peridinin-containing plastids, suggesting that the original plastid was replaced by a green algal endosymbiont on the branch leading to Lepidodinium spp. (e.g., Saldarriaga et al. 2001; Shalchian-Tabrizi et al. 2006). Thus, Takishita et al. (2008) and Minge et al. (2010) proposed that 1) the putative “green algal” genes were transferred from the green algal endosymbiont nucleus, and 2) the “typical dinoflagellate” plastid genes were present in the host nucleus prior to the acquisition of the green algal endosymbiont. On the other hand, studies analyzing plastid genes and pigment composition failed to resolve the precise origin of the Lepidodinium plastids (Takishita et al. 2008; Matsumoto, Shinozaki, et al. 2011; Matsumoto, Ishikawa, et al. 2011; Matsumoto et al. 2012). So far, a phylogenetic study of 11 plastid genes by Matsumoto, Shinozaki, et al. (2011) narrowed down the origin of the Lepidodinium plastids to core chlorophytes, comprising Ulvophyceae, Trebouxiophyceae, Chlorophyceae, Pedinophyceae, and Chlorodendrophyceae (Leliaert et al. 2012).
We report here the complete plastid genome of L. chlorophorum, along with phylogenetic analyses based on plastid genome data. Our phylogenetic analyses of 52 genes conserved across green algal plastid genomes suggests a robust affinity between L. chlorophorum and a pedinophyte, Pedinomonas minor. Thus, we conclude that the plastids in Lepidodinium spp. were derived from a pedinophyte (or an endosymbiotic alga closely related to pedinophytes). Our determination of the precise origin of Lepidodinium plastids enables us to discuss the possible evolution of the plastid genome in the endosymbiont. We further discuss the commonalities and idiosyncrasies in genome evolution between the L. chlorophorum plastid and other “noncanonical” dinoflagellate plastids lacking peridinin.
Materials and Methods
Plastid Genome Sequencing
Cells of L. chlorophorum NIES-1868 were cultivated in MNK medium (Matsumoto, Shinozaki, et al. 2011). Total DNA, extracted by the CTAB method (Kamikawa et al. 2009), was sequenced on an Illumina HiSeq 2000 platform. In total, 136 million paired-end reads were obtained. The first and last five bases of each read were trimmed using the FASTX-Tool kit (Ver.0.0.13) (http://hannonlab.cshl.edu/fastx_toolkit/, last accessed April 5, 2015). Approximately 130 million paired-end reads were kept, all of which had greater than 75% of bases above a quality score of 20. Genome contigs were generated using Ray (Boisvert et al. 2012). Genome scaffolding with preassembled contigs was done using SSPACE (Boetzer et al. 2011). In total, 18 contigs for putative plastid genome sequences were identified based on sequence similarity to previously determined plastid genomes. Gapped regions were filled by polymerase chain reaction and subsequent Sanger sequencing. Annotation was performed by MFANNOT (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl, last accessed October 30, 2014), RNAWEASEL (http://megasun.bch.umontreal.ca/RNAweasel/, last accessed September 23, 2014), tRNAscan-SE (Lowe and Eddy 1997), and BLAST (Basic Local Alignment Search Tool) searches of the GenBank database (http://www.ncbi.nlm.nih.gov/, last accessed October 30, 2014). The nucleotide sequence of the L. chlorophorum plastid genome was deposited to DNA Data Bank of Japan (accession no. LC008447).
Phylogenetic Analysis
For phylogenetic analysis assessing the origin of the L. chlorophorum plastid, we selected 52 plastid-encoded proteins shared among the plastid genomes of 7 chlorophytes, 5 trebouxiphytes, 4 ulvophytes, a pedinophyte (P. minor), 6 prasinophytes, a chlorarachniophyte (Bigelowiella natans), a euglenophyte (Euglena gracilis), and L. chlorophorum (26 taxa in total; supplementary table S1, Supplementary Material online). We considered plastids of the prasinophytes and the euglenophyte as the outgroup. All genome data analyzed here, except those of L. chlorophorum, were available as of August 2014. Amino acid sequences of each protein were aligned and trimmed manually. We analyzed 52 single protein alignments individually by the maximum-likelihood (ML) method, with the LG (Le and Gascuel 2008) + Γ + F model, and detected no apparent signal of nonvertical gene inheritance in any of the single-protein alignments (data not shown). We concatenated all the single-protein alignments into a single 52-protein alignment (8,917 amino acid positions in total), and subjected it to the ML method with the LG + Γ + F model. The ML tree was heuristically searched from ten distinct maximum parsimony (MP) trees. In ML bootstrap analyses (100 replicates), a heuristic tree search was performed from a single MP tree per replicate. The ML phylogenetic analyses described above were conducted by RAxML ver. 7.2.8 (Stamatakis 2006).
The 52-protein alignment was also analyzed by PhyloBayes ver. 3.3 under the CAT-GTR + Γ model (Lartillot et al. 2009). Two parallel Markov Chain Monte Carlo chains were run for 13,000 cycles, in which 500,000 trees and log-likelihoods (lnLs) were sampled. We considered independent runs converged when the maximum discrepancy observed across all bipartitions was less than 0.3, and the effective sample size was greater than 100 (Lartillot et al. 2009). The first 2,500 cycles were discarded as burn-in, and the remaining trees were summarized to obtain Bayesian posterior probabilities (BPPs).
We prepared two extra 52-protein alignments, and subjected them to the ML method as described above. The second 52-protein alignment was generated from the original alignment by adding two streptophytes and four prasinophytes (32 taxa and 8,917 amino acid positions in total). The third 52-protein alignment was identical to the original alignment, except that B. natans and E. gracilis were excluded (24 taxa and 8,917 amino acid positions in total).
Results
General Features of the L. chlorophorum Plastid Genome
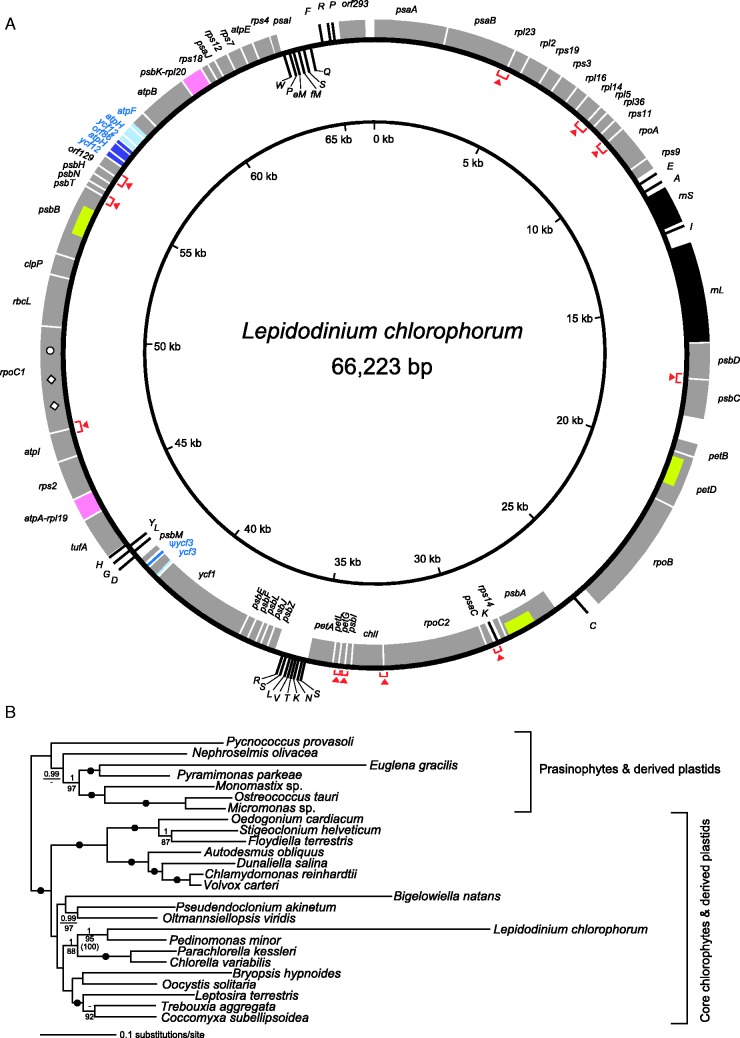
The complete sequence of the L. chlorophorum plastid genome can be mapped as a circular molecule of 66,223 bp (fig. 1A). The overall A+T content is 65.4%, and approximately 84% of the genome comprises coding region. We identified two ribosomal RNA (rRNA) genes and 27 transfer RNA (tRNA) genes, which are sufficient to translate all of the amino acid codons (supplementary table S2, Supplementary Material online). The L. chlorophorum plastid genome likely utilizes a deviant genetic code in which AUA is assigned as methionine as reported in Matsumoto, Ishikawa, et al. (2011). Out of 63 open reading frames (ORFs), putative functions for 60 ORFs were assigned based on sequence similarity (fig. 1A). Two pairs of genes appeared to be fused with each other (colored in pink in fig. 1A; see supplementary fig. S1, Supplementary Material online, for the details). All of the plastid genes listed above (except three ORFs of unknown function) had been found in green algal plastid genomes sequenced prior to this study. Notably, the L. chlorophorum plastid genome was found to possess only a single rRNA operon, contrasting to typical plastid genomes with two rRNA operons as a part of inverted repeats (IRs). We observed 11 overlapping gene-pairs (indicated by red arrowheads in fig. 1A; see supplementary fig. S2, Supplementary Material online, for the details). Three genes were found to possess introns (yellowish green boxes shown in fig. 1A), which were predicted as group II introns based on the putative secondary structure (supplementary fig. S3, Supplementary Material online; Note that no obvious ORF was found in any of the three introns, although typical group II introns carry an intronic ORF coding reverse transcriptase, maturase, and endonuclease domains [e.g., Lambowitz and Zimmerly 2004; Kamikawa et al. 2009]). We suspect that a gene-duplication has created a tandem array of ycf3-like regions (ψycf3; colored in light blue and dark blue in fig. 1A); one seemingly encodes a functional protein, and the other is most likely nonfunctional due to a large deletion (supplementary fig. S4, Supplementary Material online). Likewise, a region containing the 5′ terminal portion of atpF, and the entire atpH and ycf12 was likely duplicated in tandem (colored in light blue and dark blue in fig. 1A; Note that orf86 corresponds to the partial atpF). It remains uncertain whether rpoC1 is functional, as the corresponding ORF is interrupted by one UAG codon and two UAA codons (shown by an open circle and open diamonds, respectively, in fig. 1A; see supplementary fig. S5, Supplementary Material online, for the details). Note that both UAG and UAA codons are used as translation termination signal for other ORFs (supplementary table S2, Supplementary Material online). We confirmed that these in-frame termination codons were present on the corresponding mRNA (data not shown), indicating that these “stop” codons were not edited posttranscriptionally. Thus, rpoC1 is transcriptionally active but may be pseudogenic, raising the possibility that a functional gene copy exists in the host genome.
Fig. 1.—
Complete plastid genome of Lepidodinium chlorophorum and ML phylogeny inferred from an alignment comprising 52 plastid proteins. (A) Physical map of the L. chlorophorum plastid genome. Protein-coding and rRNA-coding regions are shown by gray and closed boxes, respectively; tRNAs and pseudogenes are shown by lines. Tandemly duplicated regions were highlighted in light blue and dark blue. Two ORFs generated by gene fusion are colored in pink (see supplementary fig. S1, Supplementary Material online, for the details). Red arrowheads indicate physically overlapping coding regions (see supplementary fig. S2, Supplementary Material online, for the details). Group II introns are depicted as yellowish green boxes (see supplementary fig. S3, Supplementary Material online, for the details). A single UAA and two UAG codons found within the rpoC1 coding region are shown by an open circle and open diamonds, respectively (see supplementary fig. S4, Supplementary Material online, for the details). (B) Phylogenetic analysis exploring the origin of the L. chlorophorum plastid. The alignment comprises 52 plastid proteins (8,917 amino acid positions in total) sampled from 26 genomes of 23 green algae and 3 green alga-derived plastids. Phylogenetic analyses were performed by both ML and Bayesian frameworks. As both methods reconstructed very similar trees, only the ML tree is shown here. BPPs and ML bootstrap values (MLBPs) are shown above and below the corresponding nodes. MLBPs less than 80% and BPPs less than 0.95 are omitted from the figure. Dots correspond to MLBPs of 100% and BPPs of 1.00. The MLBP from the analysis excluding rapidly evolving Bigelowiella natans and Euglena gracilis is shown in parentheses.
Phylogenetic Affinity between the Plastids of L. chlorophorum and P. minor
The origin of the L. chlorophorum plastids was assessed by phylogenetic analyses of the 52-protein alignment (fig. 1B). The overall phylogenetic relationship among green algal (and green alga-derived) plastids recovered in this study was compatible with those presented in previously published phylogenetic studies (e.g., Smith et al. 2011; Lemieux et al. 2014). However, our 52-protein analyses successfully resolved the precise origin of the Lepidodinium plastid genome with high statistical support. In the ML analysis with the LG + Γ + F model, L. chlorophorum grouped with P. minor with an ML bootstrap value (MLBP) of 95% (fig. 1B). The phylogenetic relationship between L. chlorophorum and P. minor was reconstructed with an MLBP of 97% in the analyses of the second 52-protein alignment expanding the outgroup taxa (supplementary fig. S6, Supplementary Material online).
We were concerned whether the position of L. chlorophorum was severely biased in tree reconstruction due to the rapidly evolving nature of its plastid-encoded proteins. To test the above possibility, we then reassessed the affinity between L. chlorophorum and P. minor by applying a site-heterogeneous substitution model claimed to be robust against long-branch attraction artifacts (Lartillot et al. 2007; Rodriguez-Ezpeleta et al. 2007). We subjected the 52-protein alignment to Bayesian analysis with a site-heterogeneous model (CAT-GTR + Γ). As the trees inferred by ML analysis with the LG + Γ + F model and by Bayesian analysis with the CAT-GTR + Γ model were very similar to each other, only BPPs were presented in figure 1B. Significantly, the Bayesian tree inferred under the site-heterogeneous model recovered the clade of L. chlorophorum and P. minor with a BPP of 1.00 (fig. 1B). Further, the relationship between L. chlorophorum and P. minor was also investigated by excluding other long-branch sequences from the 52-protein alignment. Lepidodinium chlorophorum and P. minor remained as a clade with an MLBP of 100% in the third ML analysis. As the tree topology is basically consistent with figure 1B, only the MLBP of the node uniting L. chlorophorum and P. minor is presented in parentheses (fig. 1B). As both ML and Bayesian analyses of the 52-protein alignments consistently recovered the specific affinity between L. chlorophorum and P. minor, with high statistical support, we conclude that an endosymbiotic pedinophyte or green alga closely related to pedinophytes gave rise to the current plastid in L. chlorophorum.
Discussion
The precise origin of the green-colored plastids in Lepidodinium spp. had been controversial since the initial description of these dinoflagellates (Watanabe et al. 1990). Our phylogenetic analyses, based on plastid-encoded proteins, pinpointed the progenitor of Lepidodinium plastids as a pedinophyte or an alga exclusively related to pedinophytes. Unfortunately, the results presented above cannot exclude the latter possibility. However, to our knowledge, there is little evidence implying any undescribed green alga/algae specifically related to the known pedinophytes; for this reason, we favor the explanation that Lepidodinium plastids are descended from a pedinophyte. These similar but distinguishable scenarios for the origin of Lepidodinium plastids should be revisited when plastid genome data from a broader collection of core chlorophytes become available.
Some populations of a dinoflagellate, Noctiluca scintillans, in Southeast Asian tropical seawater have been documented to possess a pedinophyte endosymbiont, Pedinomonas noctilucae (Sweeney 1971). Pedinomonas noctilucae is dispensable for N. scintillans, but supplies organic matter and facilitates host survival during starvation (Hansen et al. 2004; Saito et al. 2006). If Lepidodinium plastids are truly of pedinophyte origin, two independent dinoflagellate lineages have acquired pedinophytes as endosymbionts. Although the integration level of endosymbionts into the host system is apparently different between Lepidodinium spp. and N. scintillans, it is worth investigating whether those two independent partnerships were established on common genetic, physiological, and/or environmental grounds.
To decipher how the plastid genome was modified after the green algal endosymbiosis, we consider the P. minor plastid genome as a surrogate for the plastid genome of the green algal endosymbiont captured by the ancestral Lepidodinium species (table 1). Here, we focus on 1) the evolution of plastid genetic code and 2) genome reduction. First, the AUA codon was reassigned from isoleucine to methionine in the plastid genome after the uptake of the endosymbiont by the ancestral Lepidodinium species (Matsumoto, Ishikawa, et al. 2011), as the AUA codon specifies isoleucine in the P. minor plastid genome, along with other plastid genomes. A distinct type of deviant genetic code, in which the UGA codon assigns tryptophan, was reported in the plastid of an alveolate lineage distantly related to dinoflagellates (i.e., the chromerid C. velia and apicomplexan parasites; Moore et al. 2008). We may identify additional plastid genomes using deviant genetic codes by sequencing plastid genomes in more phylogenetically diverse eukaryotes; this would also help us to understand the evolution of plastid genetic codes more precisely. Second, plastid genome comparisons revealed postendosymbiotic genome reduction in L. chlorophorum. Overall, the P. minor plastid genome (∼98 kb) is approximately 1.5 times larger than the L. chlorophorum plastid genome (∼66 kb), regardless of the presence of group II introns and gene duplications (table 1). Gene density appeared to be higher in the L. chlorophorum plastid genome than the P. minor plastid genome; the proportion of intergenic regions in the former is smaller than in the latter (13.3 and 25.6%, respectively; Turmel, Otis, et al. 2009; this study; table 1). Eleven physically overlapping gene-pairs, as well as two gene fusions, contribute to genome compaction in the L. chlorophorum plastid genome; this is considerably more than the two overlapping gene-pairs and complete absence of gene fusion in the P. minor plastid genome (table 1). The size difference between the L. chlorophorum and P. minor plastid genomes also coincides with a difference in gene content (fig. 2; table 1). After the endosymbiosis, the L. chlorophorum plastid genome discarded 14 functionally assignable ORFs, many of which are involved in plastid division, protein/metabolite transport, and metabolism (fig. 2). Similar gene losses during secondary endosymbioses were proposed for the plastids in euglenophytes (Turmel, Gagnon, et al. 2009; Hrdá et al. 2012), chlorarachniophytes (Rogers et al. 2007; Tanifuji et al. 2014), heterokont algae (Oudot-Le Secq et al. 2007; Cattolico et al. 2008), cryptophytes (Khan et al. 2007), haptophytes (Sánchez Puerta et al. 2005), and photosynthetic lineages basal to apicomplexan parasites (Janouškovec et al. 2010). Unfortunately, our current knowledge is insufficient to clarify the precise driving force(s) for the gene losses commonly observed among these plastids. Finally, the reductive trend in the L. chlorophorum plastid genome is seemingly congruent with the absence of IRs (fig. 1A and table 1). However, it is difficult to conclude that the IRs were lost as a part of endosymbiotic genome reduction, as “IR-lacking” plastid genomes that are larger than the L. chlorophorum plastid genome have been reported. For instance, the IR-lacking plastid genome of Leptosira terrestris is 195 kb in size (de Cambiaire et al. 2007). There are also examples of plastid genomes bearing IRs that are similar in size to the L. chlorophorum plastid genome: IRs are present in the plastid genome of B. natans (69 kb; Rogers et al. 2007).
Table 1.
General Features of the Plastid Genomes in Lepidodinium chlorophorum (This Study) and Pedinomonas minor (Turmel, Otis, et al. 2009)
| Lepidodinium chlorophorum | Pedinomonas minor | |
|---|---|---|
| Length (bp) | 66,223 | 98,340 |
| IRs | Absent | Present |
| AT content (%) | 65.4 | 65.2 |
| Intergenic regions (%) | 13.3 | 25.6 |
| Protein/tRNA/rRNA genesa | 60b/27/2 | 74b/28/3 |
| Introns | 3c (psbA, psbB, and petD) | None |
| Physically overlapping gene pairs | rpl23-rpl2, rpl14-rpl5, rps11-rpoA, psaC-trnK(cuu), psbB-psbT, psbD-psbC, petL-petG, petG-psbI, chlI-rpoC2, atpI-rpoC1, and psbH-orf129 | psbD-psbC and cysA-trnG |
| Fused ORFs | rpl19-atpA, psbK-rpl20 | None |
| Duplications | ycf12-atpH-atpF and ycf3 | None |
| Genetic code | Deviant (AUA for methionine) | Standard |
aDuplicated gene copies were counted only once.
bUnconserved, species-specific ORFs were excluded.
cAll the introns found in the L. chlorophorum plastid genome are group II.
The data of the P. minor plastid genome were retrieved from Turmel, Otis, et al. (2009).
Fig. 2.—
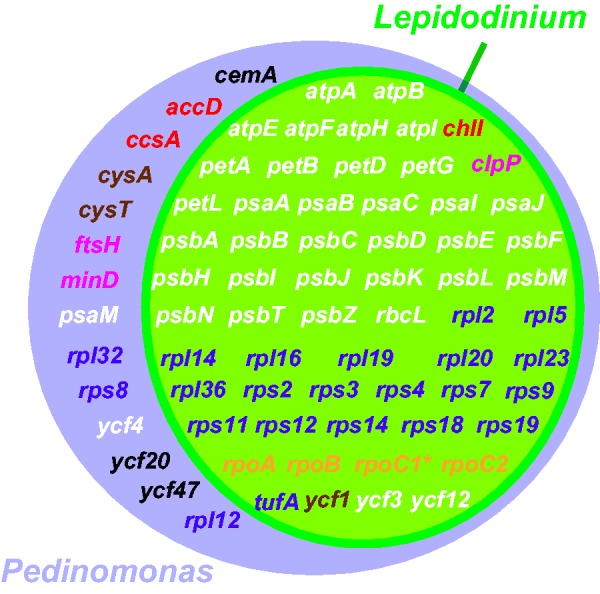
Venn diagram of the gene contents in the plastid genomes of Lepidodinium chlorophorum and Pedinomonas minor. This diagram illuminates that the protein-coding gene set in L. chlorophorum is the subset of that of P. minor. rpoC1 is highlighted by an asterisk, as this ORF is disrupted by multiple stop codons. Genes encoding proteins involved in metabolism, plastid division, translation, transcription, transport, and photosynthesis are shown in red, magenta, purple, yellow, brown, and white, respectively.
Besides the pedinophyte-derived plastids (or those derived from a green alga related to pedinophytes) in Lepidodinium spp., “noncanonical” plastids derived from haptophyte and diatom endosymbionts are known among dinoflagellates (see Introduction). Reflecting the separate origins of the three noncanonical plastids in dinoflagellates, their genomic features vary considerably. The genome of the haptophyte-derived plastid in Karlodinium veneficum lacks 39 protein-coding genes that are present in the plastid genome of the free-living haptophyte Emiliania huxleyi, suggesting that it underwent more extensive gene loss than L. chlorophorum did (Gabrielsen et al. 2011). Furthermore, mRNA editing and poly-U addition to the 3′-end of mRNA in the haptophyte-derived plastids (Dorrell and Howe 2012; Jackson et al. 2013; Richardson et al. 2014) have not been found in the green alga-derived plastid of L. chlorophorum or the diatom-derived dinoflagellate plastid (Matsumoto, Ishikawa, et al. 2011; Richardson et al. 2014). Unlike the noncanonical plastids in L. chlorophorum and K. veneficum, the genomes of diatom-derived plastids in D. baltica and Kryptoperidinium foliaceum were found to retain similar gene contents to those of free-living diatoms (Imanian et al. 2010), suggesting that no significant gene loss occurred during this endosymbiosis. As briefly overviewed above, it is difficult to extract the commonalities shared among the noncanonical dinoflagellate plastids with distinct evolutionary backgrounds. Rather, future studies need to address the reason(s) why the dinoflagellate systems can welcome diverse eukaryotic algae as endosymbionts, and integrate them as new plastids.
Supplementary Material
Supplementary tables S1 and S2 and figures S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors appreciate Dr Ryan Gawryluk (University of British Columbia) for his helpful discussion and advice on this manuscript. This work was supported by the Japanese Society for Promotion of Sciences (JSPS) awarded to R.K. (nos. 24870004, 15H05606, and 15K14591) and Y.I. (no. 23117006) and by the Institute for Fermentation, Osaka, Japan awarded to R.K.
Literature Cited
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Archibald JM. The evolution of algae by secondary and tertiary endosymbiosis. Adv Bot Res. 2012;64:87–118. [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol. 2012;13:R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattolico RA, et al. Chloroplast genome sequencing analysis of Heterosigma akashiwo CCMP452 (West Atlantic) and NIES293 (West Pacific) strains. BMC Genomics. 2008;9:211. doi: 10.1186/1471-2164-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cambiaire JC, Otis C, Turmel M, Lemieux C. The chloroplast genome sequence of the green alga Leptosira terrestris: multiple losses of the inverted repeat and extensive genome rearrangements within the Trebouxiophyceae. BMC Genomics. 2007;8:213. doi: 10.1186/1471-2164-8-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clerck O, Bogaret K, Leliaert F. Diversity and evolution of algae: primary endosymbiosis. Adv Bot Res. 2012;64:55–86. [Google Scholar]
- Dorrell RG, Howe CJ. Functional remodeling of RNA processing in replacement chloroplasts by pathways retained from their predecessors. Proc Natl Acad Sci U S A. 2012;109:18879–18884. doi: 10.1073/pnas.1212270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE. Plastid evolution: origins, diversity, trends. Curr Opin Genet Dev. 1998;8:655–661. doi: 10.1016/s0959-437x(98)80033-6. [DOI] [PubMed] [Google Scholar]
- Gabrielsen TM, et al. Genome evolution of a tertiary dinoflagellate plastid. PLoS One. 2011;6:e19132. doi: 10.1371/journal.pone.0019132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen PJ, Miranda L, Azanza R. Green Noctiluca scintillans: a dinoflagellate with its own greenhouse. Mar Ecol Prog Ser. 2004;275:79–87. [Google Scholar]
- Hrdá Š, Fousek J, Szabová J, Hampl V, Vlček Č. The plastid genome of Eutreptiella provides a window into the process of secondary endosymbiosis of plastid in euglenids. PLoS One. 2012;7:e33746. doi: 10.1371/journal.pone.0033746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanian B, Pombert JF, Keeling PJ. The complete plastid genomes of the two “dinotoms” Durinskia baltica and Kryptoperidinium foliaceum. PLoS One. 2010;5:e10711. doi: 10.1371/journal.pone.0010711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C, Gornic SC, Waller RF. A tertiary plastid gains RNA editing in its new host. Mol Biol Evol. 2013;30:788–792. doi: 10.1093/molbev/mss270. [DOI] [PubMed] [Google Scholar]
- Janouškovec J, Horák A, Oborník M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey SW, Sielicki M, Haxo FT. Chloroplast pigment patterns in dinoflagellates. J Phycol. 1975;11:374–384. [Google Scholar]
- Kamikawa R, et al. Mitochondrial group II introns in the raphidophycean flagellate Chattonella spp. suggest a diatom-to-Chattonella lateral group II intron transfer. Harmful Algae. 2009;160:364–375. doi: 10.1016/j.protis.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Diversity and evolutionary history of plastids and their hosts. Am J Bot. 2004;91:1481–1493. doi: 10.3732/ajb.91.10.1481. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot Microbiol. 2009;56:1–8. doi: 10.1111/j.1550-7408.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan H, et al. Plastid genome sequence of the cryptophyte alga Rhodomonas salina CCMP1319: lateral transfer of putative DNA replication machinery and a test of chromist plastid phylogeny. Mol Biol Evol. 2007;24:1832–1842. doi: 10.1093/molbev/msm101. [DOI] [PubMed] [Google Scholar]
- Kim E, Archibald JM. Diversity and evolution of plastids and their genomes. In: Aronsson H, Sandelius AS, editors. The chloroplast-interactions with the environment. Berlin (Germany): Springer-Verlag; 2009. pp. 1–39. [Google Scholar]
- Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(Suppl. 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Lepage T, Blanquart S. PHYLOBAYES 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25:2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Leliaert F, et al. Phylogeny and molecular evolution of the green algae. CRC Crit Rev Plant Sci. 2012;31:1–46. [Google Scholar]
- Lemieux C, Otis C, Turmel M. Six newly sequenced chloroplast genomes from prasinophyte green algae provide insights into the relationships among prasinophyte lineages and the diversity of streamlined genome architecture in picoplanktonic species. BMC Genomics. 2014;15:857. doi: 10.1186/1471-2164-15-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Ishikawa SA, Hashimoto T, Inagaki Y. A deviant genetic code in the green alga-derived plastid in the dinoflagellate Lepidodinium chlorophorum. Mol Phylogenet Evol. 2011;60:68–72. doi: 10.1016/j.ympev.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Shinozaki F, et al. Green-colored plastids in the dinoflagellate genus Lepidodinium are of core chlorophyte origin. Protist. 2011;162:268–276. doi: 10.1016/j.protis.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Kawachi M, Miyashita H, Inagaki Y. Prasinoxanthin is absent in the green-colored dinoflagellate Lepidodinium chlorophorum strain NIES-1868: pigment composition and 18S rRNA phylogeny. J Plant Res. 2012;125:705–711. doi: 10.1007/s10265-012-0486-6. [DOI] [PubMed] [Google Scholar]
- Minge MA, et al. A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum. BMC Evol Biol. 2010;10:191. doi: 10.1186/1471-2148-10-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- Oudot-Le Secq MP, et al. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol Genet Genomics. 2007;277:427–439. doi: 10.1007/s00438-006-0199-4. [DOI] [PubMed] [Google Scholar]
- Richardson E, Dorrell RG, Howe CJ. Genome-wide transcript profiling reveals the coevolution of plastid gene sequences and transcript processing pathways in the fucoxanthin dinoflagellate Karlodinium veneficum. Mol Biol Evol. 2014;31:2376–2386. doi: 10.1093/molbev/msu189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, et al. Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes. Curr Biol. 2005;15:1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, et al. Detecting and overcoming systematic errors in genome-scale phylogenies. Syst Biol. 2007;56:389–399. doi: 10.1080/10635150701397643. [DOI] [PubMed] [Google Scholar]
- Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- Saito H, Furuya K, Lirdwitayaprasit T. Photoautotrophic growth of Noctiluca scintillans with the endosymbiont Pedinomonas noctilucae. Plankton Benthos Res. 2006;1:97–101. [Google Scholar]
- Saldarriaga JF, Taylor FJR, Keeling PJ, Cavalier-Smith T. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J Mol Evol. 2001;53:204–213. doi: 10.1007/s002390010210. [DOI] [PubMed] [Google Scholar]
- Sánchez Puerta MV, Bachvaroff TR, Delwiche CF. The complete plastid genome sequence of the haptophyte Emiliania huxleyi: a comparison to other plastid genomes. DNA Res. 2005;12:151–156. doi: 10.1093/dnares/12.2.151. [DOI] [PubMed] [Google Scholar]
- Schnepf E, Elbrachter M. Dinophyte chloroplasts and phylogeny: a review. Grana. 1999;38:81–97. [Google Scholar]
- Shalchian-Tabrizi K, et al. Combined heat shock protein 90 and ribosomal RNA sequence phylogeny supports multiple replacements of dinoflagellate plastids. J Eukaryot Microbiol. 2006;53:217–224. doi: 10.1111/j.1550-7408.2006.00098.x. [DOI] [PubMed] [Google Scholar]
- Smith DR, et al. The GC-rich mitochondrial and plastid genomes of the green alga Coccomyxa give insight into the evolution of organelle DNA nucleotide landscape. PLoS One. 2011;6:e23624. doi: 10.1371/journal.pone.0023624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAXML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Sweeney BM. Laboratory studies of a green Noctiluca from New Guinea. J Phycol. 1971;7:53–58. [Google Scholar]
- Takishita K, et al. Origins of plastids and glyceraldehyde-3-phosphate dehydrogenase genes in the green-colored dinoflagellate Lepidodinium chlorophorum. Gene. 2008;410:26–36. doi: 10.1016/j.gene.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Tanifuji G, et al. Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: convergent reductive evolution and frequent recombination in nucleomorph-bearing algae. BMC Genomics. 2014;15:374. doi: 10.1186/1471-2164-15-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor FJRE. 1987 The biology of dinoflagellates. Oxford: Blackwell Scientific Publications. [Google Scholar]
- Tengs T, et al. Phylogenetic analyses indicate that the 19′-hexanoyloxy-fucoxanthin containing dinoflagellates have tertiary plastids of haptophyte origin. Mol Biol Evol. 2000;17:718–729. doi: 10.1093/oxfordjournals.molbev.a026350. [DOI] [PubMed] [Google Scholar]
- Turmel M, Gagnon MC, O’Kelly CJ, Otis C, Lemieux C. The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol. 2009;26:631–648. doi: 10.1093/molbev/msn285. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The chloroplast genomes of the green algae Pedinomonas minor, Parachlorella kessleri, and Oocystis solitaria reveal a shared ancestry between the Pedinomonadales and Chlorellales. Mol Biol Evol. 2009;26:2317–2331. doi: 10.1093/molbev/msp138. [DOI] [PubMed] [Google Scholar]
- Watanabe MM, Suda S, Inouye I, Sawaguchi T, Chihara M. Lepidodinium viride gen. et sp. nov. (Gymnodiniales, Dinophyta), a green dinoflagellate with a chlorophyll a- and b-containing endosymbiont. J Phycol. 1990;26:741–751. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.