Abstract
During a survey of human saliva by a top-down reversed-phase high-performance liquid chromatography with electrospray ionization mass spectrometry approach, two proteins eluting at 27.4 and 28.4 min, with average masses of 15 494 ± 1 and 11 142 ± 1 Da, were detected in a subject from Boston. The Δmass value (4352 Da) of the two proteins was similar to the difference in mass values between intact (150 amino acids, [a.a.]) and truncated acidic proline-rich proteins (aPRPs; 106 a.a.) suggesting an a.a. substitution in the first 106 residues resulting in a strong reduction in polarity, since under the same experimental conditions aPRPs eluted at ~22.5 min (intact) and 23.5 min (truncated forms). Manual inspection of the high-resolution high-performance liquid chromatography with electrospray ionization tandem mass spectra of the truncated isoform showed the replacement of the phosphorylated Ser-22 in PRP-3 with a Phe residue. Inspection of the tandem mass spectra of the intact isoform confirmed the substitution, which is allowed by the code transition TCT→TTT and is in agreement with the dramatic increase in elution time. The isoform was also detected in two other subjects, one from Boston (unrelated to the previous) and one from Rome. For this reason we propose to name this variant PRP-1 (PRP-3) RB (Roma-Boston) Ser22(phos)→Phe.
Keywords: Acidic proline-rich proteins, Human saliva, Saliva polymorphism, Variants
1 Introduction
Acidic proline-rich proteins (aPRPs) represent one of the major families of human salivary proteins (15–20% w/w of the total) [1]. aPRPs are absorbed selectively onto hydroxyapatite [1–3] and are involved in the modulation of oral calcium phosphate chemistry [1]. Moreover, they are probable modulators of the bacterial colonization of the teeth [4] and participate in the formation of the acquired enamel pellicle [5,6]. They are polymorphic, being secreted by two poly-allelic loci, named PRH1 and PRH2, localized on chromosome 12p13.2 [7, 8]. In the majority of the Western population the PRH2 locus is bi-allelic and responsible for the expression of the PRP-1 and PRP-2 isoforms, while the PRH1 locus is tri-allelic and responsible for the expression of Pif-s (Parotid IEF variant, slow), Db-s (double band, slow), and Pa (parotid isoforms). These names derive from electrophoretic and IEF separation of human salivary proteins and are somewhat misleading because all five isoforms (when present) are secreted both by parotid (about 75%) and submandibular/sublingual (about 25%) glands [9]. PRP-1, PRP-2, PIF-s, and Pa isoforms are 150 amino acid (a.a.) residues long, while Db-s is 171 a.a. residues long due to the insertion of a 21 a.a. repeat domain after position 81 of the other isoforms. PRP-1 differs from PRP-2 by the substitution Asp→Asn at position 50, and PIF-s, Pa and Db-s differ from PRP-2 for the substitution Asp→Asn at residue 4 [10]. Db-s and Pa differ from the other isoforms by the substitution Leu→Ile at residue 27. Moreover, the Pa isoform displays a cysteine instead of an arginine at position 103. Further aPRP polymorphisms, although structurally not defined, have been previously described [11–13].
PRP-1, PRP-2, PIF-s, and Db-s isoforms are cleaved after Arg-106 generating a 44 a.a. residue peptide, named P-C peptide, common to all the four isoforms, and giving rise to four different truncated derivatives named PRP-3, PRP-4, parotid IEF variant, fast (Pif-f) (106 a.a. residues), and Db-f (127 a.a. residues). The proteinase responsible for this cleavage recognizes the . . . Arg-Pro-Pro-Arg↓consensus sequence at position 103–106 (124–127 in the Db-s isoform) and probably belongs to the class of convertases, enzymes working in the Golgi apparatus during the secretion process of many endocrine and exocrine glands and tissues [14]. The Pa isoform is not prone to this cleavage due to the Arg-103→Cys substitution that eliminates the consensus sequence for convertase recognition. It is commonly present in human saliva as a Pa-dimer, generated by the formation of a disulfide bond between two Cys-103 residues [15].
The acidic character of aPRPs is confined to the N-terminal 30 a.a. residue sequence, where many aspartic and glutamic acid residues are present. The remaining part is basic and, similar to the basic PRPs, shows repeated sequences of prolines and glutamines, often separated by glycine residues. All aPRPs, whether intact or truncated, are phosphorylated at Ser-8 and Ser-22 by a Golgi casein kinase recently recognized as Fam20C [16, 17] and display a pyroglutamic residue at the N terminus. However, minor amounts of tri-phosphorylated (Ser-8, -17, -22), as well as mono-phosphorylated isoforms (either Ser-8 or Ser-22), are always detected in human whole saliva, while nonphosphorylated derivatives are detected sporadically and in negligible amounts [15].
The characterization of a new salivary acidic proline-rich protein isoform by high-resolution HPLC–ESI-MS and MS/MS is described in this study. The isoform shows a low frequency being detected in only two subjects living in Boston and in a subject living in Rome out of a survey of more than 200 subjects. This isoform expands the amazing number of polymorphisms within the aPRP protein family [7], and may have to be taken into account in future functional and genetic studies.
2 Materials and methods
2.1 Reagents and equipment
Chemicals and reagents were all LC–MS grade and purchased from J. T. Baker (Deventer, the Netherlands), Merck (Darmstadt, Germany) and Sigma-Aldrich (St. Louis, MO, USA). The low-resolution HPLC–ESI-IT-MS apparatus was a Surveyor HPLC system (Thermo Fisher Scientific, San Jose, CA, USA) connected via a T splitter to a PDA diode-array detector and to an Advantage mass spectrometer. The mass spectrometer was equipped with an ESI source. The chromatographic column was a Zorbax SB300 C8 (Agilent) column, with 5 μm particle diameter (column dimensions 150 × 2.1 mm). The high-resolution HPLC–ESI-MS apparatus was an Ultimate 3000 Micro HPLC apparatus (Dionex, Sunnyvale, CA, USA) equipped with a FLM-3000-Flow manager module coupled to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific). In this case a Zorbax SB300 C8 (Agilent) column (3.5 μm particle diameter; column dimension 150 ×1.0 mm) was utilized.
2.2 Sample collection
Whole saliva (from 0.2 to 1.0 mL) was collected between 10.00 and 12.00 a.m. A total of 50 samples were collected at the Boston University. Saliva was collected under stimulatory conditions. Whole saliva flow was stimulated by asking donors to chew on a 25 square inch piece of parafilm. All donors were healthy, Caucasian, 35 females and 15 males aged between 21 and 65 years. One hundred and sixty samples of resting saliva were collected from 96 healthy subjects living in Rome (54 females and 42 males aged between 23 and 60 years) and from 64 healthy subjects living in Cagliari (36 females and 28 males, aged between 21 and 58 years). All the subjects in Rome and Cagliari were Caucasian. Samples were collected at least 30 min after tooth brushing and/or food/beverage consumption. After collection salivary samples were immediately mixed with an equal volume of 0.2% 2,2,2-TFA v/v in an ice bath. After stirring, the acidic solution was centrifuged at 9000 × g for 5 min to remove the precipitate and the acidic clear solution was either immediately analyzed by HPLC–ESI-MS (100 μL, corresponding to 50 μL of saliva) or freeze-dried and stored at −80°C. The study protocol and written consent forms were approved by the Medical Ethics Committee of the Faculty of Medicine of the Catholic University of Rome and the Institutional Review Board at Boston University Medical Campus. The informed consent procedures at both universities are consistent with the latest stipulations established by the Declaration of Helsinki.
2.3 RP-HPLC–ESI-MS analysis
The following solutions were utilized for the low resolution chromatographic separation: (eluent A) 0.050% aqueous TFA and (eluent B) 0.050% TFA in acetonitrile/water 80:20 v/v. The gradient applied was linear from 0 to 55% in 40 min, at a flow rate of 0.30 mL/min. The T splitter provided a flow-rate of about 0.20 mL/min toward the diode array detector and 0.10 mL/min toward the ESI source. During the first 5 min of separation the eluate was not applied to the mass spectrometer to avoid instrument damage due to the high salt concentration. The diode array detector was set at a wavelength of 214 and 276 nm. Mass spectra were collected every 3 ms in the positive ion mode. MS spray voltage was 4.50 kV and the capillary temperature was 250°C.
High-resolution HPLC–ESI-MS/MS experiments were performed using (A) 0.1% v/v aqueous formic acid and (B) 0.1% v/v formic acid in acetonitrile. The applied gradient was: 0–4 min 5%B, 4–38 min from 5 to 50% B (linear), 38–41 min from 50 to 90% B (linear), at a flow rate of 80 μL/min. Mass spectra were collected in a data-dependent scan (MS/MSdata) mode with a capillary temperature of 250°C, a sheath gas flow of 18 arbitrary units, a source voltage of 3.6 kV and a capillary voltage of 40 V. Measurements were performed in the positive ion mode and mass accuracy was calibrated before measurements. Selected protein charge states were isolated with a width of 6–10 m/z units and activated for 30 ms using 35% normalized collision energy and an activation q value of 0.25.
2.4 Anionic gel electrophoresis
Anionic gel electrophoresis was conducted as previously described [1, 18, 19] using the mini-gel system (Bio-Rad, Hercules, CA, USA) and spacers of 1.5 mm thickness. The separating gel contained 7.5% w/v acrylamide. After electrophoresis at a constant voltage of 120 V, the proteins were stained with 0.5% Amido Black in 7% w/v glacial acetic acid for 16 h, followed by destaining for 4–8 h in 7% acetic acid.
2.5 HPLC–ESI-MS and MS/MS data analysis
ESI-MS and MS/MS spectra were deconvoluted by the Xtract procedure resident in Xcalibur 2.07 version, using the following parameters: resolution 100 000, S/N threshold 2, fit factor 44%, remainder 25%, and averagine-no sulfur isotope distribution for protein masses from 500 to 150 000. An averagine is a virtual protein that has an average natural C, H, O, N, and S content. Spectra were manually investigated.
3 Results
During a survey of human whole saliva by a top-down low-resolution HPLC–ESI-MS platform, two unknown proteins with average masses of 15 494 ± 1 and 11 142 ± 1 Da, eluting at 27.4 and 28.4 min, respectively, were detected in the chromatographic profile of a subject living in Boston (Fig. 1). Although the elution times of the two proteins were far from those displayed under the same chromatographic conditions (22.5 and 23.5 min) by the known aPRPs, the mass difference of 4352 ± 1 Da coincided with the mass difference of intact and truncated aPRPs (4352 Da), suggesting that the protein with an average mass of 11 142 ± 1 Da could derive from the larger one by the loss of the P-C peptide (C-terminal 44 residues). Building on this hypothesis, the two unknown proteins could be aPRP isoforms with a.a. substitutions in the first 106 residues. The apparent reduction in mass by 20–21 Da between the newly discovered proteins and the well known representatives of the aPRP family did not correspond to any common a.a. substitution compatible with the decrease in polarity and concomitant increase in retention time observed.
Figure 1.
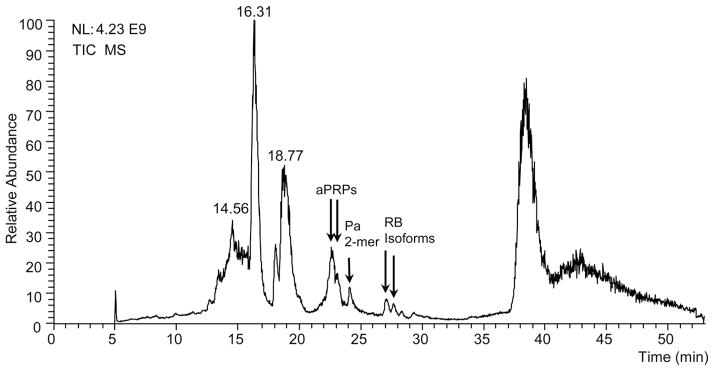
HPLC–MS total ion current (TIC) profile of the acidic-soluble fraction of saliva from a subject showing the PRP-1 RB variant. The arrows indicate the elution range of known aPRPs (in this subject: entire and truncated Pif and PRP-2 forms), of Pa 2-mer and of the RB variant (entire and truncated forms).
High-resolution HPLC–ESI-MS/MS analysis and manual inspection of the MS/MS fragmentation spectra of different multiply charged ions of the truncated isoform allowed us to establish that the a.a. substitution was Ser22 (Phos)→ Phe. This also confirmed that the two unknown proteins were aPRPs. This substitution affects one of the two phosphorylated serine residues in aPRPs, in agreement with a mass difference of 19.93 Da consistent with a substitution of Ser(Phos)→Phe and the dramatic change in polarity. The Ser→Phe substitution is caused by a single nucleotide polymorphism resulting from the code transition TCT→TTT.
Figure 2 shows the PRP3 RB variant sequence (one letter code). The bars on the sequence indicate all the fragmentation sites generating the b and y ions in the MS/MS experiments. The filled squares correspond to the b and y fragments observed in each of the seven MS/MS spectra acquired on the ions charged from +7 to +13 of the PRP3 RB variant. The m/z value of each ion is shown on the left only for the b series. The difference between experimental and theoretical data was <0.1 Da for all the ions. Data in the figure indicate that: (i) fragments of the y series were more abundant than those of the b series; (ii) different from what was expected, the less charged ions generated a richer fragmentation pattern, that is, the y85 fragment ion ([M + H]1+ (monoisotopic) exp. 8847.36 ± 0.05; theor. 8847.22 m/z), relevant for the substitution recognition, was observed in the fragmentation spectrum of the [M + 8H]8+ ion (1393.65 m/z); (iii) the isoform was a variant of the aPRP-3 isoform (PRH2 locus). In fact, the 14 fragment ions from b5 (exp. 584.22 ± 0.01, theor. 584.22 m/z) to b41 (exp. 4560.99 ± 0.02, theor. 4560.98 m/z) were in agreement with an Asp residue at position 4, and the 33 fragment ions from y57 (exp. 5849.89 ± 0.02, theor. 5849.97 m/z) to y102 (exp. 10 681.99 ± 0.04, theor. 10 691.98 m/z) were in agreement with an Asn residue at position 50. Moreover, the m/z value of the singly charged ion of the truncated isoform ([M + H]1+ 11 136.17 ± 0.04 m/z) obtained by deconvolution of the ESI spectrum perfectly agreed with the theoretical value of the PRP-3 isoform ([M + H]1+ 11 136.15 m/z); (iv) only few fragment ions (not reported in the Figure) at low intensity of the b – H3PO4 series and none of the y – H3PO4 series were detected, suggesting that in the CID fragmentation of a large phosphorylated peptide/protein the neutral loss of phosphoric acid is not a dominant event. The MS/MS spectrum generated from the multiply charged ions of the entire isoform (150 a.a. residues; not reported) was less rich, however some fragment ions confirmed the structure: 1) y129 (exp. 13 197.40 ± 0.04, theor. 13 175.39 m/z) confirmed the Phe residue in position 22; 2) 18 y fragment ions from y101 (exp. 10 200.06 ± 0.04, theor. 10 200.04 m/z) to y143 (exp. 14 689.03 ± 0.04, theor. 14 689.00 m/z) confirmed the Asn residue in position 50 and the unique phosphorylation site on Ser-8; 3) 14 b fragment ions from b5 (exp. 584.22 ± 0.01, theor. 584.22 m/z) to b41 (exp. 4561.00 ± 0.02, theor. 4560.98 m/z) confirmed the Asp residue in position 4.
Figure 2.
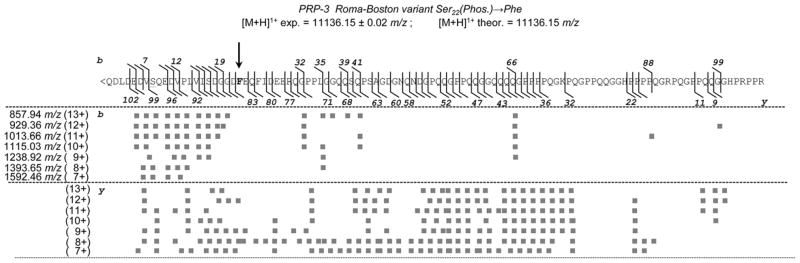
In the top of the figure the PRP3 RB variant sequence (one letter code) is reported. The bars on the sequence indicate all the fragmentation sites generating the b and y ions in the MS/MS experiments. The filled squares correspond to the b and y fragments observed in each of the seven MS/MS spectra acquired on the ions charged from +7 to +13 of the PRP3 RB variant. The m/z value of each ion is shown on the left only for the b series. The difference between experimental and theoretical data was <0.1 Da for all the ions.
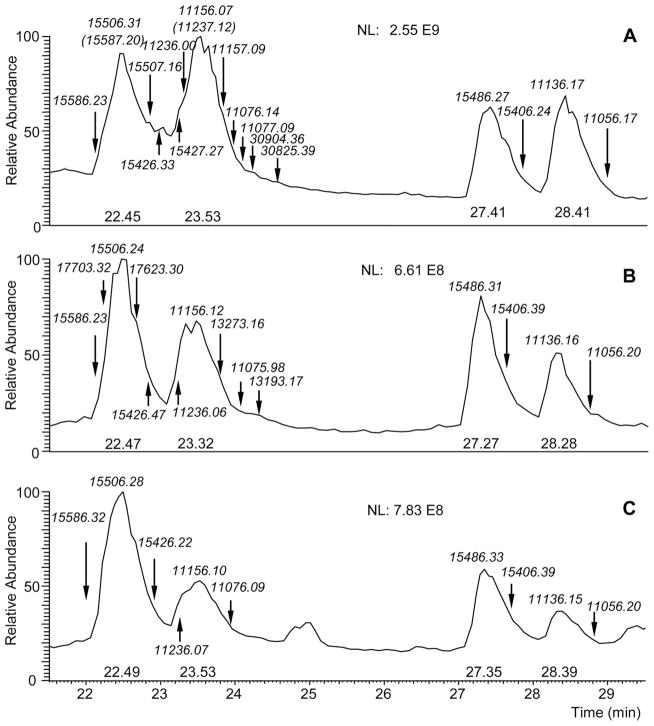
The same isoforms were subsequently detected in two more subjects, one from Boston and one from Rome. For this reason we propose to call this aPRP isoform PRP-1 (PRP-3) RB (Roma-Boston) S22(phos)→Phe. Figure 3 shows a portion of the high-resolution HPLC–ESI-MS TIC profile of the salivary samples from the three subjects, with the monoisotopic [M + H]1+ m/z values and the position of the aPRPs isoforms detected. The list of all the aPRPs isoforms and them/z values of the [M + H]1+ ions detected in the HPLC–ESI-MS profiles of the three subjects are also reported and compared with the theoretical values in Table 1. They allowed defining the allelic arrangement of the aPRPs in the three subjects as follows:
Figure 3.
Enlargement of the high-resolution HPLC–ESI-MS TIC profile of saliva from the three subjects showing the Roma-Boston variant, with the monoisotopic m/z values of the [M + H]1+ ions and the position of the aPRPs isoforms detected. Panel A: Subject Boston I; Panel B: Subject Boston II; Panel C: Subject Roma.
Table 1.
Monoisotopic m/z values of the [M + H]1+ ions of the acidic PRPs detected in three subjects carrying the PRP-1 (PRP-3) RB Ser22(Phos)→Phe variant (see also Fig. 3)
| aPRPs | Subject Boston I | Subject Boston II | Subject Roma | |
|---|---|---|---|---|
| [M + H]1+ Exp. | [M + H]1+ Exp. | [M + H]1+ Exp. | [M + H]1+ Theor. | |
| PRP-1 mono-P | – | 15 426.47 | 15 426.22 | 154 26.27 |
| PRP-1 di-P | – | 15 506.24 | 15 506.28 | 15 506.24 |
| PRP-1 tri-P | – | 15 586.23 | 15 586.32 | 15 586.21 |
| PRP-2 mono P | 15 427.27 | – | – | 15 427.26 |
| PRP-2 di-P | 15 507.16 | – | – | 15 507.22 |
| PRP-2 tri-P | 15 587.20 | – | – | 15 587.19 |
| Pif-s mono P | 15 426.33 | 15 426.47 | 15 426.22 | 15 426.27 |
| Pif-s di P | 15 506.31 | 15 506.24 | 15 506.28 | 15 506.24 |
| Pif-s tri P | 15 586.23 | 15 586.23 | 15 586.32 | 15 586.21 |
| Db-s mono P | – | – | – | 17 543.31 |
| Db-s di P | – | 17 623.30 | – | 17 623.28 |
| Db-s tri P | – | 17 703.32 | – | 17 703.25 |
| Pa 2-mer tri P | 30 825.39 | – | – | 30 824.32 |
| Pa 2-mer tetra P | 30 904.36 | – | – | 30 904.28 |
| RB (PRP-1) non P | 15 406.24 | 15 406.39 | 15 406.39 | 15 406.34 |
| RB (PRP-1) mono P | 15 486.27 | 15 486.31 | 15 486.33 | 15 486.31 |
| RB (PRP-1) di P | – | – | – | 15 566.27 |
| PRP-3 mono P | – | 11 075.98 | 11 076.09 | 11 076.11 |
| PRP-3 di P | – | 11 156.12 | 11 156.10 | 11 156.08 |
| PRP-3 tri P | – | 11 236.06 | 11 236.07 | 11 236.04 |
| PRP-4 mono P | 11 077.09 | – | – | 11 077.09 |
| PRP-4 di P | 11 157.09 | – | – | 11 157.06 |
| PRP-4 tri P | 11 237.12 | – | – | 11 237.03 |
| Pif-f mono P | 11 076.14 | 11 075.98 | 11 076.09 | 11 076.11 |
| Pif-f di P | 11 156.07 | 11 156.12 | 11 156.10 | 11 156.08 |
| Pif-f tri P | 11 236.00 | 11 236.06 | 11 236.07 | 11 236.04 |
| Db-f mono P | – | 13 193.17 | – | 13 193.15 |
| Db-f di P | – | 13 273.16 | – | 13 273.12 |
| Db-f tri p | – | – | – | 13 353.08 |
| RB (PRP-3) non P | 11 056.17 | 11 056.20 | 11 056.20 | 11 056.18 |
| RB (PRP-3) mono P | 11 136.17 | 11 136.16 | 11 136.15 | 11 136.15 |
| RB (PRP-3) di P | – | – | – | 11 216.12 |
| P-C peptide | 4369.19 | 4369.19 | 4369.19 | 4369.18 |
Subject Boston I (healthy male, 62 years old, Caucasian)
This subject, in addition to the PRP-1 RB isoform, displayed m/z values of the [M + H]1+ ions exactly matching PRP-2 and PRP-4. Furthermore, the subject displayed m/z values of the [M + H]1+ ions corresponding to Pif-s and Pif-f and the Pa 2-mer (Fig. 3, panel A). Therefore, the subject’s aPRP phenotype is:
Locus PRH2: PRP-2 and PRP1 RB heterozygous.
Locus PRH1: Pif and Pa heterozygous.
Subject Boston II (healthy male, 24 years old, Caucasian)
This subject, in addition to the PRP-1 RB isoform, displayed m/z values of the [M + H]1+ ions matching PRP-1 and PRP-3 as well as Pif-s and Pif-f. Furthermore, the subject displayed m/z values of the [M + H]1+ ions corresponding to Db-s and Db-f (Fig. 3, panel B). Therefore, the donor’s aPRP phenotype is:
Locus PRH2: PRP-1 and PRP1 RB heterozygous.
Locus PRH1: Pif and Db heterozygous.
Subject Roma (healthy female, 57 years old, Caucasian)
This subject, in addition to the PRP-1 RB isoform, displayed m/z values of the [M + H]1+ ions exactly matching PRP-1 and PRP-3 as well as Pif-s and Pif-f (Fig. 3, panel C). Therefore, the subject’s aPRP phenotype is:
Locus PRH2: PRP-1 and PRP1 RB heterozygous.
Locus PRH1: Pif homozygous.
Based on the results above, and the population of 210 subjects sampled, the allele frequency of the RB variant was estimated to be 3/420 = 0.0071.
The RB isoform was also detectable by anionic polyacrylamide gel electrophoresis as shown in Fig. 4, in which the saliva from the subject loaded in lane 5 contained the PRP RB variant (Boston II subject). The lower electrophoretic mobilities of the PRP1/PRP3 RB variants compared to PRP1/PRP3 is consistent with the substitution of the negatively charged phosphorylated Ser residue at position 22 with a neutral a.a.
Figure 4.
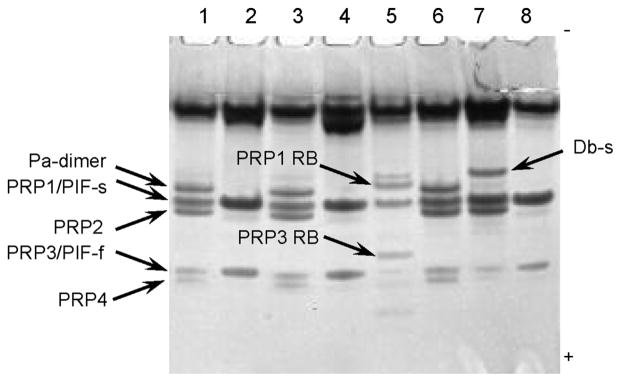
Anionic PAGE of proteins in parotid saliva from eight healthy subjects (50 μL per lane). One of the subjects in this pool (subject 5, Boston II) carried the PRP1/PRP3 RB variants. The position of the common aPRP isoforms were determined by reference to purified protein standards (not shown). The lower electrophoretic mobilities of the PRP1/PRP3 RB variants compared to PRP1/PRP3 is consistent with the substitution of the negatively charged Ser(Phos) residue at position 22 with a neutral a.a.
4 Conclusions
This study at first confirms that high-resolution ESI-MS/MS carried out on intact protein (top-down strategy) can characterize subtle modifications present in the protein primary structure [20–22]. With regard to aPRPs polymorphisms, the possible allelic combinations of the most common aPRP isoforms generate 18 different phenotypes. Concerning the products of the PRH-1 locus, the PIF allele is more frequent (about 66% in the adult population) than Pa and Db alleles (about 18 and 15%, respectively) in the Caucasian population [13, 15, 23]. Therefore, PIF homozygosis is the most common phenotype detectable in adults (about 45%), followed by PIF-Pa heterozygosis (24%), PIF-Db heterozygosis (20%), Db-Pa heterozygosis (5%), and Pa (3%) and Db (2%) homozygosis. Concerning the PRH-2 locus, the frequency of the PRP-1 allele is about 71% and of PRP-2 allele is 28%. Therefore, PRP-1 homozygosis is the most common phenotype (50%) followed by PRP-1/PRP-2 heterozygosis (40%) and PRP-2 homozygosis (8%). All subjects were heterozygous for the mutation and based on the populations studied in Rome and Boston the frequency of the mutated gene was found to be 0.0071. The frequency may obviously differ in other geographical areas.
Previous detection of aPRP polymorphisms was reported by Shintani et al. [11, 12] in the Asian population and by Hay et al. in the Western-European population [13] without characterization of the specific isoforms. It is entirely possible that the chromatographically characterized peaks of X-s and X-f by Hay et al. [13] correspond to the PRP1/PRP3 BR Ser22(phos)→Phe variants structurally characterized in this study.
The substitution involves one of the two phosphorylated serine residues of PRP-1. Elimination of one of the two phosphoserines and the resultant reduction in negative protein charge could significantly modify the functional properties of the aPRPs, which are strongly associated with the N-terminal negative domain comprising residues 1–30 [3, 5, 6]. However, in all subjects identified so far, the isoform is present only in one copy, and compensation by the common, doubly phosphorylated isoform can be expected. Moreover, acidic PRPs are represented by multiple isoforms, providing a high level of redundancy. Thus, a significant functional role for the identified single nucleotide substitution is difficult to conceive. It is interesting to note that we postulated previously that phosphorylation of the third minor site, Ser17, is hierarchical, with phosphorylation of Ser22 being mandatory for the Fem20C Golgi casein kinase recognition and subsequent phosphorylation of Ser17 [15]. Consistent with this, the diphosphorylated RB isoform with phosphorylated Ser 8 and Ser 17 was not detected in any of the three subjects.
Acknowledgments
The authors gratefully acknowledge the financial support of Cagliari University, Catholic University of Rome, MIUR, Italian National Research Council (CNR), Regione Sardegna and Nando Peretti Foundation, NIH/NIDCR/NIAID (grants DE05672 (FO), DE07652 (FO), AI087803 (EJH) and AI101067 (EJH)), according to their programs of scientific diffusion.
Abbreviations
- a.a
amino acid
- aPRP
acidic proline-rich protein
- Db-s (Db-f)
double band, slow (fast)
- Pa
parotid isoforms
- Pif-s (Pif-f)
parotid IEF variant, slow (fast)
Footnotes
The authors have declared no conflict of interest.
References
- 1.Oppenheim FG, Hay DI, Franzblau C. Biochemistry. 1971;10:4233–4238. doi: 10.1021/bi00799a013. [DOI] [PubMed] [Google Scholar]
- 2.Hay DI, Oppenheim FG. Arch Oral Biol. 1974;19:627–632. doi: 10.1016/0003-9969(74)90130-7. [DOI] [PubMed] [Google Scholar]
- 3.Moreno EC, Kresak M, Hay DI. J Biol Chem. 1982;257:2981–2989. [PubMed] [Google Scholar]
- 4.Gibbons RJ, Hay DI. Infect Immun. 1988;56:439–445. doi: 10.1128/iai.56.2.439-445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kousvelari EE, Baratz RS, Burke B, Oppenheim FG. J Dent Res. 1980;59:1430–1438. doi: 10.1177/00220345800590081201. [DOI] [PubMed] [Google Scholar]
- 6.Bennick A, Chau G, Goodlin R, Abrams S, Tustian D, Madapallimattam G. Arch Oral Biol. 1983;28:19–27. doi: 10.1016/0003-9969(83)90022-5. [DOI] [PubMed] [Google Scholar]
- 7.Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ. Ann N Y Acad Sci. 2007;1098:22–50. doi: 10.1196/annals.1384.030. [DOI] [PubMed] [Google Scholar]
- 8.Messana I, Inzitari R, Fanali C, Cabras T, Castagnola M. J Sep Sci. 2008;31:1948–1963. doi: 10.1002/jssc.200800100. [DOI] [PubMed] [Google Scholar]
- 9.Messana I, Cabras T, Pisano E, Sanna MT, Olianas A, Manconi B, Pellegrini M, Paludetti G, Scarano E, Fiorita A, Agostino S, Contucci AM, Calò L, Picciotti PM, Manni A, Bennick A, Vitali A, Fanali C, Inzitari R, Castagnola M. Mol Cell Proteomics. 2008;7:911–926. doi: 10.1074/mcp.M700501-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Hay DI, Bennick A, Schlesinger DH, Minaguchi K, Madapallimattam G, Schluckebier SK. Biochem J. 1988;255:15–21. doi: 10.1042/bj2550015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shintani M, Minaguchi K, Suzuki K, Lim KA. Biochem Genet. 1990;28:173–184. doi: 10.1007/BF00561335. [DOI] [PubMed] [Google Scholar]
- 12.Minaguchi K, Shintani M, Suzuki K. Hum Hered. 1990;40:221–230. doi: 10.1159/000153934. [DOI] [PubMed] [Google Scholar]
- 13.Hay DI, Ahern JM, Schluckebier SK, Schlesinger DH. J Dent Res. 1994;73:1717–1726. doi: 10.1177/00220345940730110701. [DOI] [PubMed] [Google Scholar]
- 14.Cai K, Bennick A. Arch Oral Biol. 2004;49:871–879. doi: 10.1016/j.archoralbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Inzitari R, Cabras T, Onnis G, Olmi C, Mastinu A, Sanna MT, Pellegrini MG, Castagnola M, Messana I. Proteomics. 2005;5:805–815. doi: 10.1002/pmic.200401156. [DOI] [PubMed] [Google Scholar]
- 16.Tagliabracci VS, Engel JL, Wen J, Wiley SE, Worby CA, Kinch LN, Xiao J, Grishin NV, Dixon JE. Science. 2012;336:1150–1153. doi: 10.1126/science.1217817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tagliabracci VS, Pinna LA, Dixon JE. Trends Biochem Sci. 2013;38:121–130. doi: 10.1016/j.tibs.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ornstein L. Ann N Y Acad Sci. 1964;121:321–349. doi: 10.1111/j.1749-6632.1964.tb14207.x. [DOI] [PubMed] [Google Scholar]
- 19.Davis BJ. Ann N Y Acad Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 20.Cabras T, Boi R, Pisano E, Iavarone F, Fanali C, Nemolato S, Faa G, Castagnola M, Messana I. J Sep Sci. 2012;35:1079–1086. doi: 10.1002/jssc.201101066. [DOI] [PubMed] [Google Scholar]
- 21.Messana I, Cabras T, Iavarone F, Vincenzoni F, Urbani A, Castagnola M. J Sep Sci. 2013;36:128–139. doi: 10.1002/jssc.201200830. [DOI] [PubMed] [Google Scholar]
- 22.Cabras T, Iavarone F, Pirolli D, De Rosa MC, Vitali A, Faa G, Cordaro M, Messana I, Ekström J, Castagnola M. J Sep Sci. 2013;36:2848–2861. doi: 10.1002/jssc.201300312. [DOI] [PubMed] [Google Scholar]
- 23.Azen EA, Maeda N. Adv Hum Genet. 1988;17:141–199. doi: 10.1007/978-1-4613-0987-1_5. [DOI] [PubMed] [Google Scholar]