Abstract
Studies on actinobacterial diversity in limestone habitats are scarce. This paper reports profiling of actinobacteria isolated from Hundung limestone samples in Manipur, India using ARDRA as the molecular tool for preliminary classification. A total of 137 actinobacteria were clustered into 31 phylotypic groups based on the ARDRA pattern generated and representative of each group was subjected to 16S rRNA gene sequencing. Generic diversity of the limestone isolates consisted of Streptomyces (15 phylotypic groups), Micromonospora (4), Amycolatopsis (3), Arthrobacter (3), Kitasatospora (2), Janibacter (1), Nocardia (1), Pseudonocardia (1) and Rhodococcus (1). Considering the antimicrobial potential of these actinobacteria, 19 showed antimicrobial activities against at least one of the bacterial and candidal test pathogens, while 45 exhibit biocontrol activities against at least one of the rice fungal pathogens. Out of the 137 actinobacterial isolates, 118 were found to have at least one of the three biosynthetic gene clusters (PKS-I, PKS-II, NRPS). The results indicate that 86% of the strains isolated from Hundung limestone deposit sites possessed biosynthetic gene clusters of which 40% exhibited antimicrobial activities. It can, therefore, be concluded that limestone habitat is a promising source for search of novel secondary metabolites.
Keywords: actinobacterial diversity, limestone habitat, Hundung, antibacterial, biocontrol, biosynthetic genes, Streptomyces
Introduction
Actinobacteria are major producers of secondary metabolites such as antimicrobial compounds, anticancer molecules and immunosuppressant agents (Takahashi and Omura, 2003). Since the beginning of antibiotic revolution, actinobacteria especially the genus Streptomyces have played major roles as antibiotic producers (Bérdy, 2005). However, the discovery of new antibiotics has not been in pace with the increase in demand for new antibiotics. The exhaustion of the usual terrestrial sources and the rise of resistant pathogens dictate the search for new antibiotics. To meet urgent clinical needs, screening for secondary metabolites from actinobacteria residing in unexplored habitats is warranted to, possibly, generate novel compounds.
Limestone habitats have high deposition of CaCO3 salts and may be considered a special habitat. Limited studies have been done for systematically exploring such habitats for novel actinobacterial strains (Kim et al., 1998; Groth et al., 1999a, 2001; Jurado et al., 2009; Nakaew et al., 2009; Niyomvong et al., 2012). Some reports are available on actinobacterial diversity in hypogean environments but the studies were focused on biodeterioration and conservation of paleolithic cave art. Actinobacteria implicated in deterioration of art work are considered serious risk factors if environmental changes promote their massive proliferation (Groth et al., 1999b; Portillo et al., 2009). To date, four new genera Beutenbergia, Fodinibacter, Hoyosella, and Knoellia have been reported from limestone habitats and related limestone ecosystems such as cave biofilms (Groth et al., 1999b, 2002; Jurado et al., 2009; Wang et al., 2009).
Manipur has a huge reserve of good quality limestone suitable for use in the manufacture of cement. The major limestone reserves have been located by Geological Survey of India near Ukhrul district, Manipur. Other limestone deposit sites include areas in Hundung, Phungyar, Meihring, Mova, Khonggoi, Lambui, and Paoyi. This paper reports the actinobacterial diversity profiling of the Hundung limestone deposit sites using ARDRA as the molecular tool for preliminary classification. ARDRA has been originally designed to decrease selection of duplicate clones in molecular analysis. It has been less frequently used in the study of bacterial diversity profiling unlike techniques such as DGGE. The paper also incorporates the results of antimicrobial screening of the Hundung actinobacterial strains.
Materials and methods
Sampling
Samples for the isolation of actinobacteria were collected from limestone deposit sites, Manipur, India (25.05°N, 94.33°E). The samples included limestones from the quarry site and rice field soil from the adjoining areas. These samples were aseptically packed in polyethylene bags and taken to the laboratory at the earliest possible time. Samples were then kept refrigerated till processing for isolation.
Isolation of actinobacteria
Two synthetic media, Gauze's Medium No. 1 (GM1, pH 5.3) (Atlas, 1997) and Starch Casein Nitrate Agar (SCNA, pH 8.5) (Kűster and Williams, 1964), were used for the isolation of actinobacteria. Isolation was done using the procedure as described earlier (Nimaichand et al., 2012). The strains were preserved as lyophilized cultures and as glycerol suspension (20% w/v) at −80°C.
Amplified ribosomal DNA restriction analysis (ARDRA; Heyndrickx et al., 1996)
Genomic DNA extraction and amplification of the 16S rRNA gene was done as described by Li et al. (2007). The amplified products were checked and purified by HiPurA™ 96 PCR product purification kit (HiMedia, India). Restriction digestion of the amplified 16S rRNA gene product was done using the enzymes HhaI and HinfI (New England Biolabs, UK). The reaction mixture containing 10 μl amplified 16S rRNA gene product, 2 μl NEB buffer 4 (10X), 1 μl restriction enzyme (10 U/μl) and 7 μl deionized water was incubated at 37°C for 2 h and inactivated by heating at 70°C for 10 min. To 20 μl of the restriction digest, 4 μl loading dye (6X) (Promega) was added. Each sample was loaded in a well in agarose gel (3%, w/v) and the gel was run at 100 V for 90 min. In another well, 1 μl DNA ladder (100 bp) (Promega) was loaded to estimate the size of the restriction fragment. The gel was visualized in a gel documentation system (BIORAD Gel Doc EZ Imager). Bands between 100 and 1000 bp were used as reference points and banding patterns were analyzed by scoring the prominent bands. ARDRA band profiles for all the strains were scored with the help of GelBuddy software (Zerr and Henikoff, 2005) for the presence or absence of restriction fragments. A dendrogram was generated using the software package NTSYSpc version 2.02. The phylogenetic relationship was determined according to the method of unweighted pair group method with arithmetic mean (UPGMA; Sneath and Sokal, 1973). Based on the similarity indices (70% and above) in the dendrogram, all the strains were clustered into different phylotypic groups.
Sequencing of 16S rRNA genes
Sequencing of a randomly-selected representative strain for each phylotypic group was done. The partial 16S rRNA gene sequence of the strain was identified using the EzTaxon-e server database (Kim et al., 2012). The phylogenetic tree of these strains based on neighbor-joining method (Saitou and Nei, 1987) along with related type species were constructed using the software package MEGA version 5.2 (Tamura et al., 2011). Distances were calculated according to Kimura's two-parameter model (Kimura, 1983). To determine the support of each clade, bootstrap analysis was performed with 1000 resamplings (Felsenstein, 1985).
Nucleotide accession numbers
The partial 16S rRNA gene sequences were deposited in GenBank with the following accession numbers: KP883248-KP883278.
Antimicrobial screening
The indicator pathogens used for antimicrobial screening were: Bacillus subtilis MTCC 121, Escherichia coli MTCC 739, Pseudomonas aeruginosa DN1, Candida albicans MTCC 227, Candida vaginitis CV, Curvularia oryzae MTCC 2605, Fusarium oxysporum MTCC 287, Helminthosporum oryzae MTCC 3717, Pyricularia oryzae MTCC 1477, Rhizoctonia oryzae-sativae MTCC 2162 and Rhizoctonia solani MTCC 4633. All the test pathogens were procured from Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh, India except for DN1 (lab collection) and CV (clinical isolate gifted from the Centre for DNA Fingerprinting and Diagnostics (CDFD), Hyderabad).
Antimicrobial assays against the bacterial and candidal strains were performed by agar well diffusion method (Hugo and Russell, 1983). Antifungal bioassay was done by dual culture technique (Khamna et al., 2009). The mycelial growth inhibition was calculated using the formula:
where, C = Radial growth of the test pathogen in the control plate, and T = Radial growth of the test pathogen in the test plate.
Screening for biosynthetic genes
Three sets of degenerate primers were used for amplification of PKS-I, PKS-II and NRPS specific domains (Metsä-Ketalä et al., 1999; González et al., 2005). The primers used are listed in Table 1. PCR amplifications were performed in eppendorf mastercycler in a final volume of 25 μl containing 5 μl reaction buffer (with Mg2+) (10x) (Bioline, USA), 0.5 μl of each primer (100 μM) (IDT, USA), 2.0 μl of dNTPs mixture (2.5 mM) (Bioline, USA), 0.15 μl of Taq DNA polymerase (2.5 U/μl) (Bioline, USA), 2.5 μl DMSO (HiMedia, India), 11.85 μl deionized water and 2.5 μl of extracted DNA. Amplification was done using the following protocol: one denaturation step of 94°C for 5 min; 30 amplification cycles of 94°C for 1 min, 57°C (for K1F-M6R and A3F-A7R) or 58°C (for KSαF-KSαR) for 1 min, and 72°C for 2 min; and a final extension at 72°C for 5 min. Amplification products were analyzed in agarose gel (1%) using DNA ladder (100 bp) (Promega) as reference.
Table 1.
PCR primers for screening the biosynthetic genes.
| Primer name | Sequence (5′–3′) | Target gene | Length of target gene fragment (bp) | References |
|---|---|---|---|---|
| K1F M6R |
TSA AGT CSA ACA TCG GBC A CGC AGG TTS CSG TAC CAG TA |
PKS-I | 1200–1400 | González et al., 2005 |
| KSαF KSαR |
TSG CST GCT TGG AYG CSA TC TGG AAN CCG CCG AAB CCG CT |
PKS-II | 600 | Metsä-Ketalä et al., 1999 |
| A3F A7R |
GCS TAC SYS ATS TAC ACS TCS GG SAS GTC VCC SGT SCG GTA S |
NRPS | 700–800 | González et al., 2005 |
Results
Description of sampling sites
For the actinobacterial isolation, six Hundung samples were collected and used. The sample collection sites included: an abandoned cement factory site (Sample 1), quarry sites (Sample 2–5) and a rice field adjoining the quarry site (Sample 6). The limestones in Hundung, with color ranging from light gray to brown, are of good quality grade which are suitable for production of cement (Bhatt and Bhargava, 2005). The estimated reserve of this Hundung limestone is about 1.88 million tons (Sadangi, 2008; Lisam, 2011). The general characteristics of the samples used for isolation are highlighted in Table 2.
Table 2.
Profile of the Hundung limestone samples and coding scheme for the actinobacterial strains.
| Sample | Sampling sites | pH of the sample | Isolation medium | No. of strains | Isolate code |
|---|---|---|---|---|---|
| 1 | Cement factory location | 9.26 | GM1 | 29 | MBRL 1–MBRL 29 |
| SCNA | 22 | MBRL 200–MBRL 221 | |||
| 2 | Quarry site 1 | 8.70 | GM1 | 33 | MBRL 30–MBRL 61 |
| SCNA | 15 | MBRL 222–MBRL 237 | |||
| 3 | Quarry site 2 | 7.45 | GM1 | – | – |
| SCNA | 10 | MBRL 238–MBRL 247 | |||
| 4 | Quarry site 3 | 6.50 | GM1 | 14 | MBRL 62–MBRL 75 |
| SCNA | 3 | MBRL 248–MBRL 250 | |||
| 5 | Quarry site 4 | 7.91 | GM1 | – | – |
| SCNA | 2 | MBRL 251–MBRL 252 | |||
| 6 | Soil sample from the adjoining rice field | 4.89 | GM1 | 6 | MBRL 76–MBRL 81 |
| SCNA | 3 | MBRL 253–MBRL 255 | |||
| Total number of strains | 137 | ||||
Actinobacterial isolation
Among the isolates obtained, 137 morphologically distinct putative actinobacterial strains were selected for further studies. These included 51 strains from Sample 1, 48 from Sample 2, 10 from Sample 3, 17 from Sample 4, 2 from Sample 5 and 9 from Sample 6. The coding scheme for the actinobacteria from the Hundung samples is shown in Table 2.
Diversity analysis of Hundung actinobacteria
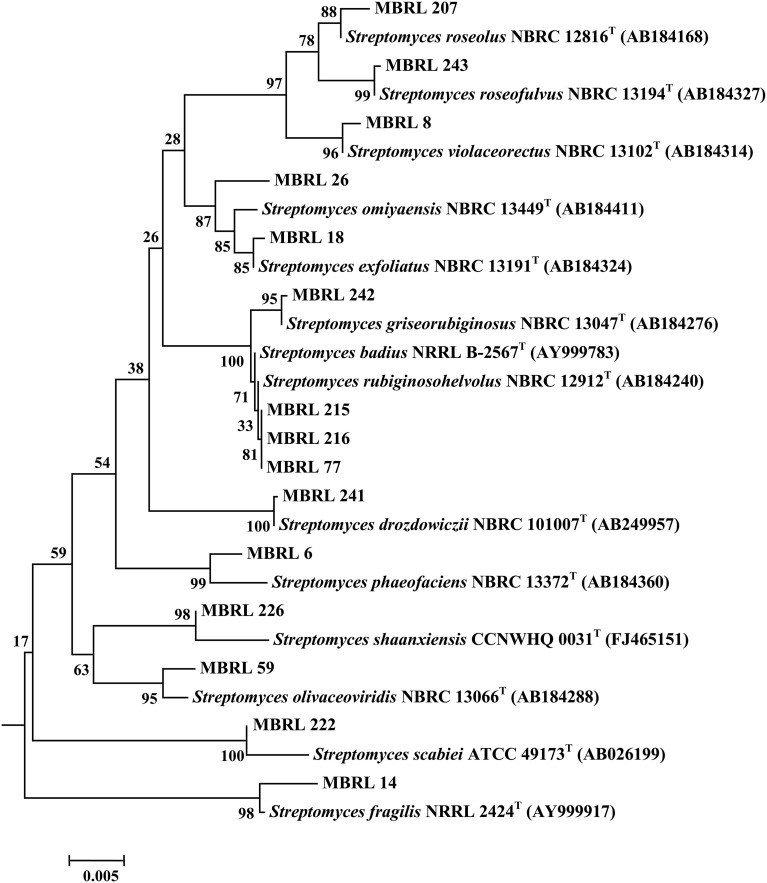
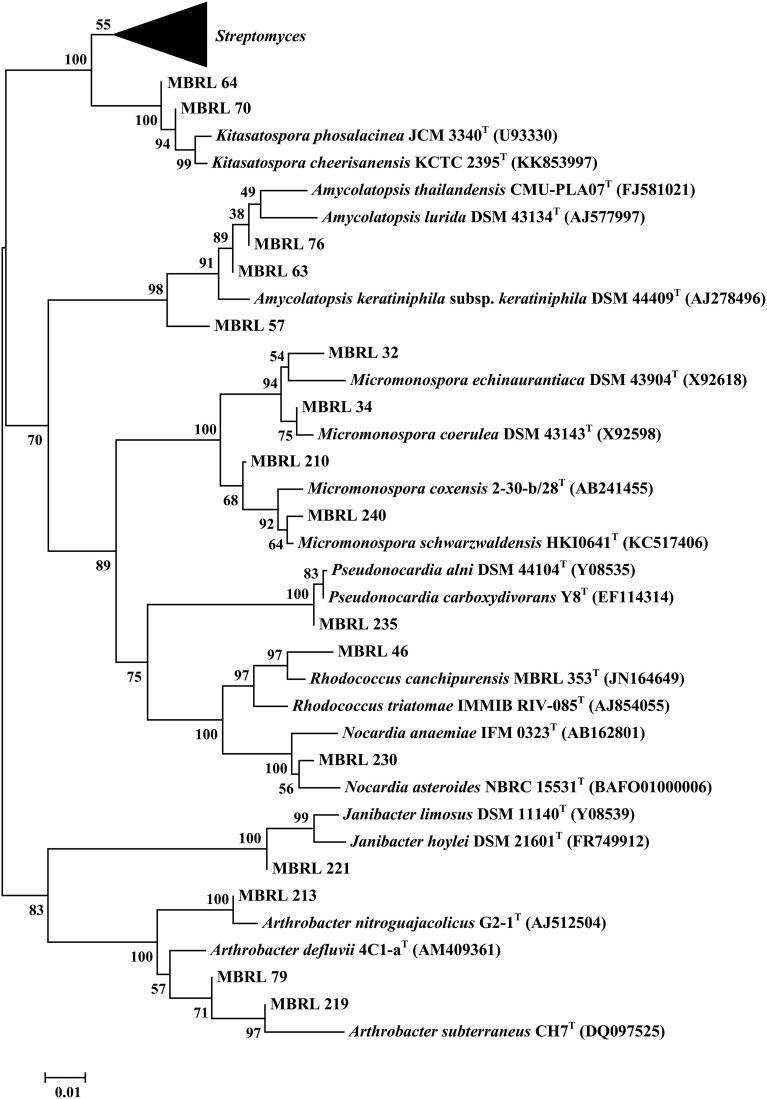
Upon analysis of the ARDRA-based dendrogram (Supplementary Figure S1), the isolates were classified into 31 phylotypic groups (see Supplementary Table S1 for classification pattern of the Hundung actinobacteria based on ARDRA-dendrogram). The 16S rRNA gene sequence profile for these 31 phylotypic groups is given in Table 3. Fifteen of these phylotypes belong to the genus Streptomyces. In addition, four phylotypes belong to the genus Micromonospora, three each to Arthrobacter and Amycolatopsis and two to Kitasatospora. Remaining phylotypes comprise of the genera Janibacter, Rhodococcus, Nocardia, and Pseudonocardia. Among the different sites, sample 2 gave the highest diversity (16 phylotypes), followed closely by sample 1 (15 phylotypic groups). Sample 1 yielded the genera Streptomyces, Janibacter, Arthrobacter, Amycolatopsis, and Micromonospora while sample 2 generated Streptomyces, Rhodococcus, Amycolatopsis, Micromonospora, Arthrobacter, Nocardia, and Pseudonocardia. Sample 3 which contained 5 phylogenetic groups yielded the genera Streptomyces and Micromonospora. Four genera viz., Streptomyces, Janibacter, Amycolatopsis, and Kitasatospora were present in sample 4 while sample 6 contained 3 genera: Streptomyces, Amycolatopsis, and Arthrobacter. Sample 5 yielded Streptomyces strains only though this may not reflect the true actinobacterial diversity in this sample, as we have selected only 2 strains from the isolates obtained from this sample. Nonetheless, overall analysis of the Hundung sites (1–6) indicated Streptomyces to be the dominant genus in these habitats. Figures 1, 2 depict the dendrograms based on the 16S rRNA gene sequences of the Streptomyces and rare actinobacterial strains obtained from Hundung limestone habitats.
Table 3.
Sequence analysis profile of representative strain of each phylotypic group.
| Phylotypic group | Strain | Accession number | Closest homolog | Pairwise similarity (%) |
|---|---|---|---|---|
| I | MBRL 216 | KP883268 | Streptomyces badius NRRL B-2567T | 100.00 |
| II | MBRL 221 | KP883270 | Janibacter limosus DSM 11140T | 99.52 |
| III | MBRL 6 | KP883248 | Streptomyces phaeofaciens NBRC 13372T | 99.20 |
| IV | MBRL 77 | KP883262 | Streptomyces rubiginosohelvolus NBRC 12912T | 100.00 |
| V | MBRL 207 | KP883264 | Streptomyces roseolus NBRC 12816T | 99.74 |
| VI | MBRL 241 | KP883276 | Streptomyces drozdowiczii NBRC 101007T | 100.00 |
| VII | MBRL 26 | KP883252 | Streptomyces omiyaensis NBRC 13449T | 99.30 |
| VIII | MBRL 243 | KP883278 | Streptomyces roseofulvus NBRC 13194T | 100.00 |
| IX | MBRL 213 | KP883266 | Arthrobacter nitroguajacolicus G2-1T | 100.00 |
| X | MBRL 219 | KP883269 | Arthrobacter subterraneus CH7T | 98.96 |
| XI | MBRL 46 | KP883255 | Rhodococcus canchipurensis MBRL 353T | 98.45 |
| XII | MBRL 57 | KP883256 | Amycolatopsis lurida DSM 43134T | 97.46 |
| XIII | MBRL 222 | KP883271 | Streptomyces scabiei ATCC 49173T | 100.00 |
| XIV | MBRL 76 | KP883261 | Amycolatopsis thailandensis CMU-PLA07T | 98.95 |
| XV | MBRL 32 | KP883253 | Micromonospora kangleipakensis MBRL 34T | 98.42 |
| XVI | MBRL 64 | KP883259 | Kitasatospora phosalacinea JCM 3340T | 99.35 |
| XVII | MBRL 70 | KP883260 | Kitasatospora cheerisanensis KCTC 2395T | 99.40 |
| XVIII | MBRL 210 | KP883265 | Micromonospora coxensis 2-30-b/28T | 99.27 |
| XIX | MBRL 240 | KP883275 | Micromonospora schwarzwaldensis HKI0641T | 99.48 |
| XX | MBRL 8 | KP883249 | Streptomyces violacerectus NBRC 13102T | 100.00 |
| XXI | MBRL 18 | KP883251 | Streptomyces exfoliatus NBRC 13191T | 100.00 |
| XXII | MBRL 14 | KP883250 | Streptomyces fragilis NRRL 2424T | 99.44 |
| XXIII | MBRL 63 | KP883258 | Amycolatopsis keratiniphila subsp. keratiniphila DSM 44409T | 99.92 |
| XXIV | MBRL 34 | KP883254 | Micromonospora coerulea DSM 43143T | 98.52 |
| XXV | MBRL 242 | KP883277 | Streptomyces griseorubiginosus NBRC 13047T | 100.00 |
| XXVI | MBRL 226 | KP883272 | Streptomyces shaanxiensis CCNWHQ 0031T | 99.52 |
| XXVII | MBRL 215 | KP883267 | Streptomyces rubiginosohelvolus NBRC 12912T | 100.00 |
| XXVIII | MBRL 79 | KP883263 | Arthrobacter defluvii 4C1-aT | 100.00 |
| XXIX | MBRL 230 | KP883273 | Nocardia asteroides NBRC 15531T | 98.64 |
| XXX | MBRL 235 | KP883274 | Pseudonocardia carboxydivorans Y8T | 100.00 |
| XXXI | MBRL 59 | KP883257 | Streptomyces olivaceoviridis NBRC 13066T | 99.49 |
Figure 1.
Dendrogram of the representative Streptomyces strains based on the 16S rRNA gene sequences. Numbers at nodes are levels of bootstrap support (%) for branch points (1000 resamplings). Bar, 0.002 substitutions per nucleotide position.
Figure 2.
Dendrogram of the representative rare actinobacterial strains based on the 16S rRNA gene sequences. Numbers at nodes are levels of bootstrap support (%) for branch points (1000 resamplings). Bar, 0.01 substitutions per nucleotide position.
Antimicrobial activities
Antibacterial and anticandidal activities
Antibacterial and anticandidal activity was assessed against a set of indicator organisms. The antibacterial and anticandidal profiles of the Hundung actinobacteria are shown in Table 4. Of 137 actinobacterial isolates, 19 exhibited antimicrobial activities against at least one of the test pathogens. In case of Bacillus subtilis, 18 strains showed inhibition, of which 5 (MBRL 5, MBRL 10, MBRL 201, MBRL 204, MBRL 251) showed inhibition zones above 17 mm diameter. Against Escherichia coli, 5 strains exhibited antagonistic activities of which 2 (MBRL 5, MBRL 10) showed inhibition zone sizes above 17 mm diameter. No strain had activity against Pseudomonas aeruginosa. Against Candida albicans, 5 strains showed inhibitory activity and 4 against Candida vaginitis (See Supplementary Table S2 for complete antibacterial and anticandidal profile).
Table 4.
Antimicrobial, biocontrol and biosynthetic genes profile of the Hundung actinobacteria.
| Phylotypic group | Total number of isolates | Genus classified | Test pathogens | Biosynthetic gene | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MTCC 121 | MTCC 739 | DN1 | MTCC 227 | CV | MTCC 2605 | MTCC 287 | MTCC 3717 | MTCC 1477 | MTCC 2162 | MTCC 4633 | PKS-I | PKS-II | NRPS | |||
| I | 29 | Streptomyces | 4 | 4 | 0 | 4 | 3 | 5 | 7 | 12 | 8 | 10 | 10 | 4 | 24 | 23 |
| II | 2 | Janibacter | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 1 | 1 |
| III | 27 | Streptomyces | 4 | 0 | 0 | 1 | 1 | 3 | 2 | 4 | 5 | 5 | 6 | 14 | 24 | 19 |
| IV | 1 | Streptomyces | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 |
| V | 2 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 |
| VI | 4 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 3 | 2 |
| VII | 1 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 1 |
| VIII | 1 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 0 |
| IX | 1 | Arthrobacter | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| X | 1 | Arthrobacter | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| XI | 1 | Rhdococcus | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| XII | 17 | Amycolatopsis | 6 | 0 | 0 | 0 | 0 | 2 | 2 | 4 | 3 | 5 | 5 | 9 | 9 | 16 |
| XIII | 1 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| XIV | 1 | Amycolatopsis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| XV | 2 | Micromonospora | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| XVI | 1 | Kitasatospora | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| XVII | 2 | Kitasatospora | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| XVIII | 1 | Micromonospora | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| XIX | 1 | Micromonospora | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| XX | 1 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| XXI | 2 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| XXII | 7 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 1 | 3 | 4 | 1 |
| XXIII | 1 | Amycolatopsis | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| XXIV | 1 | Micromonospora | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| XXV | 18 | Streptomyces | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 3 | 11 | 3 |
| XXVI | 1 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| XXVII | 2 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| XXVIII | 2 | Arthrobacter | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 |
| XXIX | 1 | Nocardia | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| XXX | 2 | Pseudonocardia | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 4 | 0 |
| XXXI | 3 | Streptomyces | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 | 2 |
MTCC 121, Bacillus subtilis; MTCC 739, Escherichia coli; DN1, Pseudomonas aeruginosa; MTCC 227, Candida albicans; CV, Candida vaginitis; MTCC 2605, Curvularia oryzae; MTCC 287, Fusarium oxysporum; MTCC 3717, Helminthosporum oryzae; MTCC 1477, Pyricularia oryzae; MTCC 2162, Rhizoctonia oryzae-sativae; MTCC 4633, Rhizoctonia solani.
Biocontrol activities
Several actinobacterial strains exhibited biocontrol potential against rice fungal pathogens. Forty five actinobacterial strains from Hundung limestone habitat showed biocontrol activities against at least one of the rice fungal pathogens. Frequencies of biocontrol activities against the indicator fungal pathogens were as follows: Helminthosporum oryzae MTCC 3717 (22.6%), Rhizoctonia solani MTCC 4633 (18.2%), Rhizoctonia oryzae-sativae MTCC 2162 (16.8%), Pyricularia oryzae MTCC 1477 (14.6%), Curvularia oryzae MTCC 2605 (10.9%) and Fusarium oxysporum MTCC 287 (9.5%) respectively. Table 4 summarizes the biocontrol profile of the Hundung actinobacteria (See Supplementary Table S3 for complete biocontrol activity profile).
Screening for biosynthetic genes
It is well known that many bioactive metabolites in actinobacteria are produced by PKS and NRPS gene clusters. Screening for genes associated with secondary metabolism is helpful in evaluating the biosynthetic potential of actinobacteria. Of 137 Hundung actinobacterial strains, 118 possessed at least one of the three biosynthetic gene clusters. A total of 43 strains had a single type of biosynthetic gene cluster (PKS-I, 5 strains; PKS-II, 27; NRPS, 11). The remaining 75 strains had two or more of the biosynthetic gene clusters: 9 strains possessed both PKS-I and PKS-II; 8 had both PKS-I and NRPS; 36 had both PKS-II and NRPS while 22 strains had all the three biosynthetic gene clusters. Table 4 shows the amplication profile for biosynthetic genes in the Hundung actinobacteria (see Supplementary Table S4 for complete PCR profile of biosynthetic genes).
Discussion
Diversity profiling focused on actinobacteria in limestone habitats started when Kim et al. (1998) reported the diversity of actinobacteria antagonistic to phytopathogenic fungi in caves of Korea. They reported the presence of Streptomyces, Micromonospora, Nocardioform actinobacteria, Actinomyces, Dactylosporangium, Saccharomonospora, and Streptosporangium in these habitats. Groth et al. (1999a) studied the actinobacterial diversity in Karstic caves (Altamira and Tito Bustillo) located in northern Spain and reported members of the genera Streptomyces, Nocardia, Rhodococcus, Nocardioides, Amycolatopsis, Saccharothrix, Brevibacterium, Microbacterium, and coccocid actinobacteria of the family Micrococcaceae. Groth et al. (2002) reported the isolation of a new genus Knoellia from limestone caves. To the repertoire of the actinobacterial diversity in caves, Nakaew et al. (2009) added the genera Nonomuraea, Actinocorallia, Catellatospora, Microbispora, and Sprillospora. Jurado et al. (2009) reported the new genus Hoyosella from cave biofilms in Spain. Niyomvong et al. (2012) found the presence of the genera Streptomyces, Actinomadura, Actinoplanes, Gordonia, Microbispora, Micromonospora, Nocardia, Nonomuraea, and Saccharopolyspora in the tropical limestone caves of Khao No-Khao Kaeo karst in Thailand.
Considering the rich diversity of actinobacteria in limestone habitats, the present study on actinobacterial diversity of limestone deposit sites in Hundung, Manipur, India, has special significance. As per our findings, the genus Streptomyces is predominantly present in these limestone habitats. This is also indicated by the presence of phylotypic group III (represented by the genus Streptomyces) in all the six samples used for actinobacterial isolation. Apart from Streptomyces, we also observed the presence of rare actinobacteria Micromonospora, Arthrobacter, Amycolatopsis, Kitasatospora, Janibacter, Rhodococcus, Nocardia, and Pseudonocardia. This work forms the first report of the isolation of Janibacter and Kitasatospora from limestone and related habitats.
ARDRA is preferable to other molecular genome typing methods for preliminary phylogenetic grouping as it is faster and more cost effective than the other approaches. Moreover, as ARDRA is based on the presence of restriction sites within the ribosomal DNA, duplicate strains will most likely have the same restriction pattern. The use of ARDRA in this study, therefore, helped reduce the number of duplicate strains among the isolates from the community, indicating the true diversity of the community even when the sample size is small.
In the course of a screening program for novel antibiotics from strains obtained from Grotta dei Cervi, a cave in Italy, Herold et al. (2004) identified a bioactive complex, Cervimycins A–D, from a strain of Streptomyces tendae. Cervimycins are potent antibiotics against multidrug resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecalis (VRE) strains (Herold et al., 2005). Quadri and Agsar (2012) have investigated antimicrobial activities of actinobacteria of limestone quarries located at Deccan traps, India. Of 63 actinobacteria from this habitat, six strains (belonging to the genera Streptomyces, Micromonospora, Nonomuraea, Kribbella, Lechevalieria, and Saccharothrix) showed potent antimicrobial activity against Bacillus subtilis, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Staphylococcus aureus, Salmonella typhi, and Candida albicans. Carlsohn (2011) found novel strains of Amycolatopsis saalfeldensis, Kribbella aluminosa, and Streptomyces strains from a mine in Germany and they were strongly inhibitory to Stapthylococcus aureus, Mycobacterium smegmatis, and Candida albicans, and moderately antagonistic to Escherichia coli. Rule and Cheeptham (2013) reported some Streptomyces strains from a volcanic cave in Canada (Cheeptham et al., 2013) as antagonistic to Micrococcus luteus, MRSA, Mycobacterium smegmatis, Pseudomonas aeruginosa, Escherichia coli and Candida albicans.
In the present study, 5 Hundung actinobacteria were found to be potent antimicrobial strains. Of these, 2 Streptomyces species MBRL 201 and MBRL 251 showed strong antimicrobial activity against Bacillus subtilis, but less bioactivity against Escherichia coli, Candida albicans, and Candida vaginitis. Besides these two, two other Hundung Streptomyces species (MBRL 5 and MBRL 10) also exhibited promising antimicrobial activities. MBRL 204, another Hundung Streptomyces strain, exhibited relatively lesser antimicrobial activity compared to the other 4 isolates (MBRL 5, MBRL 10, MBRL 201 and MBRL 251).
Soil actinobacteria have been proposed as promising biocontrol agents (Goodfellow and Williams, 1983; Chater, 1993). However, actinobacteria from limestone habitats have not been investigated for their biocontrol potential. Quadri and Agsar (2012) have reported that only 9.5% of the strains isolated from limestone habitats have antifungal activities against Aspergillus fumigates, Aspergillus niger, and Fusarium solani. In the current investigation, many strains belonging to the genera Streptomyces and Amycolatopsis (e.g., Phylotypic group I, III and XII) were found to have biocontrol activities against selected rice fungal pathogens Curvularia oryzae, Fusarium oxysporum, Helminsthosporum oryzae, Pyricularia oryzae, Rhizoctonia oryzae-sativae, and Rhizoctonia solani. Rare actinobacteria belonging to genera Janibacter and Pseudonocardia obtained from Hundung limestone habitats also exhibited significant biocontrol potential against some fungal pathogens.
The biosynthetic gene clusters play a crucial role in microbial natural product biosynthesis. The biosynthesis of cervimycin complex (metabolites reported from limestone related habitats) involved the type II PKS system. Other antibacterial metabolites such as Ravidomycins from Streptomyces rabidus are biosynthesized by type II PKS system (Kharel et al., 2010). Hence, it is imperative to screen for the presence of these biosynthetic gene clusters in the actinobacterial isolates. In our studies, 118 of the 137 actinobacterial isolates were found to have at least one of the three biosynthetic gene clusters. Of these 118 actinobacteria possessed the biosynthetic gene clusters, 47 exhibited antimicrobial and/or biocontrol activities indicating that less than 50% of the strains possessing biosynthetic gene clusters were bioactive under the screening condition. The findings of the various experiments indicate that 86% of the strains isolated from Hundung limestone rocks possessed biosynthetic gene clusters of which 40% exhibited antimicrobial activities. It can, therefore, be concluded that limestone habitats is a promising source for search of novel secondary metabolites.
Author contributions
SN planned, conducted the experiments, analyzed the data, and prepared the manuscript, AD performed, analyzed and interpreted the ARDRA data, KT performed the biocontrol assay, DN and WL supervised the experiments.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors also wish to thank Dr. Reena Haobam, Department of Biotechnology, Manipur University, for extending the gel documentation facility. SN wishes to thank the University Grants Commission (UGC), Government of India (GOI), for offering him the Rajiv Gandhi National Fellowship. AD wishes to thank UGC, GOI for Basic Scientific Research (BSR) fellowship. WL was supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014).
Glossary
Abbreviations
- ARDRA
Amplified Ribosomal DNA Restriction Analysis
- DGGE
Denaturing Gradient Gel Electrophoresis
- GM1
Gauze Medium No. 1
- NRPS
Non-Ribosomal Peptide Synthetase
- PKS
Polyketide Synthase
- RFLP
Restriction Fragment Length Polymorphism
- SCNA
Starch Casein Nitrate Agar.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.00413/abstract
References
- Atlas R. M. (1997). Handbook of Microbiological Media, 2nd Edn. New York, NY: CRC Press. [Google Scholar]
- Bérdy J. (2005). Bioactive microbial metabolites. J. Antibiot. 58, 1–26 10.1038/ja.2005.1 [DOI] [PubMed] [Google Scholar]
- Bhatt S. C., Bhargava G. K. (2005). Land and People of Indian states & Union territories. New Delhi: Kalpaz Publications. [Google Scholar]
- Carlsohn M. R. (2011). Isolation and Characterization of Mine-Dwelling Actinomycetes as Potential Producers of Novel Bioactive Secondary Metabolites. PhD thesis. Leibniz Institute for Natural Product Research and Infection Biology – Hans-Knoll-Institute, Jena. [Google Scholar]
- Chater K. F. (1993). Genetics of differentiation in Streptomyces. Annu. Rev. Microbiol. 47, 685–713. 10.1146/annurev.mi.47.100193.003345 [DOI] [PubMed] [Google Scholar]
- Cheeptham N., Sadoway T., Rule D., Watson K., Moote P., Soliman L. C., et al. (2013). Cure from the cave: volcanic cave actinomycetes and their potential in drug discovery. Int. J. Speleol. 42, 35–47 10.5038/1827-806X.42.1.5 [DOI] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- González I., Ayuso-Sacido A., Anderson A., Genilloud O. (2005). Actinomycetes isolated from lichens: evaluation of their diversity and detection of biosynthetic gene sequences. FEMS Microbiol. Ecol. 54, 401–415. 10.1016/j.femsec.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Goodfellow M., Williams S. T. (1983). Ecology of actinomycetes. Annu. Rev. Microbiol. 37, 189–216. 10.1146/annurev.mi.37.100183.001201 [DOI] [PubMed] [Google Scholar]
- Groth I., Schumann P., Laiz L., Sanchez-Moral S., Caòveras J. C., Saiz-Jimenez C. (2001). Geomicrobiological study of the Grotta dei Cervi, Porto Badisco, Italy. Geomicrobiol. J. 18, 241–258 10.1080/01490450152467778 [DOI] [Google Scholar]
- Groth I., Schumann P., Schuetze B., Augsten K., Kramer I., Stackebrandt E. (1999b). Beutenbergia cavernae gen. nov., sp. nov., an L-lysine containing actinomycete isolated from a cave. Int. J. Syst. Evol. Microbiol. 49, 1733–1740. 10.1099/00207713-49-4-1733 [DOI] [PubMed] [Google Scholar]
- Groth I., Schumann P., Schutze B., Augsten K., Stackebrandt E. (2002). Knoellia sinensis gen. nov., sp. nov., and Knoellia subterranea sp. nov., two novel actinobacteria isolated from a cave. Int. J. Syst. Evol. Microbiol. 52, 77–84. [DOI] [PubMed] [Google Scholar]
- Groth I., Vettermann R., Schuetze B., Schumann P., Saiz-Jimenez D. (1999a). Actinomycetes in Karstic caves of northern Spain (Altamira and Tito Bustillo). J. Microbiol. Methods 36, 115–122. 10.1016/S0167-7012(99)00016-0 [DOI] [PubMed] [Google Scholar]
- Herold K., Gollmick F. A., Groth I., Roth M., Menzel K. D., Möllmann U., et al. (2005). Cervimycin A-D: a polyketide glycoside complex from a cave bacterium can defect vancomycin resistance. Chemistry 11, 5523–5530. 10.1002/chem.0.200590060 [DOI] [PubMed] [Google Scholar]
- Herold K., Xu Z., Gollmick F. A., Gräfe U., Hertweck C. (2004). Biosynthesis of cervimycin C, an aromatic polyketide antibiotic bearing an unusual dimethylmalonyl moiety. Org. Biomol. Chem. 2, 2411–2414. 10.1039/B409221J [DOI] [PubMed] [Google Scholar]
- Heyndrickx M., Vauterin L., Vandamme P., Kersters K., De Vos P. (1996). Applicability of combined amplified ribosomal DNA restriction analysis (ARDRA) patterns in bacterial phylogeny and taxonomy. J. Microbiol. Methods 26, 247–259 10.1016/0167-7012(96)00916-5 [DOI] [Google Scholar]
- Hugo W. B., Russell A. D. (1983). Pharmaceutical Microbiology, 3rd Edn. Oxford: Blackwell. [Google Scholar]
- Jurado V., Kroppenstedt R. M., Saiz-Jimenez C., Klenk H. P., Mouniee D., Laiz L., et al. (2009). Hoyosella altamirensis gen. nov., sp. nov., a new member of the order Actinomycetales isolated from a cave biofilm. Int. J. Syst. Evol. Microbiol. 59, 3105–3110. 10.1099/ijs.0.008664-0 [DOI] [PubMed] [Google Scholar]
- Khamna S., Yokata A., Lumyong S. (2009). Actinomycetes isolated from medicinal plant rhizospheric soils: diversity and screening of antifungal compounds, indole-3-acetic acid and siderophore production. World J. Microbiol. Biotechnol. 25, 649–655 10.1007/s11274-008-9933-x [DOI] [Google Scholar]
- Kharel M. K., Nybo S. E., Shephered M. D., Rohr J. (2010). Cloning and characterization of the ravidomycin and chrysomycin biosynthetic gene clusters. Chembiochem 11, 523–532. 10.1002/cbic.200900673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. S., Lee J. Y., Hwang B. K. (1998). Diversity of actinomycetes antagonistic to plant pathogenic fungi in cave and sea-mud soils of Korea. J. Microbiol. 36, 86–92. [Google Scholar]
- Kim O. S., Cho Y. J., Lee K., Yoon S. H., Kim M., Na H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1983). The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Kűster E., Williams S. T. (1964). Selection of media for isolation of Streptomycetes. Nature 202, 928–929. 10.1099/ijs.0.038075-0 [DOI] [PubMed] [Google Scholar]
- Li W. J., Xu P., Schumann P., Zhang Y. Q., Pukall R., Xu L. H., et al. (2007). Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int. J. Syst. Evol. Microbiol. 57, 1424–1428. 10.1099/ijs.064749-0 [DOI] [PubMed] [Google Scholar]
- Lisam K. S. (2011). Encyclopedia of Manipur, Vol. 3 New Delhi: Kalpaz Publications. [Google Scholar]
- Metsä-Ketalä M., Salo V., Halo L., Hautala A., Hakala J., Mäntsälä P., et al. (1999). An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 180, 1–6. 10.1111/j.1574-6968.1999.tb08770.x [DOI] [PubMed] [Google Scholar]
- Nakaew N., Pathom-aree W., Lumyong S. (2009). Generic diversity of rare actinomycetes from Thai cave soils and their possible use as new bioactive compounds. Actinomycetol 23, 21–26 10.3209/saj.SAJ230201 [DOI] [Google Scholar]
- Nimaichand S., Zhu W. Y., Yang L. L., Ming H., Nie G. X., Tang S. K., et al. (2012). Streptomyces manipurensis sp. nov., a novel actinomycete isolated from a limestone deposit site in Manipur, India. Antonie van Leeuwenhoek 102, 133–139. 10.1007/s10482-012-9720-4 [DOI] [PubMed] [Google Scholar]
- Niyomvong N., Pathom-aree W., Thamchaipenet A., Duangmal K. (2012). Actinomycetes from tropical limestone caves. Chiang Mai J. Sci. 39, 373–388. [Google Scholar]
- Portillo M. C., Saiz-Jimenez C., Gonzalez J. M. (2009). Molecular characterization of total and metabolically active bacterial communities of “white colonizations” in the Altamira Cave, Spain. Res. Microbiol. 160, 41–47. 10.1016/j.resmic.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Quadri S. R., Agsar D. (2012). Antimicrobial attributes of rare actinobacteria detected from limestone quarries. Int. J. Pharma. Bio. Sci. 3, 137–147. [Google Scholar]
- Rule D., Cheeptham N. (2013). The effects of UV light on the antimicrobial activities of cave actinomycetes. Int. J. Speleology. 42, 147–153 10.5038/1827-806X.42.2.7 [DOI] [Google Scholar]
- Sadangi H. C. (2008). Emergent North East India: a Way Forward. New Delhi: Isha Books. [Google Scholar]
- Saitou N., Nei M. (1987). The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Bio. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sneath P. H. A., Sokal R. R. (1973). Numerical Taxonomy. San Francisco, CA: Freeman. [Google Scholar]
- Takahashi Y. O., Omura S. (2003). Isolation of new actinomycete strains for the screening of new bioactive compounds. J. Gen. Appl. Microbiol. 49, 141–154. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Bio. Evol. 28, 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. G., Wang Y. X., Liu J. H., Chen Y. G., Zhang X. X., Wen M. L., et al. (2009). Fodinibacter luteus gen. nov., sp. nov., a novel actinobacterium isolated from a salt mine in Yunnan. Int. J. Syst. Evol. Microbiol. 59, 2185–2190 10.1099/ijs.0.006882-0 [DOI] [PubMed] [Google Scholar]
- Zerr T., Henikoff S. (2005). Automated band mapping in electrophoretic gel images using background information. Nucleic Acids Res. 33, 2806–2812. 10.1093/nar/gki580 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.