Abstract
Recent research has shown that Twitter data analytics can have broad implications on public health research. However, its value for pharmacovigilance has been scantly studied – with health related forums and community support groups preferred for the task. We present a systematic study of tweets collected for 74 drugs to assess their value as sources of potential signals for adverse drug reactions (ADRs). We created an annotated corpus of 10,822 tweets. Each tweet was annotated for the presence or absence of ADR mentions, with the span and Unified Medical Language System (UMLS) concept ID noted for each ADR present. Using Cohen’s kappa1, we calculated the inter-annotator agreement (IAA) for the binary annotations to be 0.69. To demonstrate the utility of the corpus, we attempted a lexicon-based approach for concept extraction, with promising success (54.1% precision, 62.1% recall, and 57.8% F-measure). A subset of the corpus is freely available at: http://diego.asu.edu/downloads.
Introduction
Prescription drug usage continues to grow as part of health care strategies to increase health and improve quality of life. The National Center for Health Statistics (NCHS) reported that over a 10 year period, the percentage of those who were taking either ‘one’, ‘two or more’, or ‘five or more’ drugs increased 4%, 6% and 5% respectively.2 While these drugs are prescribed for their therapeutic properties, their use may result in unintended or adverse effects.3 An adverse drug reaction (ADR), as defined by the World Health Organization (WHO), is: “Any response to a drug which is noxious and unintended, and which occurs at doses normally used in man for the prophylaxis, diagnosis, or therapy of disease, or for the modifications of physiological function.”4 The morbidity and mortality rates associated with ADRs are considerable.
Pharmacovigilance is defined as “the science and activities relating to the detection, assessment, understanding and prevention of adverse effects or any other possible drug-related problems”.5 ADRs are the primary focus of pharmacovigilance. ADRs may be detected in either the pre-marketing clinical trials or post-marketing surveillance of a drug. Post-marketing surveillance has traditionally been dependent on spontaneous reporting systems (SRS), which are maintained and monitored by various regulatory agencies, such as the FDA’s Adverse Event Reporting System (FAERS). The FAERS provides consumers and providers with a system to report suspected ADRs. One of the main problems with SRS is that they are generally under-utilized due to the voluntary nature of reporting. A systematic review, conducted in 2006, estimated an under-reporting rate of 85–94%.6 Recognizing the limitations of the SRS, the FDA launched an active surveillance initiative in 2008.
One of the FDA’s Sentinel Initiative’s aims is to use existing health data, such as electronic health records and claims data, to actively monitor for adverse event signals.7 In addition to using existing health data to automatically detect ADRs, research has explored other data sources for this information. One such source is online social networks, in particular health related networks such as DailyStrength or PatientsLikeMe. Here, patients can freely discuss and share their experience with drug products. One of the first efforts to find ADRs in user posted comments was made by Leaman et al.7 They used a modified lexicon based approach to extract drug and ADR relationships from user comments in the health networking and forum site DailyStrength.com. The authors concluded that user comments do contain extractable information regarding ADRs and these consumer reported effects do overlap with the known adverse effects. Generic social networking sites, such as Twitter, have also been found to contain valuable health related information. Recent research has shown that Twitter data analytics can have broad implications on public health research. Paul and Dredze demonstrated that public health topics, including syndromic information, geography based risk factors, and information about symptoms and medication, could be extracted from Twitter.8 One limitation they note is the need for more data in order to produce better results.
Twitter is a valuable resource for researchers because the discussions are publicly available and can be accessed through the Twitter’s streaming application programming interface (API). This unfettered access to volumes of up-to-date information is not without its challenges for natural language processing (NLP) researchers looking to automatically extract relevant information. One difficulty is the paucity of relevant data, preliminary analysis demonstrated that less than 1% of tweets that included a drug name had an actual ADR mention. In contrast, our recent analysis of data from the health forum DailyStrength.com shows approximately 24% of the comments mention an ADR. Twitter would seem too sparse for exploration. However, with over 645 million users, 58 million tweets per day9, and 80% of users posting tweets about themselves,10 the value of Twitter as a source of similar data calls for careful reconsideration, even if one has to collect data over a longer period to get a large data volume.
Another challenge with Twitter is in the lack of structure in tweets. Tweets are restricted in character count, forcing users to condense their sentiments while still conveying the intended meaning. This leads to highly unstructured text that contains many abbreviations, some of which are unique to Twitter. They contain frequently misspelled words, colloquialisms, idioms, and metaphors that make automatic processing more difficult. For the task of mining for ADR mentions, these issues compound an already challenging problem.
Ritter et al. examined the performance of standard natural language processing tools (NLP) for named entity recognition (NER) in tweets.11 They found that standard NLP tools performed poorly due to the compressed nature of the tweets, which eliminates the context needed to find an entity’s type, and the relatively infrequent use of unique entity types, which makes it difficult to amass sufficient training data. They developed a set of NLP tools, specific to Twitter that outperformed the standard tools by a margin of 25%.
Research into the use of Twitter for detection of drug and adverse effect events has been sparse. Bian et al. utilized a data set of 2 billion tweets to explore the use of Twitter for real-time pharmacovigilance.12 They chose five cancer drugs and mined the data set for mentions of those drugs. They then developed a SVM classifier to identify adverse effects caused by the drugs. Their results were qualified by the authors as “rather low”, which they ascribe to: the nature of the tweets (unstructured, abbreviated words), the use of NLP tools that were created for more structured text and the use of nonmedical terms by the users that made matching difficult.
In this paper, we used a specifically selected corpus to evaluate the viability of Twitter as a source of ADR mentions and its potential value for pharmacovigilance. We anticipate that our findings could help augment other signals used to detect relationships between a drug and an adverse reaction.
Methods
To create a corpus of highly relevant data for our task, we collected tweets for 74 drugs (target drugs). A total of over 187,000 tweets that contained a mention of one of our target drugs were collected. After filtering out likely advertisements and balancing the data set, our final corpus consisted of 10,822 tweets. The corpus was annotated in two stages. The first was a binary annotation of each tweet indicating whether it contained an ADR mention or not. A combination of the tweets identified as containing an ADR and a randomly selected sample of 1,000 tweets that were marked as not containing a mention of an ADR were then annotated for specific concept spans and UMLS concept IDs. We annotated the spans and UMLS IDs for each adverse effect or indication mentioned, such that one tweet could have multiple mentions. We distinguished mentions about adverse reactions (an unexpected, negative effect resulting from the adequate use of the drug) from mentions about indications (the sign or symptom for which the drug was prescribed in the first place). This distinction is important, and it is a hard distinction to make even for annotators, let alone automated systems. A detailed description of these steps follows.
Data Acquisition
The 74 drugs included in this study are broadly used drugs whose adverse effects are well known (truth set), plus drugs released between 2007 and 2010, for which not all adverse effects are yet known and will only become visible as the drugs are more widely used (test set). Drugs in the truth set were selected on the basis of their widespread use, as demonstrated by their presence in the Top 200 products by volume in the US market published by IMS Health; and also based on whether they had a known adverse effect in at least one of the categories of interest. Many of the drugs for the truth set have come into widespread use recently, which allows for testing the capability of the NLP methods to confirm signals that are now known, but not so at the time of release. For the newer drugs, going back to 2007 allows for market growth leading to common prescribing and thus, likely presence in tweets. The list was narrowed based upon forecasts for widespread use, the prevalence of disease states and conditions, and on whether the drug was new in class. Major categories include drugs for central nervous system and mental health conditions such as Alzheimer’s disease and schizophrenia. Treatments for age-related diseases (such as diabetes, cardiovascular diseases, urinary dysfunction, and musculoskeletal disorders) also met the criterion for potential widespread use, considering increased life expectancy. Additionally, biologics are an important class that was included because adverse events have traditionally been difficult to detect. Given the lag between FDA approval and recognition of serious adverse events in this category, we selected four biologics to follow as part of the experimental set, and one is included in the truth set.
The drugs names, both the generic and brand, were used as keywords in the Twitter Search API. One of the first challenges we faced with acquiring the relevant tweets was accounting for a wide variety of drug name misspellings. To ensure that as much data as possible was obtained, we also used projected misspellings of drug names as keywords. The misspellings were created using a phonetic spelling filter that generates variants based on the phenomes of the correct spelling.13 The filter generated many variants, and about 18% of them were added to the keyword list for reasonable coverage. Those were selected based on their Google search web prevalence.
The expanded drug list was used to search Twitter via the API. A total of 187,450 tweets were collected over a seven month period (August 2013 – February 2014). These required further filtering to remove advertisements and to balance the dataset among the drugs. This was accomplished by first removing texts containing URLs. Retweets were not removed. The number of tweets was thus reduced to 71,571. Next, to balance the number of tweets for each drug in the corpus, a maximum of 500 to 800 tweets for each drug name variant was randomly selected. This was necessary due to the vast imbalance of tweets containing mentions of some popular drug names as compared to others. Also, maintaining a soft limit, rather than a hard one, ensured that drugs that are highly commented upon also contain a higher number of samples in the corpus. This reduced the final corpus to 10,822 tweets. This process is summarized in Figure 1. A full description of the corpus appeared in Ginn et al.14 A subset of the corpus is available online at http://diego.asu.edu/downloads.
Figure 1.
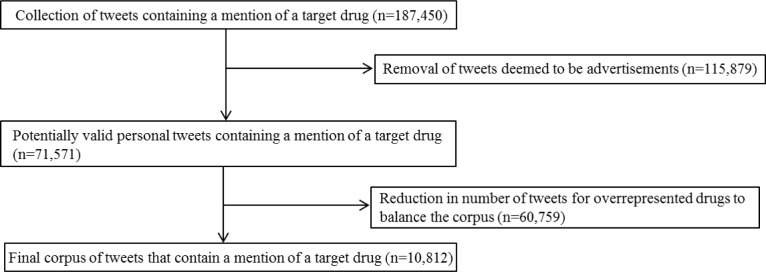
Summary of tweets collected and subsequent reductions to obtain the final corpus.
Annotation
For annotation, we defined an adverse reaction as “an undesired effect of the drug experienced by the patient.” This included mentions where the patient expressed the notion that the drug worsened their condition. An indication was defined as “the sign, symptom, syndrome, or disease that is the reason or the purpose for the patient taking the drug or is the desired primary effect of the drug. Additionally, the indication is what the patient, or prescriber believes is the main purpose of the drug.” The annotated spans were mapped to UMLS concept IDs found in the lexicon.
Our lexicon was derived from our earlier work, presented in Leaman et al.,7 and includes terms and concepts from four resources: the Coding Symbols for a Thesaurus of Adverse Reaction Terms (COSTART) vocabulary created by the U.S. Food and Drug Administration for post-market surveillance of adverse drug reactions (a subset of the UMLS Metathesaurus), which contains 3,787 concepts15; the side effect resource (SIDER), which contains 888 drugs linked with 1,450 adverse reaction terms extracted from pharmaceutical insert literature16; the Canada Drug Adverse Reaction Database, or MedEffect, which contains associations between 10,192 drugs and 3,279 adverse reactions.17 We added terms from SIDER 216 and the Consumer Health Vocabulary (CHV)18, which includes more colloquialisms.
The corpus was manually annotated in two stages by two expert annotators. The entire corpus was initially annotated for binary classification of the tweets as either ‘hasADR’ for those with a mention of an adverse reaction, following the definition described above, or ‘noADR’ for those without. The purpose of the binary classification was to validate our assumption that people would discuss their personal experiences with prescription drugs via tweets.
In the second stage of annotation, a combination of all tweets from the corpus classified as ‘hasADR’ and a random selection of 1,000 tweets from the corpus that were classified as ‘noADR’ were annotated to capture the spans in the text that expressed either an adverse reaction or an indication. The spans of the mentions were annotated by the same annotators who performed the binary annotations following the rules set forth in the annotation guidelines. Some general guidelines include: only capturing mentions if the concept was experienced by the patient; if the patient reported the concept as the reason for taking the drug than that mention would be labeled as an indication; all concepts were to be mapped to the UMLS id that most closely matched the meaning of the span; and when annotating a span, the smallest number of words needed to convey the meaning were chosen. Table 1 illustrates a sample of tweets and their annotations.
Table 1.
A sample of the tweets collected from Twitter and their annotations
| Sample Comments | Classification | Annotations |
|---|---|---|
| 20s 8th day with #Effexor still experiencing some side effects (drowsiness,sleepiness,GI effects). Moderate improvement in mood #depression | hasADR | “drowsiness” - drowsiness: adverse effect, “sleepiness” - sleepiness: adverse effect, “GI effect” – gastro intestinal reaction: adverse effect, “depression” – depression: indication |
| Over-eaten AGAIN just before bed. Stuffed. Good chance I will choke on my own vomit during sleep. I blame #Olanzapine #timetochange #bipolar | hasADR | “over-eaten” – increased appetite: adverse effect, “bipolar” – bipolar disorder: indication |
| @brokenmind_ Quetiapine was horrific for me in relation to wait gain. Such a horror story. But the weight will come off one day at a time. | hasADR | “wait gain” – weight gain: adverse effect |
| Tomorrow, my second infusion of Tysabri! Good luck for me! #Godblessme #MSLife | noADR | “MS” multiple sclerosis:indication |
| Do not take Cymbalta if you breathe – stolen from Tay | noADR | |
| Rules of Prozac: 1: You can never sleep, ever again. NEVER EVER 2: No you may NOT switch your brain off. Ever. 3: Exhaustion is your friend. | hasADR | “never sleep” – insomnia: adverse effect, “not switch your brain off” – racing thoughts: adverse effect, “exhaustion” – exhaustion: adverse effect |
| @FriarDanny I appreciate it. I gained over 30lbs with Paxil so I’m trying something different, tired of the appetite side effects. | hasADR | “gained over 30lbs” – weight gain: adverse effect, “appetite” – increased appetite: adverse effect |
| This cipro is totally “killing” my tummy.. hiks.. | hasADR | “”killing” my tummy” – gastric pain: adverse effect |
| Well played tysabri…kicking butt #nosleep. | hasADR | “nosleep” – insomnia: adverse effect |
| @Sectioned_ @bipolarlife7 @BBCWomansHour ah yes, I’m starting to think my paroxetine turns panic attacks into fat. | hasADR | “panic attacks” – panic attack: indication, “fat” – weight gain: adverse effect |
The annotation of both adverse drug reactions and indications was necessary due to the fact that these two categories can have the same concepts associated with them. For example, a user may state “I take it for insomnia”, while another may say, “Had to stop treatment, it was causing insomnia.” The first mention is an indication and the second an adverse reaction but both would be mapped to the same UMLS concept id for insomnia. Thus, the type of the mention (adverse effect or indication) can vary, while the concept id can be the same.
Automatic Concept Extraction
We aimed at measuring the utility of lexicon-based techniques for the automatic concept extraction from Twitter data. Our proposed concept extraction method is based on an information retrieval technique that is flexible enough to deal with term variability in user posts. We used Apache Lucene1 for both indexing and retrieval of the ADR lexicon concepts. A Lucene index was built from concepts and the associated UMLS IDs in the lexicon. Before indexing, we preprocessed the concepts which included removal of stop words and lemmatization. The lexicon entries were lemmatized to WordNet2 roots using the Dragon toolkit3.
To identify the indexed concepts present in a tweet, we generated a Lucene search from preprocessed tokens in the tweet sentences. We split tweets into sentences using Stanford Tokenizer4. The preprocessing of the sentences included spelling corrections, stop word removal, and lemmatization. For spelling corrections, we used Lucene SpellChecker that was customized to suggest a list of correct spellings for a given word based on ADR lexicon and a list of common English words from SCOWL2 (Spell Checker Oriented Word Lists).
The spans of the mentions associated with the retrieved lexicon concepts were then identified in the sentences using string comparison with regular expressions. The technique is flexible enough to identify both single and multi-word concepts, regardless of the order of the words in sentences or presence of other words in between.
Since the focus of this study is to extract adverse effects from the tweets, a manual filtering method was implemented to remove indications. This method filters the terms based on verbs that precede the term, such as “helps” or “works” for indications.
Results
Annotation statistics
After filtering out advertisements, the corpus contained tweets for 54 of the 74 target drugs. The number of tweets returned for four of these drugs was considerably higher than the rest and their numbers were reduced by making a random selection of tweets from each in order to balance out the dataset. From the resulting corpus of 10,822 tweets, 1,008 were determined during the binary phase of annotation to contain at least one mention of an adverse effect. These tweets were from 31 of the target drugs. The total number of adverse effects annotated was 1,285. The top ten drugs in order of the number of adverse effect mentions annotated are listed in Table 2.
Table 2.
Top 10 drugs by sorted by the number of ADR mentions annotated. The generic name is indicated for those that were added to the original drug list for searching.
| Drug Brand/Generic Name | ADR Mentions Annotated per Drug | Total Number of Tweets in Corpus |
|---|---|---|
| Seroquel/quetiapine | 237 | 1,082 |
| Effexor/venlafaxine | 176 | 461 |
| Vyvanse | 122 | 800 |
| Paxil/paroxetine | 92 | 683 |
| Prozac/fluoxetine | 76 | 1,307 |
| Lamictal/lamotrigine | 73 | 395 |
| Zyprexa/olanzapine | 68 | 377 |
| Humira | 64 | 560 |
| Cymbalta/duloxetine | 63 | 832 |
| Trazodone | 58 | 530 |
Agreement between the annotators was measured by calculating inter-annotator agreement (IAA). We calculated IAA for span using a partial matching criterion. This means that the annotators were considered to be in agreement if there was some overlapping portion in the span selected by each annotator. IAA for concept ID and binary annotations was calculated using an exact matching criterion. The precision, recall and F-measures19 associated with IAA of the annotations are shown in Table 3. For binary annotations, a kappa value of 0.69 was calculated.
Table 3.
IAA for span, concept ID and binary classification
| IAA Type | Precision | Recall | F-measure |
|---|---|---|---|
| Span | 0.8099 | 0.9802 | 0.8869 |
| Concept ID | 0.7278 | 0.9688 | 0.8311 |
| Binary Classification | 0.8155 | 0.8829 | 0.8448 |
Automatic concept extraction and identification
The system was evaluated on 1,873 annotated tweets, for adverse reactions only. We defined true positives as those extracted by the machine that match with the mentions labeled by annotators using partial matching.. Our system achieved 54.1% precision, 62.1% recall, and 57.8% F-measure for this experiment.
Error Analysis
For error analysis of the system, we chose a random sample of 50 false positive (FP) and 50 false negative (FN) results to review. We analyzed each group separately and categorized the errors by error type (Figure 2). For FPs, the errors fell into three main categories: the extraction of indications as ADRs, the extraction of terms from the tweets that were in the lexicon but not being used to discuss an ADR, and extraction of ADR mentions that were not experienced directly by the user. FN results were separated in to two main categories: the ADR was expressed using colloquial or descriptive terminology, or the ADR mention was expressed in a similar but not a direct match to the lexicon entry.
Figure 2.
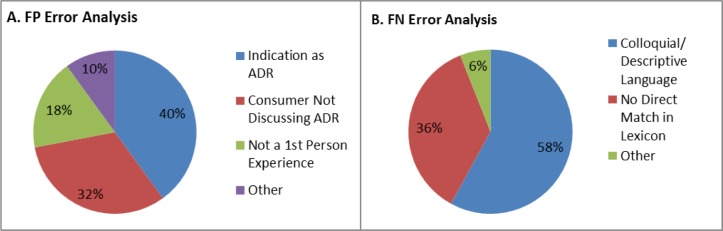
2A shows the category and prevalence of the 50 FP errors analyzed. 2B shows the category and prevalence of the 50 FN errors analyzed.
Discussion
To assess the validity of finding ADRs in tweets, we compared the adverse effects that were found in the tweets to known and documented adverse effects. The results are reported in Table 4 for the top ten drugs as determined by the number of ADR mentions annotated. We found at least one corresponding adverse effects in all ten. In a few instances, the most frequent adverse event in the tweets aligned with the most frequently found adverse event in the clinical trials. We only assessed the relationship between the most frequent adverse reactions. Given the known adverse effects were from clinical trials, it may be interesting to examine less frequently occurring events in the trials as compared to the mentions. The relatively small sample size and short duration of the trial may not adequately represent such rare events, but these may get elucidated in the wide spread, long term Twitter evidence.
Table 4.
List of drugs included in preliminary analysis, with their most common adverse reactions, frequency of incidence in adults, as listed in the FDA’s online drug library.22 The last column shows the most frequent adverse effects extracted from the Twitter data using our automated system. Effects found in both are highlighted in bold.
| Drug Brand/Generic Name | Primary Indications | Documented Adverse Effects (no order) | Adverse Effects Found in Tweets (Frequency) |
|---|---|---|---|
| Seroquel/Quetiapine | Schizophrenia, Bipolar I Disorder: manic episodes, Bipolar Disorder | somnolence, dry mouth, headache, dizziness, asthenia, constipation, fatigue | somnolence (22.2%), abnormal dreams (9.6%), feel like a zombie (8.1%), weight gain (6.6%), restless leg syndrome (6.6%), increased appetite (5.9%), sleep paralysis (2.9%), dizziness (2.2%), psychosis (2.2%), tremors (2.2%) |
| Effexor/venlafaxine | Major Depressive Disorder (MDD) | nausea, headache, somnolence, dry mouth, dizziness | withdrawal syndrome (21.3%), insomnia (11.1%), headache (4.3%), malaise (4.3%), abnormal dreams (4.3%), nausea (3.4%), shaking (3.4%), fatigue (3.4%) |
| Vyvanse | ADHD | decreased appetite, insomnia, dry mouth, diarrhea, nausea | insomnia (38.2%), OCD (9.3%), anger (5.6%), heart racing (5.6%), depression (3.6%), psychosis (3.6%), headache (3.6%), feel weird (3.6%) |
| Paxil/Paroxetine | MDD, Obsessive Compulsive Disorder (OCD), Panic Disorder, Social Anxiety Disorder, Generalized Anxiety Disorder (GAD), PTSD | nausea, somnolence, abnormal ejaculation, asthenia, tremor, insomnia, sweating | withdrawal syndrome (27.7%), weight gain (12.8%), depression (8.5%), headache (6.4%), somnolence (6.4%), allergic (6.4%), feel sick (6.4 %), emotional (6.4%) |
| Prozac/Fluoxetine | MDD, OCD, Bulimia Nervosa Panic Disorder | nausea, headache, insomnia, nervousness, anxiety, somnolence | somnolence (22.2%), withdrawal syndrome (8.9%), feeling ill (8.9%), abnormal dreams (6.7%), suicidal thoughts (6.7%), tremors (6.7%), allergic reaction (4.4%) |
| Lamictal lamotrigine | Epilepsy, Bipolar Disorder | vomiting, coordination abnormality, dizziness, rhinitis, dyspepsia, nausea, headache, diplopia, ataxia, insomnia, fatigue, back pain | insomnia (17.9%), rash (12.8%), lethargy (7.7%), joint pain (5.1%), feel like a zombie (5.1%), feel sick (5.1%) |
| Zyprexa/olanzapine | Schizophrenia, Bipolar I Disorder | dizziness, constipation, personality disorder, weight gain, akathisia, somnolence, dry mouth, asthenia, dyspepsia | weight gain (40.0%), somnolence (11.4%), increased appetite (8.6%), dependence (5.7%) |
| Humira | Rheumatoid Arthritis, Juvenile Idiopathic Arthitis, Psoriatic Arthitis, Crohn’s Disease, Ulcerative Colitus, Plaque Psoriasis | upper respiratory infection, rash, headache, sinusitis, accidental injury | somnolence (24%), feel sick (8%), palpitations (8%), ache/pains (8%), joint pain (4%), headache (4%), rash (4%), respiratory disorder (4%) |
| Cymbalta/Duloxetine | MDD, GAD, Diabetic Peripheral Neuropathy, Fibromyalgia, Chronic Muscoskeletal Pain | nausea, headache, dry mouth, fatigue, somnolence | withdrawal syndrome (16.3%), fatigue (14.0%), somnolence (7.0%), dizziness (7.0%), dry mouth (4.7%), depression (4.7%), rash (4.7%), migraine (4.7%) |
| Trazodone | MDD | somnolence, headache, dry mouth, dizziness, nausea | somnolence (24.3%), abnormal dreams (16.2%), hangover effect (8.1%), headache (5.4%), insomnia (5.4%), withdrawal syndrome (5.4%) |
Compared to the 78.3% precision and 69.9% recall we reported in Leaman et al.7, for the DailyStrength corpus for concept extraction, we see that the performance against the Twitter dataset is much lower. This could be due to the inherent characteristics of a tweet, with greater presence of “creative” expressions that do not match even the augmented lexicon. There is evidence of this when we look at the sources of FN errors. The largest source of these errors was the language used to describe the ADR.
Some of these issues stem from using inexact terminology to describe the effect, which has no match in the lexicon; examples include ‘in a haze’ or ‘wired’. Not surprisingly, this issue also arises when the user uses an idiomatic expression or metaphors to describe the effect. For example, to express the adverse effect of dry mouth, a user tweeted “….it feels like the Sahara desert in my mouth”. This was also the largest reason for errors reported by Leaman et al.7, but with a slightly smaller percentage of their associated FNs (55%). In order to mitigate these errors, other approaches that are less reliant on lexicon matching are being explored. Nikfarjam and Gonzalez21 have proposed using association rule mining to extract ADR mentions from colloquial text, extracting frequent patterns to find mentions instead of using a dictionary match. Pattern-based extraction showed improvements over the lexicon based approach, but the method requires a large amount of training data.
The FP errors were mainly due to misclassifying extracted terms as ADRs when they were not used as such. The biggest source of these errors was instances where an indication was misclassified as an ADR. There were also FP errors caused by the extraction of terms that were not used for discussing either an ADR or an indication. Some examples of this include a user name that contains a lexicon term (‘TScpCancer’) or the use of a term that is not necessarily related to an ailment (‘Sick of meds’). Although the post-processing rules were effective in filtering some of the non ADR mentions, machine learning classification is needed in future research to examine the effectiveness of modeling the contextual and semantic features of the tweets in distinguishing different semantic types. In the future, we will explore the effectiveness of training a sequence labelling classifier such as Conditional Random Fields (CRFs) for this extraction task.
Another smaller, though notable, source of errors for FPs was the system’s extraction of adverse effects that were not experienced by the user. Our guidelines for annotation stipulated that mentions of adverse effects were to be annotated only if they were experienced by the patient, in order to prevent annotations of song/movie quotes, social gossip, or potential advertisements. This presents a challenge when utilizing a social networking site that is not devoted solely to health topics. The unstructured, informal, and isolated nature of the tweets compounds this challenge by making some tweets unclear. It can be difficult for annotators to discern the relationship of the person writing the tweet with the drug they are mentioning. There are tweets that seem to be a commentary or report on the drug and its side effects (“Effexor XR side effects : - sucidal thoughts - insomnia - feeling high & unresponsible. -Dry mouth.”), or of a song lyric (“I’m too numb to feel, blow out the candle, blindness #np #cymbalta”) but without any context of why the person is tweeting about it, it is difficult to know. It could be a way to relay a personal experience or a repetition of something they heard. The determination of whether these are first person accounts requires the annotator to make a judgment call or the use of specialized knowledge about commonly used conventions in Twitter. In the song example above, using a common abbreviation to express a song you want to share on Twitter is #np (‘now playing‘) and this is often followed by #name_of_artist. However, the user who used #Cymbalta possibly indicated they are ascribing the meaning of the lyric to the drug. Training a system on the nuances of Twitter conventions, or being able to discern 3rd person from 1st person accounts, is a very significant challenge — especially when tweets are preprocessed with techniques that affect symbol and punctuation placement.
From Table 2, we can see that the number of tweets collected for a drug is not necessarily indicative of the number of ADRs that we can expect to find. This may be attributed to the fact that Twitter is a general social networking site and will contain a lot of noise when attempting to utilize its data for a specific research purpose. The prevalence of tweets may be due to the drug being the news, or the drug may have a catchy slogan used in its advertisement that gets tweeted frequently (such as Cymbalta’s “Depression hurts, Cymbalta can help”). To try to eliminate some of the noise, the binary data in the annotated corpus will be used in future work to refine our methods for searching and filtering for tweets that may contain ADRs.
In this study we aimed at exploring Twitter as a resource for the automatic extraction of ADRs, and the effectiveness of the commonly used lexicon-based techniques for ADR extraction from tweets. In the future, with more annotated data available, we will examine the utility of state-of-the-art machine learning classifiers such as CRF for concept extraction. Our current study has a number of limitations, which are as follows:
- Modified distribution of comments: Although we attempted to enforce a soft limit on the number of comments for each drug in the annotation set, their distributions in the annotation set are not representative of their actual distribution in real life data. A thorough analysis is required to identify if this will affect the performance of our system when applied to real life data.
- For some drugs, we were only able to collect a small number of tweets. More tweets associated with those drugs will be required to make reliable predictions in future tasks.
- We have not performed detailed experimentation to assess how the data imbalance problem posed by Twitter data affects performance of ADR detection systems. We will address this aspect of research in the future.
Conclusion
We have shown that people do tweet about their adverse effects experiences with their prescription drug use. In these tweets, they mention the drug name, along with the adverse effect(s), making it possible to automatically extract the drug and adverse effect relationship. We were, however, only able to achieve moderate success with the lexicon based system used to automatically extract the adverse effect mentions. A large part of this difficulty is due to the problem of using a formal lexicon to match to colloquial text. Another significant problem is the large data imbalance associated with data collected from Twitter. In the future, we will also explore machine learning algorithms that take into account the data imbalance issue with Twitter data. Our continuing efforts at annotation of data makes the application of supervised learning approaches a lucrative future possibility.
Footnotes
References
- 1.Carletta J. Assessing agreement on classification tasks: the kappa statistic. Comput Linguist. 1996;22(2):249–254. [Google Scholar]
- 2.Gu Q, Dillon CF, Burt V. Prescription Drug Use Continues to Increase: U.S. Prescription Drug Data for 2007–2008. [Accessed March 8, 2014]. Available at: http://www.cdc.gov/nchs/data/databriefs/db42.pdf. [PubMed]
- 3.Edwards IR, Aronson JK. Adverse drug reactions: definitions, diagnosis, and management. Lancet. 2000;356(9237):1255–9. doi: 10.1016/S0140-6736(00)02799-9. [DOI] [PubMed] [Google Scholar]
- 4.Safety of Medicines - A Guide to Detecting and Reporting Adverse Drug Reactions - Why Health Professionals Need to Take Action: References. World Heal Organ. 2002. [Accessed March 9, 2014]. Available at: http://apps.who.int/medicinedocs/en/d/Jh2992e/12.html.
- 5.Lindquist M. The Need for Definitions in Pharmacovigilance. Drug Safety. 2007;30(10) doi: 10.2165/00002018-200730100-00001. [DOI] [PubMed] [Google Scholar]
- 6.Hazell L, Shakir SA. Under-Reporting of Adverse Drug Reactions: A Systematic Review. Drug Safety 2006. 2006;29(5):385. doi: 10.2165/00002018-200629050-00003. 12p. 4 Charts. [DOI] [PubMed] [Google Scholar]
- 7.Platt R, Wilson M, Chan KA, Benner JS, Marchibroda J, McClellan M. The New Sentinel Network — Improving the Evidence of Medical-Product Safety. N Engl J Med. 2009;361(7):645–647. doi: 10.1056/NEJMp0905338. [DOI] [PubMed] [Google Scholar]
- 8.Paul M, Dredze M. You are what you tweet: analyzing Twitter for public health. [Accessed March 9, 2014];Proceedings of the Fifth International AAAi Conference on Weblogs and Social Media. 2011:265–72. Available at: http://www.aaai.org/ocs/index.php/ICWSM/ICWSM11/paper/viewFile/2880/3264. [Google Scholar]
- 9.Twitter Statistics | Statistic Brain. [Accessed March 9, 2014]. Available at: http://www.statisticbrain.com/twitter-statistics/
- 10.Naaman M, Boase J, Lai CH. Proceedings of the 2010 ACM conference on Computer supported cooperative work - CSCW ’10. Vol. 2010. New York, New York, USA: ACM Press; Is it really about me? p. 189. [DOI] [Google Scholar]
- 11.Ritter A, Clark S, Mausam, Etzioni O. Named entity recognition in tweets: an experimental study. 2011. [Accessed March 13, 2014]. pp. 1524–1534. Available at: http://dl.acm.org/citation.cfm?id=2145432.2145595.
- 12.Bian J, Topaloglu U, Yu F. Proceedings of the 2012 international workshop on Smart health and wellbeing - SHB ’12. New York, New York, USA: ACM Press; 2012. Towards large-scale twitter mining for drug-related adverse events; p. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pimalkhute P, Patki A, Nikfarjam A, Gonzalez GH. Phoenitic Spelling Filter for Keyword Selection in Drug Mention Mining from Social Media. AMIA Summit. 2014 [PMC free article] [PubMed] [Google Scholar]
- 14.Ginn R, Pimalkhute P, Nikfarjam A, et al. Mining Twitter for adverse drug reaction mentions: A corpus and classification benchmark. BioTexM. 2014 [Google Scholar]
- 15.Coding Symbols for Thesaurus of Adverse Reaction Terms (COSTART) Source Information. [Accessed April 9, 2014]. Available at: http://www.nlm.nih.gov/research/umls/sourcereleasedocs/current/CST/
- 16.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MedEffect Canada - Health Canada. [Accessed April 11, 2014]. (n.d.). Available at: http://hc-sc.gc.ca/dhp-mps/medeff/index-eng.php.
- 18.Zeng-Treitler Q, Goryachev S, Tse T, Keselman A, Boxwala A. Estimating consumer familiarity with health terminology: a context-based approach. J Am Med Inform Assoc. 2008;15(3):349–56. doi: 10.1197/jamia.M2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc. 2013;20(5):806–13. doi: 10.1136/amiajnl-2013-001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leaman R, Wojtulewicz L, Sullivan R, Skariah A, Yang J, Gonzalez G. Towards Internet-Age Pharmacovigilance: Extracting Adverse Drug Reactions from User Posts to Health-Related Social Networks. Proc 2010 Work Biomed Nat Lang Process. 2010 Jul;:117–125. [Google Scholar]
- 21.Nikfarjam A, Gonzalez GH. Pattern mining for extraction of mentions of Adverse Drug Reactions from user comments. [Accessed March 5, 2014];AMIA Annu Symp Proc. 2011 2011:1019–26. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3243273&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- 22.FDA Drugs@FDA. [Accessed March 13, 2014]. Available at: http://www.accessdata.fda.gov/scripts/cder/drugsatfda/


