Abstract
Twenty-six million Americans are estimated to have chronic kidney disease (CKD) with increased risk for cardiovascular disease and end stage renal disease. CKD is frequently undiagnosed and patients are unaware, hampering intervention. A tool for accurate and timely identification of CKD from electronic medical records (EMR) could improve healthcare quality and identify patients for research. As members of eMERGE (electronic medical records and genomics) Network, we developed an automated phenotyping algorithm that can be deployed to identify rapidly diabetic and/or hypertensive CKD cases and controls in health systems with EMRs It uses diagnostic codes, laboratory results, medication and blood pressure records, and textual information culled from notes. Validation statistics demonstrated positive predictive values of 96% and negative predictive values of 93.3. Similar results were obtained on implementation by two independent eMERGE member institutions. The algorithm dramatically outperformed identification by ICD-9-CM codes with 63% positive and 54% negative predictive values, respectively.
Introduction
Chronic kidney disease (CKD) affects an estimated 10% to 15% of individuals in the United States, Europe and Asia. (1) CKD is a largely asymptomatic (‘silent’) yet serious condition associated with premature mortality, decreased quality of life, and increased health care expenditure. Approximately two thirds of CKD are attributable to diabetes (40% of CKD cases) and hypertension (28% of cases). (2) CKD is defined in most cases clinically by loss of kidney function as estimated by glomerular filtration rate (eGFR) below a threshold of 60 ml/min/1.73m2 (normal eGFR range 90 to 120 ml/min/1.73m2) and/or persistent increased urinary albumin excretion lasting more than 90 days. (2) Thus, identification of the vast majority of cases of diabetes- and/or hypertension-attributable CKD rests on appropriate and timely ordering and interpretation of eGFR and/or urinary albumin excretion laboratory results.
Untreated CKD can result in end-stage renal disease (ESRD) and necessitate dialysis or kidney transplantation in 2% of cases. (3) CKD is also a major independent risk factor for cardiovascular disease, all-cause mortality including cardiovascular mortality. (2) Current practice guidelines recommend tight control of blood pressure and/or hyperglycemia in particular in the presence of albuminuria to reduce ESRD and CVD risks in CKD patients. (4) However, CKD is alarmingly under diagnosed in affected primary care patients, even those with diabetes and/or hypertension. For example, among 122,502 adults enrolled in Kidney Early Evaluation KEEP, only 20% of participants with CKD stage 3 and 50% with stage 4–5 were aware of their disease. (5) Alarmingly, 43% or patients with newly diagnosed ESRD had not received specialist nephrology care and of those who did, only 25% had done so for more than one year. (5) As a result of the systematic and widespread failure to establish the diagnosis of CKD with inexpensive routine laboratory tests in primary care, affected yet unaware patient populations may not benefit from preventive measures to reduce major outcomes. The Electronic Medical Records and Genomics (eMERGE) Network is a National Human Genome Research Institute (NHGRI)-funded consortium tasked with developing methods and best-practices for the utilization of the Electronic Medical Record (EMR) as a tool for genomic research. (6) The Network’s phenotyping workgroup established best practices to develop and use phenotyping algorithms processing EMR data from disparate sources such as diagnosis and procedure codes, laboratory data, medication use, and imaging studies in order to identify cases and controls with a high degree of accuracy and confidence. The Network’s PheKB is a repository for phenotype algorithms. Phenotype algorithms on PheKB are validated at the creating site as well as at least 2 other Network institutions.
Here we describe the development of an automated algorithm as part of the eMERGE phenotyping framework that combines data from various EMR sources to identify diabetic and/or hypertensive patients with CKD. To the best of our knowledge, there have been no previous attempts to combine disparate sources of EMR data to identify CKD cases/controls. Positive and negative predictive values of the algorithm were validated extensively at the primary development site, Mount Sinai Medical Center, and significantly outperformed common recorded International Classification of Diseases, Ninth Revision, Clinical Modification (ICD-9-CM) codes associated with CKD. Finally, the CKD algorithm was successfully implemented and validated at two additional eMERGE member institutions. Thus, our novel phenotyping tool can be readily deployed by modern health systems to identify undiagnosed cases together with established cases of CKD associated with diabetes and/or hypertension for population management, quality improvement or research initiatives.
Materials and Methods
Participating Site Overview
Three institutions participated in the validation of this algorithm. Each institution uses an EMR and the key features of this are presented in Table 1. All participants represent the population receiving routine clinical care at the study institutions.
Table 1.
Overview of participating institutions’ EMR and recruitment models
| Institution | EMR Overview | Recruitment Model |
|---|---|---|
| Icahn School Of Medicine at Mount Sinai (New York, NY, USA) | Comprehensive vendor-based inpatient and outpatient EMR (Epic Systems, Verona, Wisconsin, USA) since 2003 with 10+ years ICD-9 data | Clinical population based |
| Marshfield Clinic Research Foundation (Marshfield, Wisconsin, USA) | Comprehensive internally developed EMR since 1985 75% participants have 20+ years medical history | Clinical population based |
| Columbia University Medical Center (New York, NY, USA) | Comprehensive vendor-based and self-developed inpatient and outpatient EMRs (Allscripts and iNYP) since 1990s with 22+ years of structured and unstructured data | Clinical population based |
Algorithm development
We used the 2012 Kidney Disease: Improving Global Outcomes (KDIGO) criteria for defining CKD stage 3 or higher (eGFR<60 ml/min/1.73m2 for duration ≥3 months, based on documentation or inference. The approach utilized common EMR data including diagnostic codes, medications and laboratory results. All sites utilized the International Classification of Diseases, 9th revision, clinical modification (ICD9-CM) diagnostic codes. Table 2 shows the ICD9 diagnosis and procedure codes applied in the algorithm along with the stages in the algorithm where they were used. ICD9 codes for diabetic/hypertensive kidney disease (A1, B2); kidney transplant (B1); dialysis (B3) were used for inclusion while codes for acute kidney failure (B5) and other causes of kidney disease including HIV, immunologic and developmental causes (B9) were used for exclusion criteria. We also implemented text searches to identify terms in physician observation reports for exclusion criteria. These text searches were picked by GNN and EPB using professional expertise as nephrologists and are listed at the end of Table 2. We chose to utilize a rule-based methodology because this approach has been deployed at eMERGE member institutions collaborating on phenotype development and has proven to be replicable across institutions and to produce strong predictive values.
Table 2.
ICD-9 Diagnosis, procedure codes and text searches applied in algorithm
| A1: 585.xx | Chronic kidney disease |
| B1: 55.6x | Transplant of kidney (procedure) |
| B1: V42.0 | Organ or tissue replaced by kidney transplant |
| B2: 250.4x | Diabetes with renal manifestations |
| B2: 403.xx | Hypertensive chronic kidney disease |
| B2: 404.xx | Hypertensive heart and chronic kidney disease |
| B3: V45.1 | Renal dialysis status |
| B3: V56.xx | Encounter for dialysis and dialysis catheter care |
| B3: 996.73 | Complications of renal dialysis |
| B3: 38.95 | Venous catheter for renal dialysis |
| B5: 584.xx | Acute kidney failure |
| B9:042.xx-044.xx | Human immunodeficiency virus (HIV) infection |
| B9: 282.6 | Sickle cell disease |
| B9: 581.xx | Nephrotic syndrome |
| B9: 582.xx | Chronic glomerulonephritis |
| B9: 583.xx | Nephritic and nephropathy |
| B9: 446.xx | Polyarteritis nodosa and allied conditions |
| B9: 447.6 | Vasculitis |
| B9: 753.xx | Renal agenesis and dysgenesis |
| Text search terms used in exclusion criteria | HIVAN/HIV associated nephropathy; Congenital [within 2 words of) kidney(s)]; APKD [adult polycystic kidney disease]; Sickle Cell Disease; IgA Nephropathy; Nephrotic Syndrome; Nephritic Syndrome; Glomerulonephritis; Glomerulosclerosis; Lupus Nephritis; Wegener’s granulomatosis; Goodpasture’s syndrome |
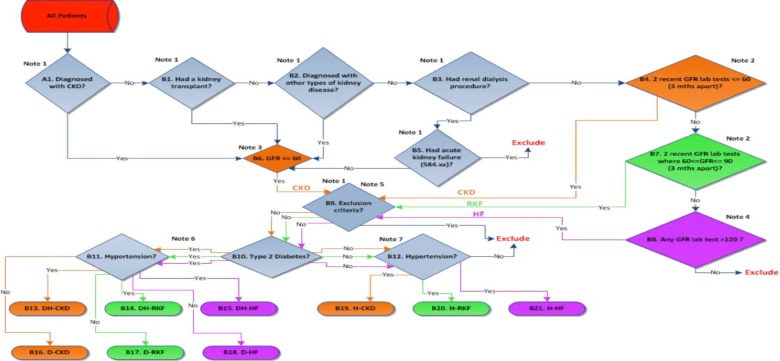
With respect to laboratory results, since various formulae are used to estimate the GFR leading to non-uniformity of eGFR results reported by clinical laboratories to the EMR. We overcome the apparent non-uniformity of eGFR lab results calculation and reporting in EHR by recalculating eGFR de novo with the CKD-EPI formula. (7) The CKD-EPI creatinine equation is based on the same four variables as the MDRD Study equation, but uses a 2-slope “spline” to model the relationship between estimated GFR and serum creatinine, and a different relationship for age, sex and race. The equation was reported to perform better and with less bias than the Modification of Diet in Renal Disease (MDRD) Study equation. We did not allow patients that had serum creatinine tests on consecutive days or within the same day as we assumed these were inpatient encounters. This was done to exclude acute kidney injury (AKI) events from the laboratory tests used for the algorithm. For patients that had duplicate serum creatinine tests meaning they had two tests at the same date/time but different values we kept the test with the maximum value in the algorithm. The patients fulfilling the criteria were then filtered through eMERGE Network’s type 2 diabetes (8) and a hypertension algorithm that was developed at Icahn School of Medicine at Mount Sinai. These validated algorithms are shown in Figure 1 and 2. The patients then were classified into diabetic CKD (DCKD), hypertensive CKD (HCKD) or diabetic/hypertensive CKD (DHCKD) cases. The complete algorithm for cases is shown in Figure 3.
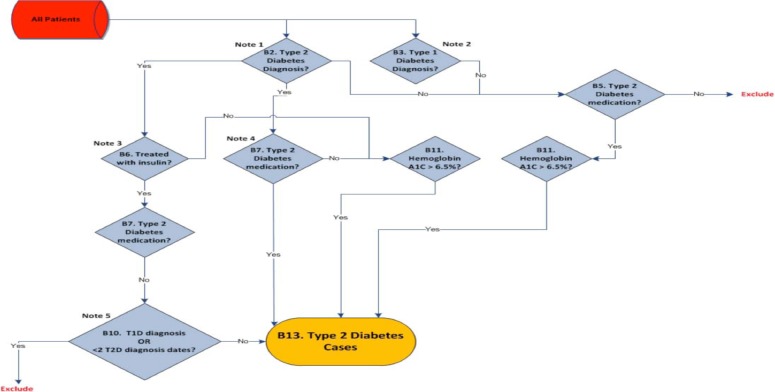
Figure 1.
Type 2 Diabetes case algorithm from the eMERGE Network
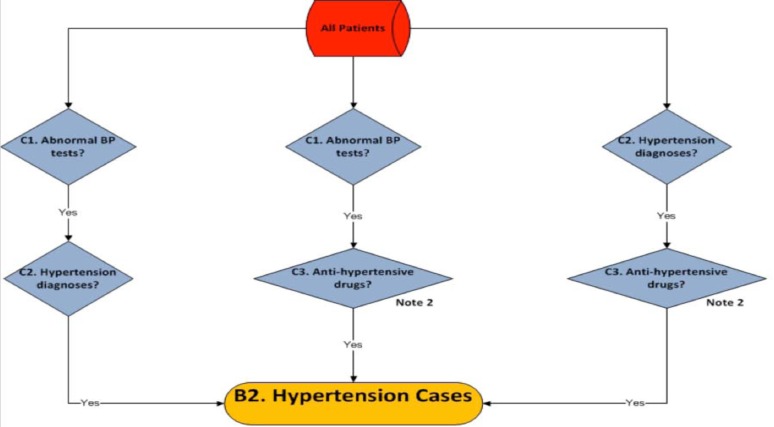
Figure 2.
Hypertension case algorithm developed at Icahn School of Medicine at Mount Sinai
Figure 3.
Phenotyping algorithm for chronic kidney disease cases
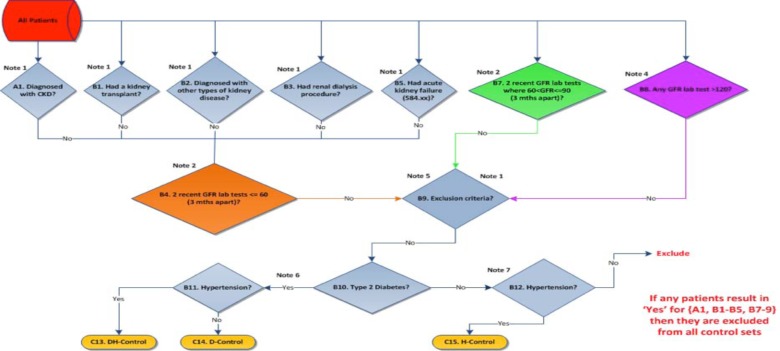
A similar approach was also adopted for the CKD controls algorithm with identical diagnostic codes, laboratory tests and text searches for exclusion criteria. Participants with 2 eGFR values from 60–89 ml/min/1.73m2 (reduced kidney function) and at least one eGFR>120 ml/min/1.73m2 (hyperfiltration) were also identified during this process. Since both reduced kidney function and hyperfiltration indicate an unrecognized early stage of kidney disease, including diabetic and hypertensive CKD they were excluded from all control sets. Once controls were identified, they were then filtered through eMERGE networks type 2 diabetes and hypertension algorithms and then classified as diabetic, hypertensive or diabetic/hypertensive controls. The algorithm for CKD controls is shown in Figure 4.
Figure 4.
Phenotyping algorithm for chronic kidney disease controls
Implementation
Data elements needed for identification of cases or controls were extracted from the EMR and stored in a shadow relational database. Deidentificaton, pruning (elimination of redundant data points), transformations (for example, separating units from numeric values when they come from the EMR as a single string) and data cleansing were performed as needed. SQL queries were developed and unit-tested for each object in the cases and controls algorithms. The queries were then combined to implement the logic of the algorithms. In the Mount Sinai implementation, the subjects and observations qualifying at each step of the algorithm were saved, facilitating subsequent validation reconciliation and algorithm refinements. A similar process of extracting EMR data to a shadow database and developing standalone queries was used at the validating sites. The decision logic for this algorithm and the use of specific terms extracted from textual clinical documentation (Table 2) exceed the clinical decision support capabilities embedded in commercial EMR’s. However Mount Sinai has developed an external CDS engine that is capable of executing complex coded decision support logic and is integrated with the Epic EMR, thereby enabling the automated CKD algorithm to be integrated into clinical workflows to provide alerts to clinicians of patients at risk for CKD. (9) The validating sites have internally developed EMR’s into which they can embed complex CDS code to provide alerts within existing clinical workflows. Sites using commercial EMR’s would most likely not be able to implement the CKD algorithm with the available technology.
Data collection
The algorithm was deployed to randomly select 600 each of CKD cases and controls from the EMR. Two independent physician reviewers (GNN and SF) at Mount Sinai Hospital manually reviewed each medical record. The gold standard for a case or a control was considered to be manual review by the physician reviewers. Any differences in agreement were arbitrated after discussion between the two reviewers. Identification of CKD in the EMR was ascertained using the CKD hierarchy of ICD9 codes (Table 3). A control was considered as correctly identified by ICD-9 codes if there was a diagnostic code identifying hypertension and/or diabetes as shown without an accompanying code for CKD (Table 3). While reviewing the charts, urine protein measurements as microalbumin/creatinine ratio and/or urine protein/creatinine ratios were also abstracted. When multiple measurements were available, the most recent measurement was recorded. In addition, documented referral to a nephrologist was abstracted. The algorithm was then deployed at Marshfield Clinic and Columbia University for secondary validation. Chart review by physician reviewers was used to validate 50 cases and 50 controls at each secondary site
Table 3.
ICD-9 codes used for identifying cases and controls
| Disease | ICD-9 code |
|---|---|
| End stage renal disease | 585.1 to 585.9 |
| Hypertensive chronic kidney disease, unspecified, with chronic kidney disease stage I through stage IV, or unspecified | 403.90 |
| Hypertensive nephropathy | 403.10 |
| Hypertensive renal disease | 403 |
| Hypertensive heart and renal disease | 404 |
| Diabetic nephropathy | 583.81 |
| Diabetic nephrosis | 581.81 |
| Diabetes with renal manifestations, type II or unspecified type, not stated as uncontrolled | 250.40 |
| Diabetes with renal manifestations, type II or unspecified type, uncontrolled | 250.42 |
| Diabetes with other specified manifestations, type II or unspecified type, not stated as uncontrolled | 250.80 |
| Diabetes with other specified manifestations, type II or unspecified type, uncontrolled | 250.82 |
| Intercapillary glomerulosclerosis | 581.81 |
| Kimmelstiel-Wilson syndrome | 581.81 |
| Hypertension | 401–405 excluding 403 and 404 |
| Diabetes Mellitus Type 2 | 250.00 to 250.93 excluding 250.40, 250.42,250.80 and 250.82) |
Data analysis
After manually reviewing the cases, the inter-rater agreement/kappa statistic was calculated. Summary statistics (positive and negative predictive values) for identification of cases and controls with the algorithm and ICD-9 codes with manual as the gold standard were estimated. Medians and interquartile ranges and proportion of missing values for the urine protein measurements and the percentage of patients that had been referred to a nephrologist were also calculated for the primary site. Positive and negative predictive values were then calculated individually for both secondary sites. All analyses were performed using STATA SE version 12, College Station, TX.
Results
A total of 1200 medical records were reviewed. Out of these, 14(1.16%) were excluded due to confidential status or missing data, leaving 1186 patients included in the final analysis. The inter-rater agreement/kappa statistic between the two independent reviewers (SF and GNN) was 92%. After arbitration of disagreements there were a total of 609 cases (202 for Diabetic CKD [DCKD], 207 for Hypertensive CKD [HCKD] and 200 for Diabetic and Hypertensive [DHCKD]) and 577 controls (190 for DCKD, 190 for HCKD and 197 for DHCKD) by manual review. The summary of the comparison between the algorithm and ICD9 codes are presented in Table 4 for the primary site. The comprehensive algorithm correctly identified 569/609(93.43%) of cases and 553/577(95.84%) of controls. In contrast, conventional screening with ICD9 codes only identified 244/609(40.06%) of cases and 433/577(75.04%) of controls. The positive predictive value (PPV) for the algorithm was 95.95% and the negative predictive value (NPV) was 93.25% compared to a PPV of 62.89% and a NPV of 54.26% compared to identification using ICD9-CM diagnostic codes. The algorithm performed similarly at secondary sites with a PPV of 92% and a NPV of 100%[Table 5].
Table 4.
Comparison of phenotyping algorithm and ICD-9 codes at primary site (Mount Sinai Hospital)
| Manual chart review | Manual chart review | ||||||
|---|---|---|---|---|---|---|---|
| Phenotyping Algorithm | Case | Control | Total | ICD-9 Codes | Case | Control | Total |
| Case | 569 | 24 | 593 | Case | 244 | 144 | 593 |
| Control | 40 | 553 | 593 | Control | 365 | 433 | 593 |
| Total | 609 | 577 | 1186 | Total | 609 | 577 | 1186 |
| Positive Predictive Value (95% Confidence Interval) | 95.95 (93.95–97.33) | Positive Predictive Value (95% Confidence Interval) | 62.89 (57.84–67.67) | ||||
| Negative Predictive Value (95% Confidence Interval) | 93.25 (90.85–95.08) | Negative Predictive Value (95% Confidence Interval) | 54.26 (50.73–57.75) | ||||
Table 5.
Performance of phenotyping algorithm at secondary sites (Marshfield Clinic and Columbia University Medical Center
| Marshfield Clinic | Columbia University Medical Center | |||||
|---|---|---|---|---|---|---|
| Manual chart review | Manual chart review | |||||
| Case | Control | Total | Case | Control | Total | |
| Phenotyping Algorithm | ||||||
| Case | 46 | 4 | 50 | 46 | 4 | 50 |
| Control | 0 | 50 | 50 | 0 | 50 | 50 |
| Total | 46 | 54 | 100 | 46 | 54 | 100 |
| Negative Predictive Value (95% Confidence Interval) | 92 (79.89–97.41) | 92 (79.89–97.41) | ||||
| Negative Predictive Value (95% Confidence Interval) | 100 (91.11–100) | 100 (91.11–100) | ||||
As part of secondary analysis the urine protein/creatinine or the urine microalbumin/creatinine values for DCKD, HCKD and DHCKD cases (Table 6). For DCKD, the median microalbumin/creatinine ratio was 39 microgram/mg of creatinine. Similarly for HCKD the median microalbumin/creatinine ratio was 5.5 microgram/mg of creatinine and the protein/creatinine ratio and for DHCKD, the median microalbumin/creatinine and protein/creatinine ratios were 35 microgram/mg of creatinine and 15 mg/mg of creatinine, with 37% and 61% of patients respectively lacking measurements at any point of time. However, there was no record of a urine albumin or protein excretion for a significant proportion of patients (ranging from 30–98%) depending on the subcategory of CKD. (Table 6) We also determined the proportion of participants that were referred to a nephrologist during any point of their clinical course in the EMR. Out of a total of 599 cases, only 112(18.7%) were referred to a nephrologist at any point during their course.
Table 6.
Microalbuminuria and proteinuria measurements in CKD cases and controls
| Diabetic CKD | ||
|---|---|---|
| Median (IQR) | N (%) of missing values | |
| Microalbumin/Creatinine | 39(10–215) | 59/200(30) |
| Hypertensive CKD | ||
| Median (IQR) | N (%) of missing values | |
| Urine microalbumin/creatinine | 5.5(3–28) | 196/200(98) |
| Urine protein/creatinine in mg/gm | 30(30–300) | 173/200(87) |
| Diabetic and hypertensive CKD | ||
| Median (IQR) | N (%) of missing values | |
| Urine microalbumin/creatinine | 35(8–127) | 74(37) |
Discussion
In the near future, EMRs will become one of the most important sources of data for both clinical and genomic association studies. Since data are present longitudinally, it may facilitate studying natural history of a disease process as well as the response to treatment in a “real world” scenario. However, identification of particular phenotypes, especially chronic, complex diseases, is challenging because of the complexity of data itself and the way in which it is recorded in EMR. However, with government interest driving the widespread use and adoption of EMR’s, this provides a vast and as-yet relatively untapped resource. (10) If robust phenotypes were constructed using meaningful information from various EMR sources, it would provide significant value for identifying patient cohorts that satisfy complex criteria. There has been significant debate about the optimal way to identify phenotypes in the EMR. (11) Automated approaches using electronic phenotyping and statistical analyses are popular as compared to simpler rule based systems. The utility of such phenotyping algorithms is manifold, including discovering novel genetic associations of complex diseases, tracking their natural history, isolating patients for clinical trials, and ensuring quality control in large institutions by ensuring that standard of care guidelines are met in these patients.
Kidney disease is a complex, common problem challenging modern healthcare. It is a major independent risk factor for all-cause mortality including cardiovascular mortality and adjusted rates of all-cause mortality are seven times greater for dialysis patients than for individuals in the general population. (3,12,13) As CKD is a significant health problem, accurate identification of diabetic and/or hypertensive CKD cases and controls for both research and clinical purposes is imperative. Accurate identification of individuals satisfying specific criteria from a large institutional population allows us to enroll for randomized trials, predict/track outcomes/progression, and perform retrospective cohort studies. (14,15) Studying the progression of complex diseases such as CKD is difficult as the recruitment of cohorts is a laborious process that creates a bottleneck in both clinical and translational research. In order to streamline this process, there has been an impetus to create EMR linked biobanks to enroll individuals in routine clinical care settings. The push from healthcare regulatory agencies for electronic medical records (EMRs) that provide a large amount of information available for research purposes has also been integral in improving the formation of research cohorts. (16) With appropriate patient consent and de-identifying data, the EMRs of patients are available and allow the studying of evolution and progression of disease. (17,18) In clinical care settings, a wealth of data is available through ICD-9 codes, discrete laboratory results, test reports, patient demographics, and notes written by the treating physicians. All of these data are available in a longitudinal form with multiple patient visits over several years. If matched to biobanks, the EMR can be used to identify traits/phenotypes in a large number of patients for biomarker/genomics research, thereby substantially reducing the effort and time needed to identify markers or variants that influence disease development, progression, or medication response. (18–20)
Although there are novel genetic associations including UMOD, APOL1 and SHROOM3, there are other potential genetic associations that explain the differential rates of CKD in different ethnic populations. (21,22) Clinical decision making is challenging due to variability in the rates of progression and lack of widely-accepted guidelines to identify patients most at risk of progression to ESRD. (23,24) For studies to assess progression over the course of the patient’s history in the EMR, accurate identification of large numbers of patients is needed. Currently the only way of identifying CKD cases/controls is by manually reviewing laboratory values, which is cumbersome, or through ICD9 codes. To accomplish these goals, researchers need robust phenotyping algorithms to effectively leverage disparate data sources in the EMR. To the best of our knowledge, this algorithm is one of the first automated phenotyping algorithms for diabetic/hypertensive CKD. It significantly outperformed conventional screening with ICD9 codes and could be deployed to different EMRs in various healthcare institutions. Thus, an integrated approach using diagnostic codes, medications, and laboratory tests yielded significant improvement over non-integrated approaches.
Although there are no recommended guidelines for nephrologist referral in CKD stage 3, there are studies suggesting that such referrals may improve prognosis. (25,26) However, we demonstrated that only 18.7% of confirmed CKD cases were referred to a nephrologist at any point during their EMR course. Thus, identifying appropriate patients for referral to a nephrologist is one of the many clinical applications of this algorithm.
Limitations
This algorithm does not include proteinuria or albuminuria measurements that are used to diagnose and stage CKD in addition to eGFR. This decision was based on the observation that in many instances urine albumin or protein excretion results are not recorded in the EMR. This approach was developed to classify coexisting prevalent CKD and T2D and/or HTN because in the vast majority of cases, T2D and/or HTN precede CKD, especially when other etiologies of kidney dysfunction are excluded (as they are in the algorithm). In a sub-analysis, we found that ≥ 95% of patients had a prior diagnosis of HTN and/or T2D before CKD, thus validating that the number of patients that might have CKD due to other etiologies are likely minimal. There was also a consideration of overfitting due to the high dimensionality of this problem under consideration. However, since the algorithm was replicated at multiple sites with near-identical results, this is likely to be negligible. Also, considering the low referral rate, it is possible that patients without EMR documented referral may have been referred to nephrologists outside the EMR.
Conclusions
In summary, we describe the development and validation of an automated algorithm for identifying diabetic/hypertensive CKD cases and controls and also demonstrate its superiority over traditional identification using ICD-9 diagnostic codes. We believe that this algorithm could be used to accurately and rapidly identify a specific target cohort within the EMR for both research and clinical purposes.
Acknowledgments
The eMERGE Network is funded by NHGRI, with additional funding from NIGMS through the following grants: U01HG004438 to Johns Hopkins University; U01HG004424 to The Broad Institute; U01HG004438 to CIDR; U01HG004610 and U01HG006375 to Group Health Cooperative; U01HG004608 to Marshfield Clinic; U01HG006389 to Essentia Institute of Rural Health; U01HG04599 and U01HG006379 to Mayo Clinic; U01HG004609 and U01HG006388 to Northwestern University; U01HG04603 and U01HG006378 to Vanderbilt University; U01HG006385 to the Coordinating Center; U01HG006382 to Geisinger Clinic; U01HG006380 to Icahn School of Medicine at Mount Sinai; U01HG006830 to The Children’s Hospital of Philadelphia; and U01HG006828 to Cincinnati Children’s Hospital and Boston Children’s Hospital.
References
- 1.Levey AS, Stevens LA, Coresh J. Conceptual model of CKD: applications and implications. Am J Kidney Dis Off J Natl Kidney Found. 2009 Mar;53(3 Suppl 3):S4–16. doi: 10.1053/j.ajkd.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 2.Levey AS, Coresh J. Chronic kidney disease. Lancet. 2012 Jan 14;379(9811):165–80. doi: 10.1016/S0140-6736(11)60178-5. [DOI] [PubMed] [Google Scholar]
- 3.National Institutes of Health, editor. Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: 2011. USRDS 2011 Annual Data Report. [Google Scholar]
- 4.Appel LJ, Wright JT, Greene T, Agodoa LY, Astor BC, Bakris GL, et al. Intensive blood-pressure control in hypertensive chronic kidney disease. N Engl J Med. 2010 Sep 2;363(10):918–29. doi: 10.1056/NEJMoa0910975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agrawal V, Jaar BG, Frisby XY, Chen S-C, Qiu Y, Li S, et al. Access to health care among adults evaluated for CKD: findings from the Kidney Early Evaluation Program (KEEP) Am J Kidney Dis Off J Natl Kidney Found. 2012 Mar;59(3 Suppl 2):S5–15. doi: 10.1053/j.ajkd.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med Off J Am Coll Med Genet. 2013 Oct;15(10):761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, Feldman HI, et al. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009 May 5;150(9):604–12. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc JAMIA. 2012 Apr;19(2):212–8. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman O, Scott SA, Ellis SB, Overby CL, Ludtke A, Hulot J-S, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther. 2013 Aug;94(2):214–7. doi: 10.1038/clpt.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Office of the National Coordinator for Health Information Technology Electronic Health Records and Meaningful Use. 2011. [Internet]. Available from: http://healthit.hhs.gov/portal.
- 11.Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, et al. A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc JAMIA. 2014 Apr;21(2):221–30. doi: 10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chronic Kidney Disease Prognosis Consortium. Matsushita K, van der Velde M, Astor BC, Woodward M, Levey AS, et al. Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: a collaborative meta-analysis. Lancet. 2010 Jun 12;375(9731):2073–81. doi: 10.1016/S0140-6736(10)60674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gansevoort RT, Matsushita K, van der Velde M, Astor BC, Woodward M, Levey AS, et al. Lower estimated GFR and higher albuminuria are associated with adverse kidney outcomes. A collaborative meta-analysis of general and high-risk population cohorts. Kidney Int. 2011 Jul;80(1):93–104. doi: 10.1038/ki.2010.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathias JS, Gossett D, Baker DW. Use of electronic health record data to evaluate overuse of cervical cancer screening. J Am Med Inform Assoc JAMIA. 2012 Jun;19(e1):e96–101. doi: 10.1136/amiajnl-2011-000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strom BL, Schinnar R, Jones J, Bilker WB, Weiner MG, Hennessy S, et al. Detecting pregnancy use of non-hormonal category X medications in electronic medical records. J Am Med Inform Assoc JAMIA. 2011 Dec;18(Suppl 1):i81–6. doi: 10.1136/amiajnl-2010-000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med. 2010 Aug 5;363(6):501–4. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 17.McCarty CA, Nair A, Austin DM, Giampietro PF. Informed consent and subject motivation to participate in a large, population-based genomics study: the Marshfield Clinic Personalized Medicine Research Project. Community Genet. 2007;10(1):2–9. doi: 10.1159/000096274. [DOI] [PubMed] [Google Scholar]
- 18.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinforma Oxf Engl. 2010 May 1;26(9):1205–10. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll RJ, Eyler AE, Denny JC. Naïve Electronic Health Record phenotype identification for Rheumatoid arthritis. AMIA Annu Symp Proc AMIA Symp AMIA Symp. 2011;2011:189–96. [PMC free article] [PubMed] [Google Scholar]
- 20.Birman-Deych E, Waterman AD, Yan Y, Nilasena DS, Radford MJ, Gage BF. Accuracy of ICD-9-CM codes for identifying cardiovascular and stroke risk factors. Med Care. 2005 May;43(5):480–5. doi: 10.1097/01.mlr.0000160417.39497.a9. [DOI] [PubMed] [Google Scholar]
- 21.Köttgen A, Glazer NL, Dehghan A, Hwang S-J, Katz R, Li M, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009 Jun;41(6):712–7. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsa A, Kao WHL, Xie D, Astor BC, Li M, Hsu C, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013 Dec 5;369(23):2183–96. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keane WF, Zhang Z, Lyle PA, Cooper ME, de Zeeuw D, Grunfeld J-P, et al. Risk scores for predicting outcomes in patients with type 2 diabetes and nephropathy: the RENAAL study. Clin J Am Soc Nephrol CJASN. 2006 Jul;1(4):761–7. doi: 10.2215/CJN.01381005. [DOI] [PubMed] [Google Scholar]
- 24.Keith DS, Nichols GA, Gullion CM, Brown JB, Smith DH. Longitudinal follow-up and outcomes among a population with chronic kidney disease in a large managed care organization. Arch Intern Med. 2004 Mar 22;164(6):659–63. doi: 10.1001/archinte.164.6.659. [DOI] [PubMed] [Google Scholar]
- 25.Orlando LA, Owen WF, Matchar DB. Relationship between nephrologist care and progression of chronic kidney disease. N C Med J. 2007 Feb;68(1):9–16. [PubMed] [Google Scholar]
- 26.Kim DH, Kim M, Kim H, Kim Y-L, Kang S-W, Yang CW, et al. Early referral to a nephrologist improved patient survival: prospective cohort study for end-stage renal disease in Korea. PloS One. 2013;8(1):e55323. doi: 10.1371/journal.pone.0055323. [DOI] [PMC free article] [PubMed] [Google Scholar]