Abstract
ClinicalTrials.gov presents great opportunities for analyzing commonalities in clinical trial target populations to facilitate knowledge reuse when designing eligibility criteria of future trials or to reveal potential systematic biases in selecting population subgroups for clinical research. Towards this goal, this paper presents a novel data resource for enabling such analyses. Our method includes two parts: (1) parsing and indexing eligibility criteria text; and (2) mining common eligibility features and attributes of common numeric features (e.g., A1c). We designed and built a database called “Commonalities in Target Populations of Clinical Trials” (COMPACT), which stores structured eligibility criteria and trial metadata in a readily computable format. We illustrate its use in an example analytic module called CONECT using COMPACT as the backend. Type 2 diabetes is used as an example to analyze commonalities in the target populations of 4,493 clinical trials on this disease.
Introduction
An important area for clinical research standards development is participant selection for clinical studies. Many clinical studies are designed to emphasize internal validity, and some design decisions may compromise external validity, which is also referred to as generalizability. When a clinical trial has limited generalizability, the study results can be difficult to translate to the real-world population to which the study results are meant to apply. This is a concern of both the public and the clinical research community [1–3], and has significantly impaired the cost-benefit ratio of many clinical studies. Meanwhile, many clinical investigators prefer to reuse existing participant selection methods [4]. However, there is no well-accepted standard for participant selection.
To supplement the top-down model of traditional research standard development, we propose to use a data-driven approach to analyzing commonalities in participant selection for clinical trials. Leveraging the timely information on the Web, community-generated information has been shown to facilitate public health, e.g., real time prediction of epidemic disease [5]. We hypothesize that this method can identify frequent eligibility features that are in use in the clinical research community and represent de facto standards for reusable eligibility features. Understanding these practices can help the design and adoption of standards derived “from researchers and for researchers”.
The official public registry of clinical trials and a valuable resource created by the National Library of Medicine of the United States [6], ClinicalTrials.gov presents a great opportunity for identifying frequently used criteria for research participant selection without expensive knowledge engineering by domain experts. Since September 2007, all clinical trials sponsored or conducted in the United States must be registered in ClinicalTrials.gov. As of 01/27/2014, 159,891 clinical trials with sites in more than 180 countries are registered, including 129,193 interventional studies and 29,061 observational studies. Strictly speaking, only interventional studies are called “clinical trials”. In this paper, we use the term “clinical trials” to represent both interventional and observational studies. Trial summaries are semi-structured so that descriptive characteristics such as study title, sponsor, and target populations’ ethnicity and gender are organized in structured fields, whereas eligibility criteria are written in free text, separated into inclusion criteria and exclusion criteria sections.
We have used ClinicalTrials.gov to identify common eligibility features for clinical research participant selection [7, 8]. As an extension to our previous work, this study integrated our previously developed parsing methods [9–11] to analyze commonalities in clinical trial participant selection. In particular, we designed and built a database called “Commonalities in Target Populations of Clinical Trials” (COMPACT) with searchable information for ClinicalTrials.gov’s 159,891 entries as of 1/27/2014. COMPACT enables analysis modules, such as the CONECT (Commonalities in Target Populations in Eligibility Criteria) module described in this paper, to surface commonalities in clinical trial target populations from COMPACT. A use-case driven approach was employed to demonstrate how commonalities in clinical trial target populations can be derived from disease specific trials’ inclusion and exclusion criteria and be leveraged to inform future clinical trial designs. This paper contributes to the research community (1) a novel method for analyzing commonalities in clinical trial target populations on the fly, and (2) a new database to inform knowledge reuse for future clinical trial eligibility criteria designs. Both resources will be made open source in the near future.
Methods
In our design for COMPACT, we formulated the commonalities of target populations as common eligibility features, including numeric features, categorical features, and their attributes. Numeric features are eligibility criteria with a numeric value range requirement for participants, such as “HbA1c > 7%”. Categorical features are eligibility criteria that accept one of a set of value options, such as “past history of stroke” (yes or no). We treat dichotomous (binary) criteria as a case of categorical criteria. To demonstrate the analytic utility of COMPACT, we developed an example analytic module called CONECT, which enables a user to mine contextual common eligibility features for trials on a certain disease from COMPACT. Next we will describe the details of both.
1. The Conceptual Design of COMPACT
The database Commonalities in Target Populations of Clinical Trials (COMPACT) includes four entities: metadata of clinical trials, structured clinical trial eligibility criteria, and common eligibility features (i.e., numeric and categorical features) and their properties indexed by disease topics.
Metadata of trials defines indexing characteristics of clinical trials provided by ClinicalTrials.gov, such as study type, intervention, medical condition, and study design (i.e., intervention model for interventional studies, allocation of participants to intervention group for interventional studies, and time perspective for observational studies).
Common eligibility features can be numeric or categorical. Each common numeric feature has five properties: numeric feature concept (e.g., HbA1c, BMI), the Unified Medical Language System (UMLS) semantic group [12] for the feature, the collective permissible value ranges derived from all the trials using this feature (e.g., [6.5%–7%] for HbA1c), the width of distinct mutually exclusive value intervals (e.g., 0.5 for [6.5%–7%] for HbA1c) and a salient modal boundary value if applicable (e.g., more diabetes trials use HbA1c of 7% as either a lower or an upper bound than any other threshold so that 7% is the modal boundary value).
Each common categorical feature has two properties: categorical feature concept and its UMLS semantic group. For example, the semantic group of “malignant neoplasm” is “Disorders”. According to the NLM document [13], all 133 semantic types of the UMLS are grouped into 15 general semantic groups. “Disease or Syndrome” and “Neoplastic Process” along with 10 other semantic types are grouped into the semantic group “Disorders”.
Figure 1 illustrates the information flow for a user, who can be a policy researcher, a clinical investigator, a trial sponsor, or others interested in such a system to interact with the COMPACT database. In this figure, the rectangle blocks represent data input and output when interacting with COMPACT. The round-corner blocks represent properties of input and output. The arrow lines represent processes when interacting with COMPACT.
Figure 1.
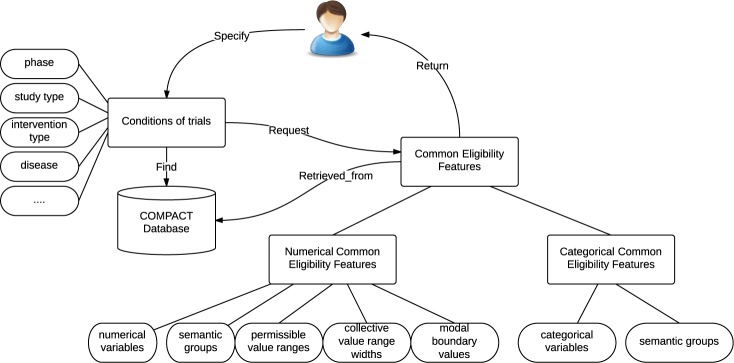
The information flow for a user to interact with the COMPACT database.
For example, when a user specifies certain queries of trials when interacting with the COMPACT database, e.g., Type 2 diabetes trials recruiting patients with HbA1c >= 7.0 %, he or she will retrieve the common eligibility features with their attributes for these trials. Contextual attributes for common eligibility features present to a user how the clinical research community uses the feature HbA1c collectively in Type 2 diabetes trials. From COMPACT, three attributes for numeric features can be generated: collective value ranges, collective value range widths, and modal boundary values. Note that the data set for analyzing these three attributes comprises all the clinical trials of the given disease in the query.
Collective value ranges give the frequently used value ranges of a numeric eligibility feature for all the trials on a certain disease. We itemize all the permissible value ranges of a numeric feature in all the trials of a certain disease and count the number of trials that use a specific value range to get the collective value ranges. For example, in the inclusion criteria of the trial NCT00035984, the criterion “HbA1c value between 7.5% and 11%” gives the value range of HbA1c in this criterion: [7.5, 11.0], in which “[ ]” means being inclusive.
Collective value range widths present the distribution of value range widths of a numeric eligibility feature for all the trials on a certain disease. To get the collective value range widths, we counted the number of trials of a certain disease with the same value range width of a numeric feature. For example, the value range width of HbA1c derived from the same criterion “HbA1c value between 7.5% and 11%” is: 11 – 7.5 = 3.5.
Modal boundary values are defined as the “most-used boundary values” for a numeric feature for eligibility determination. For example, because more diabetes trials use HbA1c of 7% as either a lower or an upper bound, “7.0%” is the modal boundary value of HbA1c for Type 2 diabetes trials. Similarly, “140 mm Hg” is a modal boundary value for systolic blood pressure in the same data set. Note that for some numeric features, there might not be a modal boundary value.
2. COMPACT Database Construction
Figure 2 presents the workflow for constructing the COMPACT database. To enable agile discovery of common eligibility features, we downloaded all the trial summaries in ClinicalTrials.gov as of January 2014, excluding those with no or non-informative eligibility criteria text, such as “please contact site for information”. The trial summaries were parsed and saved in the relational database (RDBMS) MySQL. For each trial, using previous developed parsing methods [9–11], we extracted, parsed, and stored the metadata of the trial (e.g., title, location, type, etc.), numeric features, and categorical features in three database tables “Metadata”, “Numeric_features”, and “Categorical_features”, respectively. Ross et al. found that approximately 23% of eligibility criteria in ClinicalTrials.gov entries are numeric [14], such as for HbA1c, BMI, blood glucose, and creatinine. To unify these criteria, we negated the meaning of exclusion criteria and converted them to inclusion criteria without changing or losing meaning by replacing “<”, “<=”, “>”, “>=” with “>=”, “>”, “<=”, “<”, respectively. The rationale of this conversion is that the complement of a numeric expression in an exclusion criterion indicates an inclusion criterion.
Figure 2.
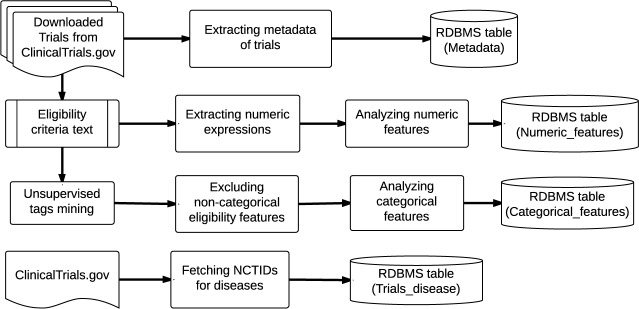
The pipeline for constructing the COMPACT database.
We applied unsupervised text mining on free-text eligibility criteria to automatically extract all frequent (i.e., appearing in at least 5% of all trials) eligibility features [9]. In this step, n-grams were generated for each criterion after noise reduction by a part-of speech (POS) tagger. The n-grams can be identified as eligibility features if they contain a substring that can be annotated using a UMLS concepts assigned one of 27 semantic types relevant to the clinical trial domain [15]. We excluded all the features whose UMLS semantic type was “Body Part, Organ, or Organ Component”. For example, in the inclusion criteria of trial NCT00934414, from the sentence “Smoking: BMI of = 40 kg/m2”, two eligibility features “smoking” and “body mass index” were extracted and retained.
Valx, a numeric expression extraction and normalization tool [10, 11] developed in our lab, was employed to extract and parse complex numeric expressions in free-text eligibility criteria. For example, from the inclusion criterion “type 2 diabetes mellitus diagnosed at least 3 months with fpg level <=240mg/dl and hba1c between 6.5% and 10% inclusive”, Valx extracted two numeric features “glucose” (“fpg” is short for “fasting plasma glucose”) and “HbA1c”, and generated the expressions “[["Glucose", "<=", 240, "mg/dL"], [HbA1c, ">=", 6.5, "%"], [HbA1c, "<=", 10.0, "%"]]”. For each numeric feature, the permissible value range, the width of the value range, and the boundary values were calculated to support common eligibility feature attributes mining. The value range of the numeric feature “HbA1c” in the criterion above is “[6.5, 10.0]” (“[ ]” means being inclusive, while “()” means being non-inclusive). Its value range width is 10.0 – 6.5 = 3.5. Its boundary values are 6.5 and 10.0 since they both appear once in this criterion. Heterogeneous semantic representations for the same numeric feature were recognized. For example, HbA1c can be written as “hemoglobin A1c” or “A1c”. Using synonyms in the UMLS and manually defined heuristics, we mapped these different representations to the same concept, HbA1c. Also, a rule-based algorithm was employed to recognize different representations for common comparison operators in eligibility criteria statements, such as “>” and “greater than”, or “>=” and “greater or equal to”. Various measurement units were harmonized. For example, blood glucose can be quantified by mg/dL or mmol/l.
Figure 3 illustrates the database schema for COMPACT. For the table “trials_disease”, we used all medical conditions defined by ClinicalTrials.gov (http://www.clinicaltrials.gov/ct2/search/browse?brwse=cond_alpha_all) and created trial indexes for the selected conditions that were each studied by at least 50 clinical trials. The four tables in Figure 3 can be joined by a common attribute “NCTID”, which is the unique identifier for a trial in the ClinicalTrials.gov. This knowledge base supports the mining of common eligibility features and their attributes in trials with various characteristics, e.g., a certain disease, a certain study design, recruiting patients of a certain gender, etc.
Figure 3.
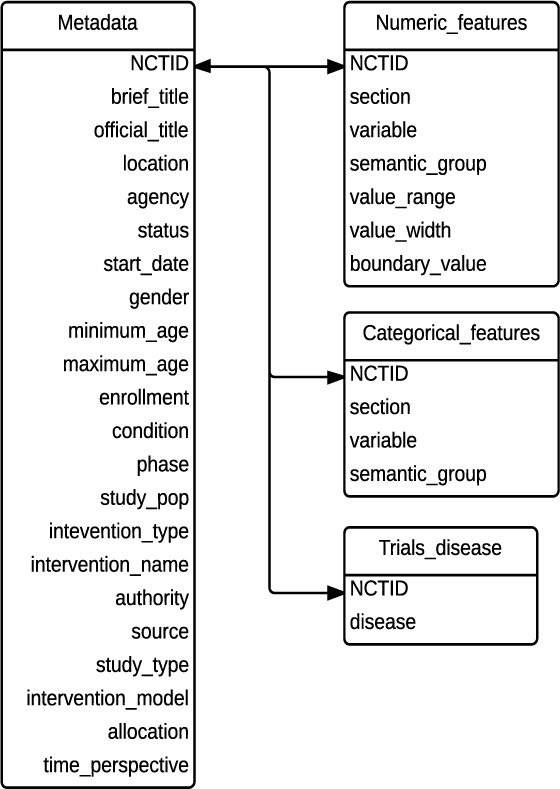
The database schema for COMPACT.
3. An Example Use of COMPACT
COMPACT enables flexible queries of sets of clinical trials for exploring the participant selection patterns in ClinicalTrials.gov. To illustrate such a use case, we developed an example common eligibility feature analytic module CONECT, leveraging RDBMS’ capability of handling a wide range of queries to mine common eligibility features from COMPACT. Figure 4 shows the workflow of CONECT using a concrete query example. The light blue rectangle boxes represent steps in the process. The ovals represent the output of each step.
Figure 4.
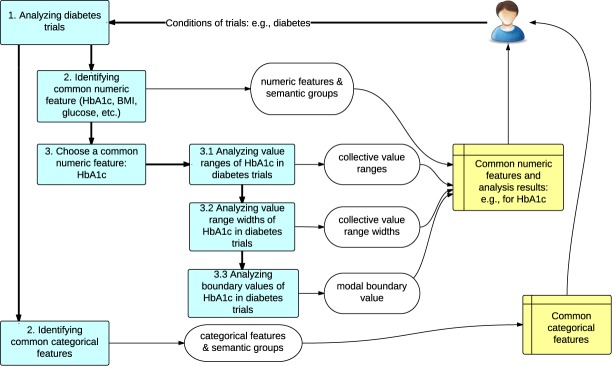
Workflow of CONECT with a concrete example of HbA1c patterns in diabetes trials.
In the example shown in Figure 4, a user may submit a query specifying the condition of trials to be “diabetes”. In Step 1, CONECT will retrieve all the diabetes trials from COMPACT using the “trials_disease” table. In Step 2, CONECT will identify common numeric features (e.g., HbA1c, BMI, and glucose) and common categorical features in diabetes trials. In Step 3, the user may choose a numeric feature “HbA1c” for further analysis. Then all the precomputed value ranges, value widths of the intervals, and boundary values of HbA1c in diabetes trials will be retrieved from COMPACT and aggregated upon the number of trials with the same value ranges, value widths, and boundary values. The output of the analysis including common numeric features, common categorical features, and the detail analysis of HbA1c in diabetes trials will be visualized and returned to the user.
Results
1. The Descriptive Statistics of COMPACT
To build the COMPACT database, we downloaded all 159,891 clinical trial records in XML format in ClinicalTrials.gov as of 01/27/2014. After excluding trials with no or non-informative eligibility criteria text, 159,187 trials were retained. The metadata fields of these trials were extracted and saved in the “Metadata” table. In order to quickly identify trials of a certain disease, we also created indexes for trials for 1,543 diseases with more than 50 trials in ClinicalTrials.gov. 229 out of these 1,543 diseases are listed as condition in more than 1,000. The trials of uncommon diseases can be retrieved by querying on the “condition” field using ClinicalTrials.gov’s API. Valx extracted 1,045,893 numeric expressions for 176,986 unique features from 429,551 sentences in the inclusion criteria and 283,885 sentences in the exclusion criteria.
2. The Sample Query
In this section, we will use “Type 2 diabetes trials that recruit patients with HbA1C >= 7.0%” as a sample query to illustrate how common eligibility features are discovered from COMPACT. HbA1c is “a lab test that shows the average level of blood sugar (glucose) over the previous 3 months. It shows how well you are controlling your diabetes” [16]. In this paper, HbA1c refers to the test result. WebMD gives the normal ranges of HbA1c. For people without diabetes, the normal value range for HbA1c is between 4% and 5.6%. HbA1c levels between 5.7% - 6.4% indicate increased risk of diabetes, and 6.5% or higher indicate diabetes [17]. The American Diabetes Association recommends a HbA1c goal of less than 7.0% [18]. In COMPACT, 4,370 trials specify the value range of a patient’s HbA1c in their eligibility criteria.
3. Common Eligibility Features and their UMLS Semantic Groups
In COMPACT, there are 4,493 Type 2 diabetes trials, out of which 700 trials (15.6%) are recruiting patients whose HbA1c must be >= 7.0%. According to the result of the analysis, five numeric features are used in the inclusion criteria of more than 50 such trials, whereas seven numeric features are used in the exclusion criteria of more than 50 such trials. Due to space limitations, Table 1 shows the top five numeric features for each section, the semantic group, the number of trials, and the percentage of the qualifying trials (700 trials for the sample query) using the numeric feature. “HbA1c” is the only feature listed as one of the top five for both inclusion criteria and exclusion criteria of such trials, which conforms to the literature [18]. “HbA1c” and “Creatinine” are the most frequently used common numeric features in the inclusion and exclusion criteria of the qualifying trials, respectively.
Table 1.
Top five numeric features frequently used in inclusion and exclusion criteria of Type 2 diabetes trials that recruit patients whose HbA1c >= 7.0%.
| Numeric features used in the inclusion criteria | Numeric features used in the exclusion criteria | ||||||
|---|---|---|---|---|---|---|---|
| Numeric features | Semantic group | # Trials | Perc. | Numeric features | Semantic group | # Trials | Perc. |
| HbA1c | Physiology | 663 | 94.7% | Creatinine | Chemicals & Drugs | 114 | 16.3% |
| BMI | Physiology | 370 | 52.8% | Systolic blood pressure | Physiology | 85 | 12.1% |
| Age | Physiology | 327 | 46.7% | Diastolic blood pressure | Physiology | 84 | 12% |
| Glucose | Chemicals & Drugs | 114 | 15.9% | ALT | Chemicals & Drugs | 73 | 10.4% |
| C-peptide | Chemicals & Drugs | 56 | 8.0% | HbA1c | Physiology | 71 | 10.1% |
For common categorical features, nine are used in inclusion criteria of more than 50 trials, whereas 46 are used in exclusion criteria of more than 50 trials. Table 2 shows the top five categorical features for each section, their sematic group, their use in inclusion or exclusion section, and their frequency. “Diabetes mellitus non-insulin-dependent” (Type 2 diabetes) and “diabetes mellitus insulin-dependent” (Type 1 diabetes) are the most frequently used categorical features in the inclusion and exclusion criteria, respectively. This is reasonable because usually Type 2 diabetes trials exclude Type 1 diabetes patients.
Table 2.
Top five categorical features frequently used in inclusion and exclusion criteria of Type 2 diabetes trials that recruit patients whose HbA1c >= 7.0%. The names of categorical features are UMLS terms.
| Categorical features used in the inclusion criteria | Categorical features used in the exclusion criteria | ||||||
|---|---|---|---|---|---|---|---|
| Categorical features | Semantic group | # Trials | Perc. | Categorical features | Semantic group | # Trials | Perc. |
| diabetes mellitus non-insulin-dependent | Disorders | 520 | 74.3% | diabetes mellitus insulin-dependent | Disorders | 236 | 33.7% |
| sulfonylurea compounds | Chemicals & Drugs | 118 | 16.9% | pharmacologic substance | Chemicals & Drugs | 229 | 32.7% |
| antidiabetics | Chemicals & Drugs | 94 | 13.4% | allergy severity -severe | Disorders | 224 | 32.0% |
| pharmacologic substance | Chemicals & Drugs | 91 | 13.0% | gravidity | Disorders | 223 | 31.9% |
| contraceptive methods | Procedures | 83 | 11.9% | malignant neoplasm | Disorders | 190 | 27.1% |
4. Collective Value Ranges of Numeric Eligibility Features
Among the top 5 numeric features discovered for the sample query, we chose the two most frequently used numeric features “HbA1c” and “BMI” to illustrate collective value ranges, collective value range widths, and modal boundary values. Table 3 shows the five most frequently used value ranges of HbA1c and BMI in Type 2 diabetes trials. “[ ]” means being inclusive, while “()” means being non-inclusive; −∞ refers to negative infinity, whereas +∞ refers to positive infinity. Out of 4,493 Type 2 diabetes trials, 2,058 trials (45.8%) use HbA1c, whereas 1,859 trials (41.4%) use BMI in their eligibility criteria. According to the analysis (after unifying inclusion and exclusion criteria), the most frequently used permissible value range for HbA1c is [7.0, 10.0], while the most frequently used value range for BMI is (−∞, 45.0].
Table 3.
Top five collective value ranges of HbA1c and BMI in Type 2 diabetes trials.
| HbA1c value ranges | Number of trials | BMI value ranges | Number of trials |
|---|---|---|---|
| [7.0, 10.0] | 228 | (−∞, 45.0] | 113 |
| (7.0, +∞) | 97 | (−∞, 40.0] | 104 |
| (−∞, 7.0] | 88 | [25.0, 40.0] | 72 |
| [7.0, 11.0] | 75 | (−∞, 40.0) | 61 |
| [7.0, +∞) | 57 | (−∞, 35.0] | 58 |
5. Collective Value Range Widths of Numeric Eligibility Features
In this study, when analyzing the collective value range widths of numeric features, we excluded all the numeric features without an upper bound or a lower bound and defer their analysis to future work.
Out of 2058 Type 2 diabetes trials with HbA1c in their eligibility criteria, 1156 trials (56.2%) specify a bounded value range for HbA1c, i.e., with both an upper bound and a lower bound. Figure 5 (a) shows the distribution of collective value range widths of HbA1c in Type 2 diabetes trials. We can see that the value range width “3” is used by 370 Type 2 diabetes trials for HbA1c. Out of 1859 Type 2 diabetes trials using BMI as a numeric feature, 981 trials (52.8%) specify a bounded value range for BMI. Figure 5 (b) shows that “15” is the most frequent value range width of BMI used by 130 Type 2 diabetes trials.
Figure 5.
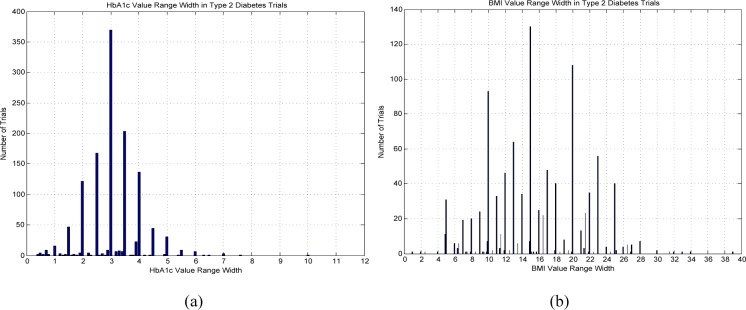
Collective value range widths for (a) HbA1c and (b) for BMI in Type 2 diabetes trials.
6. Modal Boundary Values of Numeric Eligibility Features
For analyzing the modal boundary value of a numeric feature in trials of a certain disease, we aggregated the number of trials using a certain value for eligibility determination. Figure 6 (a) below shows the distribution of Type 2 diabetes trials using a specific HbA1c value in the eligibility criteria, i.e., below or above this value. The peak in Figure 6 (a) corresponds to the modal boundary value of HbA1c, which is 7.0% for Type 2 diabetes trials, whereas this modal boundary value of BMI is 40 kg/m2, as shown in Figure 6 (b).
Figure 6.
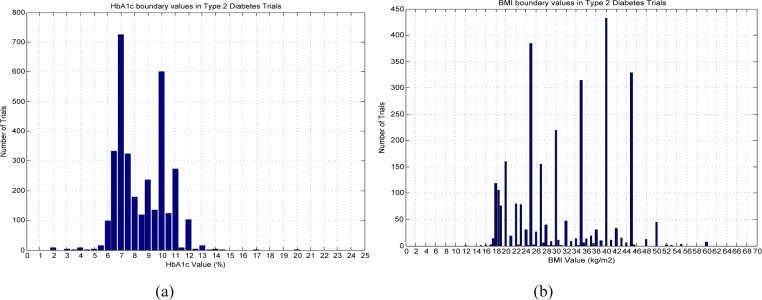
Collective boundary values for (a) HbA1c and (b) BMI, respectively, for Type 2 diabetes trials.
7. The Interface Design of CONECT (Commonalities in Target Populations in Eligibility Criteria)
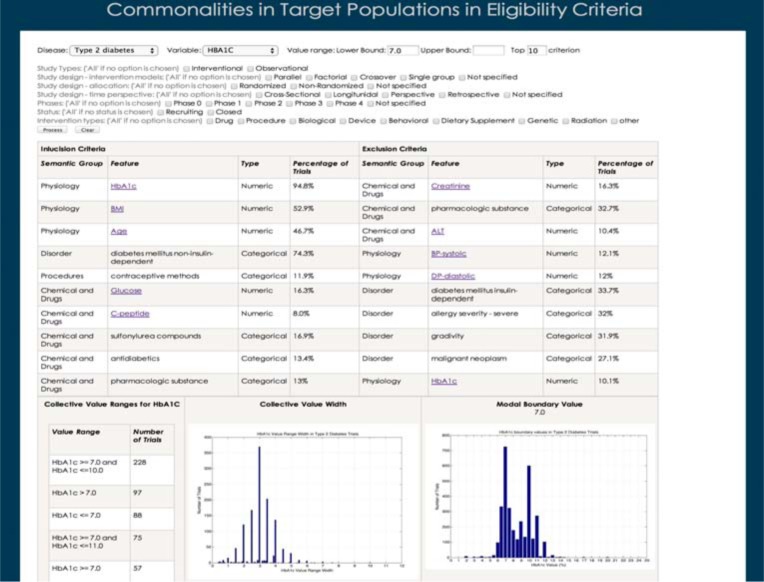
We designed an interface for CONECT so that clinical investigators will be able to utilize the COMPACT database to inform the design of new trials. Figure 7 shows the interface of the CONECT prototype. In this interface, an investigator can specify various characteristics of trials, e.g., disease, study type, study design (including intervention model, allocation, and time perspective), status, intervention type, phase, permissible values for specific numeric eligibility criteria features, and the number of common eligibility features to be retrieved. Based on the query of the investigator, CONECT will retrieve common eligibility features grouped by their semantic groups. When an investigator clicks on a certain numeric feature, its attributes, i.e., collective value ranges, collective value widths, and collective boundary values with a modal value if applicable, will be visualized in the lower space. CONECT informs the investigator what threshold other investigators have defined for the same variable in similar contexts. The analysis for contextual attributes for the categorical features is still under construction.
Figure 7.
The interface of CONECT (Commonalities in Target Populations in Eligibility Criteria) prototype.
Discussion
This paper introduces a method for analyzing commonalities in eligibility criteria and a new data-driven approach to supporting actionable knowledge reuse for clinical trial eligibility criteria design. The COMPACT database built using this method should enable its target users to learn from the distributed clinical research community. Since its launch in 1997, ClinicalTrials.gov has accumulated about 160,000 studies, among which 48.6% are completed, and 27.3% are enrolling participants. With the large amount of information about clinical trial designs available, there is a compelling need to examine the commonalities in patient selection in this repository to inform the development of new clinical trials. Previously, there was no method going to this level of detail to parse the attributes of frequent eligibility features, e.g., their permissible value ranges, modal boundary values, etc. By coupling this information with descriptive characteristics of clinical trials, our method presents a new way for clinical trial sponsors and investigators to analyze fine-grained commonalities in clinical trial target populations and understand design patterns of eligibility criteria. Leveraging existing clinical trial records, our method intends to discover popular content used in the research community to facilitate future clinical trial design.
Utility of the COMPACT Database
The most important two design principles of the COMPACT database are generalizability and extensibility. In this paper, we demonstrate the utility of the COMPACT database with an example analytic module that supports disease-specific trial design. Analytic modules can be developed for other purposes. COMPACT has additional data such as drug name and location that have not been used by CONECT. Thus, one may develop a module to find common eligibility features in trials of a certain drug. COMPACT enables flexible selection of clinical trials of various characteristics, e.g., randomized or non-randomized, starting after a certain date, with a certain eligibility feature, recruiting only male patients, etc. For example, a clinical trial designer may be interested in discovering common eligibility features for breast cancer trials starting after January 2009, or for heart disease trials that recruit patients with systolic blood pressure greater than 140 mmHg, etc. The normalization of different units of the same numeric feature enables their aggregate analysis among different clinical trials.
Comparison with Other Databases Derived from ClinicalTrials.gov
The most notable effort related to our method is a database called Aggregate Analysis of Clinical Trials (AACT). It was developed by Tasneem et al. using descriptive characteristics, e.g., conditions and intervention, to aggregate clinical trials from ClinicalTrials.gov [19]. Our method and AACT both use RDBMS to store clinical trial summaries from ClinicalTrials.gov. However, AACT does not analyze eligibility criteria in detail. In contrast, our method derives commonalities in participant selection from parsed eligibility criteria. Another database LinkedCT transformed the data of clinical trials on ClinicalTrials.gov into RDF to discover semantic links between clinical trial records [20]. However, it does not contain discrete eligibility features and hence does not support the analysis of eligibility criteria as COMPACT does. Thus, COMPACT can potentially make important contributions by facilitating knowledge reuse during eligibility criteria designs for new clinical trials.
Difference between Common Eligibility Features and Common Data Elements
Luo et al. used unsupervised machine learning to identify disease-specific Common Data Elements (CDEs) from clinical trial eligibility criteria [7]. Common eligibility features and CDEs can be both disease specific, but are different in the following three aspects: (1) CDEs are generated in a semi-automated fashion whereas common eligibility features generation is interactive and contextual, depending on the query; (2) Common eligibility features can be generated for trials of various characteristics on the fly, e.g., trials in a certain phase, eligibility criteria with a certain numeric feature in a certain range, etc., while CDEs are usually identified for a specific disease domain, and (3) Common eligibility features have contextual attributes. Numeric features have disease-specific value ranges, disease-specific value range widths and disease-specific modal boundary values. Investigators will get contextual knowledge on how these numeric features are used to define eligibility criteria for participant selection. This method overcomes the following limitations in traditional knowledge base development, including (1) centered around one or a small group of domain experts, (2) limited to one or few disease domains, (3) laborious and costly knowledge management processes, and (4) lack of scalability.
Limitations
This study has limitations. The accuracy of the commonalities in the target populations depends on the accuracy of the parsing of the eligibility criteria text. In a preliminary evaluation of the parser Valx, the precision, recall, and F-measure for extracting numeric expressions with the feature “HbA1c” were 99.6%, 98.1%, 98.8% for Type 1 diabetes trials, and 98.8%, 96.9%, 97.8% for Type 2 diabetes trials, respectively. The results of the measures for extracting numeric expressions with the feature “Glucose” were 97.3%, 94.8%, 96.1% for Type 1 diabetes trials, and 92.3%, 92.3%, 92.3% for Type 2 diabetes trials, respectively [10]. In the future, a comprehensive evaluation of Valx is necessary to assess its performance for other numeric features for their uses in clinical trials of various medical conditions. Our text mining method serves our goals presented here, but still has significant room for improvement. It currently does neither exhaustively recognize all the abbreviations nor normalize all the measurement units for every numeric feature in eligibility criteria text. Categorical features without contextual information (e.g., within 12 hours of the onset of chest pain) are sometimes not specific enough for direct reuse. Therefore, further collaborative research on natural language processing of free-text eligibility criteria is desired. In this study, we used one sample query on a common disease (i.e., Type 2 diabetes) for demonstration purposes. More studies are warranted to test how this method works for other eligibility features and other diseases.
Future Work
The COMPACT database needs to be updated on a regular basis. We will develop CONECT as a Web-based system and formally assess its value for clinical research stakeholders. To provide acceptable user experience when interacting with the system, it is imperative to improve the performance of our query processing method, which requires costly table join and value aggregation for numeric features. The current long waiting time, i.e., two minutes, to retrieve common eligibility features for some queries would impair the user experience of the system. We will create a repository to store common eligibility features for popular queries. When a popular question is asked again, the saved results can be returned to the user promptly. We will also analyze the queries submitted by the users, which may reveal new trends in trial design. With continuously improving natural language processing techniques, we will provide finer-grained common eligibility features. Temporal usage patterns will also be explored to show how commonalities in participants evolve over time and across disease domains. In the future, to enhance the analytic utility of COMPACT, we will add a table containing real world patient data and perform comparative analysis between clinical trial target populations and real world patient populations [21].
Conclusions
We introduced a novel data resource for analyzing common numeric and categorical features in eligibility criteria from public clinical trials records. We designed and built a database called COMPACT using all the clinical trial records on ClinicalTrials.gov. CONECT was introduced to illustrate example visualization of the COMPACT content and the interactions between users and COMPACT. The query “Type 2 diabetes trials that recruit patients with HbA1c >= 7.0%” was used to illustrate the method. This research can potentially help clinical investigators understand frequently used eligibility criteria and how they have been shared across studies and can inform the design for participant selection for new clinical trials. Our future work includes further improvement of visual queries using COMPACT and user evaluation of this new decision aid for clinical research stakeholders.
Acknowledgments
We thank Dr. Riccardo Miotto for contributing parsed clinical trial summaries for building the COMPACT database. This study was sponsored by the National Library of Medicine grant R01LM009886 (PI: Weng) and National Center for Advancing Translational Science grant UL1 TR000040 (PI: Ginsberg).
References
- 1.Rothwell PM. External validity of randomised controlled trials: “to whom do the results of this trial apply?”. Lancet. 2005;365(9453):82–93. doi: 10.1016/S0140-6736(04)17670-8. [DOI] [PubMed] [Google Scholar]
- 2.Fuks A, Weijer C, Freedman B, Shapiro S, Skrutkowska M, Riaz A. A study in contrasts: eligibility criteria in a twenty-year sample of NSABP and POG clinical trials. National Surgical Adjuvant Breast and Bowel Program. Pediatric Oncology Group. J Clin Epidemiol. 1998;51(2):69–79. doi: 10.1016/s0895-4356(97)00240-0. [DOI] [PubMed] [Google Scholar]
- 3.Van Spall HG, Toren A, Kiss A, Fowler RA. Eligibility criteria of randomized controlled trials published in high-impact general medical journals: a systematic sampling review. JAMA. 2007;297(11):1233–40. doi: 10.1001/jama.297.11.1233. [DOI] [PubMed] [Google Scholar]
- 4.Hao T, Rusanov A, Boland MR, Weng C. Clustering clinical trials with similar eligibility criteria features. J Biomed Inform. 2014:pii: S1532-0464(14)00011-2. doi: 10.1016/j.jbi.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim EK, Seok JH, Oh JS, Lee HW, Kim KH. Use of hangeul twitter to track and predict human influenza infection. PLoS One. 2013;8(7):e69305. doi: 10.1371/journal.pone.0069305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ClinicalTrials.gov. Feb, 2014. Available from: http://www.clinicaltrials.gov/
- 7.Luo Z, Miotto R, Weng C. A human-computer collaborative approach to identifying common data elements in clinical trial eligibility criteria. J Biomed Inform. 2013;46(1):33–9. doi: 10.1016/j.jbi.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boland MR, Miotto R, Weng C. A method for probing disease relatedness using common clinical eligibility criteria. Stud Health Technol Inform. 2013;192:481–5. [PMC free article] [PubMed] [Google Scholar]
- 9.Miotto R, Weng C. Unsupervised mining of frequent tags for clinical eligibility text indexing. J Biomed Inform. 2013;46(6):1145–51. doi: 10.1016/j.jbi.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao T, Weng C. Valx: A Knowledge-based System for Extracting and Structuring Numeric Comparison Statements in Clinical Research Eligibility Criteria Text. J Biomed Inform. Under review [Google Scholar]
- 11.Hao T, Weng C. Valx - Numeric Expression Extraction and Normalization Tool 2013 [February 2014] Available from: http://columbiaelixr.appspot.com/valx.
- 12.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The UMLS Semantic Group Feb, 2014. Available from: http://semanticnetwork.nlm.nih.gov/SemGroups/
- 14.Ross J, Tu S, Carini S, Sim I. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits Transl Sci Proc. 2010;2010:46–50. [PMC free article] [PubMed] [Google Scholar]
- 15.Luo Z, Johnson SB, Weng C. Semi-Automatically Inducing Semantic Classes of Clinical Research Eligibility Criteria Using UMLS and Hierarchical Clustering. AMIA Annu Symp Proc. 2010;2010:487–91. [PMC free article] [PubMed] [Google Scholar]
- 16.MedlinePlus. Feb, 2014. Available from: http://www.nlm.nih.gov/medlineplus/
- 17.HbA1c value range. Feb, 2014. Available from: http://www.webmd.com/diabetes/guide/glycated-hemoglobin-test-hba1c.
- 18.American Diabetes Association Website. Feb, 2014. Available from: http://www.diabetes.org/
- 19.Tasneem A, Aberle L, Ananth H, Chakraborty S, Chiswell K, McCourt BJ, Pietrobon R. The Database for Aggregate Analysis of ClinicalTrials.gov (AACT) and Subsequent Regrouping by Clinical Specialty. PLoS ONE. 2012;7(3):e33677. doi: 10.1371/journal.pone.0033677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassanzadeh O. LinkedCT. Mar, 2014. Available from: http://linkedct.org.
- 21.Weng C, Li Y, Ryan P, Zhang Y, Gao J, Liu F, Bigger JT, Hripcsak G. A Distribution-based Method for Assessing The Differences between Clinical Trial Target Populations and Patient Populations in Electronic Health Records. Applied Clinical Informatics. 2014;5(2):463–79. doi: 10.4338/ACI-2013-12-RA-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]