Abstract
Natural language processing (NLP) technologies provide an opportunity to extract key patient data from free text documents within the electronic health record (EHR). We are developing a series of components from which to construct NLP pipelines. These pipelines typically begin with a component whose goal is to label sections within medical documents with codes indicating the anticipated semantics of their content. This Clinical Section Labeler prepares the document for further, focused information extraction. Below we describe the evaluation of six algorithms designed for use in a Clinical Section Labeler. These algorithms are trained with N-gram-based feature sets extracted from document sections and the document types. In the evaluation, 6 different Bayesian models were trained and used to assign one of 27 different topics to each section. A tree-augmented Bayesian network using the document type and N-grams derived from section headers proved most accurate in assigning individual sections appropriate section topics.
Introduction
A key focus in Biomedical Informatics is the use of computerized patient data to impact medical care at the bedside. A variety of tools have been developed that consume electronic data and produce information that can be used by clinicians to gain insight and to direct diagnostic and therapeutic interventions. These include systems that organize and summarize clinical information as well as decision support applications that deliver alerts, suggestions, and otherwise support the delivery of consistent, high-quality care.
To be compatible with the tools that mediate these information interventions, data in the electronic health record is best managed in a structured form whose semantics are captured through reference to standardized medical terminologies. Unfortunately, a large subset of the clinical documentation in electronic medical records consists of free-text reports. A recurring challenge in the use of electronic medical data to support clinical care is the need to extract relevant medical facts from these clinical documents. This can be accomplished through natural language processing (NLP) technologies1. However, the use of these technologies is made more difficult by the heterogeneous nature of these documents. Not only do different types of documents focus on different clinical information, but documents are typically divided into sections each of which focuses on a different category of medical data.
For these reasons, an initial step in clinical NLP is to identify these sections and to label each with a concept code representing the principal topic of that particular section. Labels such as “History of Present Illness”, “Family History”, “Allergies”, and “Discharge Disposition” are used to represent these topics.
Typically, at this stage in processing a document, an initial parse based on structural features results in a collection of document sections each consisting of a section header followed by one or more paragraphs. The text in these paragraphs reflects those clinical facts relevant to a particular medical topic. Unfortunately, while clinicians generally organize clinical documents using semantically similar component sections, they do not necessarily use a standardized collection of section headers to distinguish among these sections. The topics involved must be discovered through an initial processing effort. In the document collection we describe below, sections containing similar concepts are entitled with a surprising number of different headers. Both the text from each section’s header and the text from within the section content can be used as input to a system that assigns a topic as a canonical label for each section. This system is called the “Clinical Section Labeler”.
The algorithms described below are designed to assign a coded descriptor to each section. This descriptor tells what the section is about (i.e. the section’s topic). Section topic labeling is the initial step in information extraction for many types of documents. When NLP systems are focused on extracting particular types of information, this information will typically be located in specific sections within the document. If each document is initially processed by a section labeler that can effectively assign a standardized topic to each section, further processing can be restricted to only those sections in which the required information is likely to be found. This can result in more accurate overall output since, for instance, an NLP system extracting current medications will look in the “Medications” section and not in the “Allergies” section, thereby avoiding mistakes and reducing processing effort.
The character of sections and section topics has been, to some extent, formalized in the HL7 Draft Standard for Trial Use (DSTU) describing the Consolidated Clinical Document Architecture (CDA)2. This standard describes approximately 60 different section types and their representation through section templates. Identifiers for these sections generally come from the Logical Observation Identifiers Names and Codes (LOINC) system3.
The challenges and goals described above have been addressed. Several approaches to automatically assigning canonical section topics have been described in the literature. Denny, et al developed a complete system design to automatically identify section boundaries and to label the sections.4 This system was designed to find sections with section headers as well as “implied” sections where section headers were absent. The algorithm used a combination of NLP techniques. These included spelling correction, Bayesian components (used to score section headers), and terminology-based rules.
In a more focused effort, Ying et al described a tool specifically for identifying sequences of section types5. They applied Hidden Markov Models (HMMs) to this task. They used simple bigrams as features and compared the HMM-based approach to models restricted to a naïve Bayesian algorithm. The version of the system based on HMMs proved significantly more accurate than the naïve Bayesian process.
Approach
In order to develop and test a system to assign topic labels to the sections of clinical documents, we began by collecting a group of medical reports. These reports were chosen to support training and testing for a section topic recognition system. This dataset consisted of 3483 clinical reports extracted from Intermountain Healthcare’s Enterprise Data Warehouse (EDW). The distribution of the reports used is indicated in table 1.
Table 1:
Reports used in Section Topic analysis.
| Report Type | Number of Reports |
|---|---|
| Consultation Report | 491 |
| Discharge Summary | 499 |
| Operative Report | 499 |
| Surgical Pathology Reports | 499 |
| History & Physical Report | 497 |
| ED Physician/LIP Report | 499 |
| XR Chest 2 Views (Frontal/Lateral) | 499 |
We used a combination of automated and manual annotation to break the text in these reports up into four categories. These were 1) section headers, 2) section content, 3) labels, and 4) values. Labels and values generally occurred as pairs such as labeled dates (“Date of Service: 12/5/2012”), although values occasionally appeared independently.
Combinations of section header and section content define the individual sections and were the targets for further analysis. Occasionally, headers did not have complementary sections. This typically occurred when a header was followed by sub-headers each of which could then have independent content. In these cases, we collapsed all subordinate sub-headers and their content into a single instance of section content.
The sections defined this way were manually labeled with topic identifiers designed to represent the basic documentation goals of each of these report components. These named identifiers were expected to express the principal documentation goal of each section. To accomplish this, identical section headers were grouped together with links back to section content. A physician reviewed the section groupings using the section content as necessary to confirm membership of different headers in a semantic class. This effort provided the annotated dataset.
Section Topic Identifiers
Once we had identified the topics that we would target, we developed and compared a group of algorithms designed to assign an appropriate topic to each combination of section header and section content found. These systems were alike in two aspects: each used features generated through extraction of N-grams (uni-grams, bi-grams, and trigrams) from the text of the section headers and/or section content and each employed models for identifying topics which were constructed using Bayesian-network-based approaches6. These Bayesian networks (BNs) were designed to use both the document type and the generated N-grams in assigning a topic for each section.
The Bayesian models differed in two aspects. First, for two of these models, N-grams were derived exclusively from the text of the section content in each section’s header/content pair; for two models, N-grams were derived exclusively from the text of the section header in each section’s header/content pair; and for two models, N-grams were derived from a combination of the text in both the header and content of each section.
The second way in which these models differed was that three were produced using an extended version of the naïve Bayes paradigm and three were developed using an extended version of tree-augmented naïve (TAN) Bayesian networks7. In developing these networks we extended the standard Bayesian paradigm by adding a node representing the document type. This allowed for the differing distributions of section topics in the various document types to be easily captured by the algorithm. Table 3 (below) indicates the different algorithms tested.
Table 3:
Preliminary results of analysis of 6 topic-identification models.
| Topic Identification Algorithms | Accuracy | AROC | Pairwise AROC | F-Measure |
|---|---|---|---|---|
| Naïve BN/Header Only | 95.83% | 0.9986 | 0.9992 | 0.9590 |
| Naïve BN/Content Only | 61.77% | 0.9317 | 0.9551 | 0.6245 |
| Naïve BN/Header + Content | 84.08% | 0.9801 | 0.9869 | 0.8400 |
| TAN BN/Header Only | 98.96% | 0.9996 | 0.9997 | 0.9869 |
| TAN BN/Content Only | 67.55% | 0.9578 | 0.9714 | 0.6792 |
| TAN BN/Header + Content | 90.90% | 0.9911 | 0.9950 | 0.9116 |
The use here of Bayesian network techniques reflects our continuing interest in this technology. We have previously used BN-based systems in a number of NLP experiments. These include systems to extract findings from chest x-ray reports8,9, systems to extract interpretations from ventilation/perfusion lung scan reports10, and tools for syndromic detection11. A description of a system (MPLUS) that uses this approach to NLP can be found in Christensen et al12. Our enduring enthusiasm for this technology reflects its ability to incorporate information from multiple linguistic sources and to provide graphical tools through which to develop and inspect the resulting semantic models.
Analysis
We analyzed the different algorithms described above by employing a 10-fold cross validation technique. In this approach, the sample was divided into 10 parts, and for each part, the algorithm was trained with 90% of the sample and tested with the remaining 10%. The results are aggregated at the end of the procedure.
In this testing we looked at three different measures of parsing accuracy. We calculated the F-statistic and the standard one-versus-all area under the ROC curve (AROC) (for each topic, the analysis compares it to the combination of all competitors). However, when choosing a topic with this method, we actually choose the most likely topic from a list of 28. Therefore, we also calculated the pair-wise AROC13 designed for multiclass classification problems.
Results
Within the initial dataset, 27,645 sections were identified. This had been reduced from 40,441 sections by the collapse of sub-sections into the parent sections. Ninety-eight different topics were assigned during annotation. These ranged in frequency from two instances (Nutrition Status, and Post-Operative Course) to 2933 instances (History of Present Illness). We determined to focus our efforts on topics with a frequency greater than or equal to 1% in the total corpus. Those topics whose frequency was less than 1% were collected into a category called “Other”. This reduced the number of distinct topics to 27 plus the broad category of “Other”. These are listed in table 2.
Table 2:
Section topic selected for analysis.
| Section Concept | Case Count |
|---|---|
| Allergies | 1269 |
| Ancillary Data | 1189 |
| Anesthesia | 417 |
| Assessment | 1865 |
| Chief Complaint | 1286 |
| Course of Care | 407 |
| Diagnoses | 817 |
| Discharge Diagnoses | 538 |
| Discharge Disposition | 418 |
| Discharge Medications | 378 |
| Family History | 934 |
| History of Present Illness | 2933 |
| Indications | 341 |
| Intraoperative Pathology Exam | 502 |
| Medications | 1355 |
| Microscopic Exam | 448 |
| Other | 2319 |
| Past Medical History | 2020 |
| Pathologic Specimens | 564 |
| Physical Exam | 1455 |
| Plan | 703 |
| Post-Operative Diagnoses | 473 |
| Pre-Operative Diagnoses | 695 |
| Procedural Findings | 663 |
| Procedure Description | 538 |
| Procedures Performed | 723 |
| Review of Systems | 1180 |
| Social History | 1215 |
In this way we narrowed the number of distinct section topics for consideration to 27. As anticipated, a review of the documents showed that these semantic labels were associated with a broad range of section header text. The 27 topics corresponded to 584 different text strings used as headers for sections from these clinical documents. For example, the single topic, “Medications”, was assigned to 1355 different sections; 53 different textual representations were found among these section headers ranging from “ADMISSION MEDICATIONS” to “She has been taking the following medications:” Across the range of topics we noted high variability in the headers produced during routine medical documentation. Interestingly, although it was not our intent, 23 of the 27 topics lineup well semantically with the section types described in the Consolidated CDA DSTU.
The goal of the Clinical Section Labeler is to use the N-grams from the section headers and content to appropriately label each section with a topic representing a principal category for the medical facts represented within that section. To accomplish this, we developed a tool that 1) extracted N-grams from section content and/or section headers, 2) executed a feature selection algorithm to identify subsets of N-grams able to discriminate among the different section topics, and 3) trained the Bayesian models described above to identify the appropriate topic for a section from among the 28 choices. The feature selection algorithm applies the Chi-square statistic to identify and discard the subset of features whose contribution to topic assignment is anticipated to be minimal. Researchers can inspect the distribution of Chi-squares and choose a threshold that will exclude irrelevant features.
Inspection of the N-grams generated by processing the section headers indicated that all useful information for topic detection could be gleaned using the top 800 N-grams. However, for topic detection using text from the section content or the text from the combination of section headers and contents, the number was larger. Over 1 million N-grams were generated from these sources and Chi-square testing suggested that a large subset of these could contribute to topic detection. We chose to use the strongest 4000 of the N-grams produced for the topic identifier based on the section content text and the strongest 3000 of the N-grams produced for the identifier designed to process the combined section header and content. (Initial efforts with the combination of header and content N-grams (approximately 5000 total N-grams) proved disappointing; reducing the combined number appeared to give better results.)
Six section topic identifiers were built and tested using the 10-fold cross validation procedure described above. They ranged in raw accuracy from 61.77% to 98.96%. Table 3 displays results from these 6 different models. Shown are their accuracy (percentage of section topics correctly classified), the one-versus-all area under the receiver operating characteristic curve (AROC), the pairwise AROC, and the F-measure.
Based on these results, it appears that, for this population of documents, processing the section header with an extended, tree-augmented naive Bayesian model is most likely to provide an appropriate topic for the section. However, the measures reported are averages (weighted for AROC and F-measure) across all topics. For a tool of this sort, one hopes for consistent accuracy across the range of topics included in the model.
Therefore, to further characterize the accuracy of the TAN BN using header text only, we evaluated its accuracy across the 27 individual topics (plus “Other”). Table 4 shows the most (“Pathologic Specimens”) and least (“Course of Care”) accurate topics identified with the TAN BN-based model using features from the section headers alone. We had anticipated that “Other” would be the least accurately identify topic, but were mistaken as indicated in the table.
Table 4:
Statistics for the most and least accurate topics identified using the TAN BN model and the section header text.
| Topic | Recall | Precision | F measure (95% confidence Intervals) | AROC (95% confidence Intervals) |
|---|---|---|---|---|
| Pathologic Specimens (Most accurately detected topic) | 0.9982 | 1.0 | 0.9991 (0.9966, 1.0) | 1.0 (1.0, 1.0) |
| Course of Care (Least accurately detected topic) | 0.9165 | 0.9739 | 0.9443 (0.9266–0.9607) | 0.9999 (0.9998–0.9999) |
| Other | 0.9621 | 0.9339 | 0.9477 (0.9411, 0.9539) | 0.9977 (0.9968, 0.9986) |
Discussion
The documents used in this evaluation represent a typical collection of the kinds of reports produced when clinicians operate in a flexible authoring environment. Transcription and dictation, speech recognition, and manual authoring with and without templates all played a part in creating the medical documentation represented here. We expect a high degree of variability in the content of many of the sections that appear in medical reports. Patient characteristics are highly variable and this will be reflected in descriptions of their conditions.
However, we had originally hoped to take advantage of consistent section headers to help us find those locations in the document where specific types of information can consistently be located. Unfortunately, section headers also showed wide variability. We therefore chose to treat them like other medical concepts, which must be derived from strings of text in medical documents.
In electronic record systems where clinicians compose their text within a standard report template, section headers can be restricted to those provided by the template system. In these cases, where clinicians used standardized headers for sections, the challenges of automated section topic recognition are reduced. But in EHRs with flexible authoring systems, different wordings and formats occur frequently for section headers. Yet these variable representations can still be mapped to a common set of underlying medical concepts. This is the goal of the Clinical Section Labeler: to assign topics to sections that guarantee the semantic character of similarly labeled sections to be consistent from report to report. Information about medications is to be grouped in a “Medications” section, and information about physical exam or past medical history tends to be found in “Physical Exam” or “Past Medical History” sections.
The Clinical Section Labeler succeeds in this to a degree. However, a review of the documents in this collection suggests that this modeling effort failed to accommodate a valuable group of document components. Many of the documents sections contained subsections, each with its own sub-heading. The initial semi-automated annotation tagged these sub-headings as headings, but then appropriately generated a pointer back to the relevant parent section heading. During the initial annotation, commonly encountered subsections (such as those representing the components of the physical exam) were assigned their own topic labels. The goal was to be able to independently identify subsections such as “Cardiovascular Exam” or “Eye Exam”. However, subheadings and subsections were used erratically in many of the documents. For example, in some of the documents discharge diagnoses were used as subheadings and the subsections were descriptions of the evaluation and course of the individual diseases. As a result we decided to focus on labeling top-level sections. Future efforts will need to accommodate more complex document models where subheadings may be either identifiers for the subcomponents of a typical section or may represent individual clinical conditions used as alternative organizing foci within the documents.
Additional observations include the following:
N-grams have limitations in large and complex document collections. We have anticipated this and plan to approach section labeling with other feature generation techniques in the future. None-the-less, we continue to find N-gram-based feature generation useful as an initial, brute force technique for configuring NLP systems.
The use of Bayesian networks looks promising. In this experiment, we were able to develop both naïve Bayesian and TAN Bayesian models from our annotated data with relative ease. Extending the model with a network node reflecting document type was also simple. In the future, additional opportunities to extend the model are available. These include incorporation of new ways to combine information from the section headers and content using a different BN structure or, perhaps, adding an HMM component to the model to take advantage of the typically consistent sequences of section topics seen in medical documents.
Our current section topic annotations will require revision. The current collection of topics represents an initial categorization driven in part by inspection of the documents extracted from our enterprise data warehouse. The goal was to investigate the existing “wild-type” section authoring process. In future work, we will refine this system. The focus will be on an organization of these concepts in the way that best supports extraction of key clinical information for specific care delivery and research activities. The section topics suggested as a part of the Consolidated CDA will help guide this revision. We envision capturing this organization in an ontology that can assist in future natural language processing efforts.
Any approach that standardizes the use of section headers will ease the section labeling problem. Our next generation of EHR tools is expected to allow us to standardize many of our templates for collecting this data. None-the-less, research that wishes to exploit the several decades worth of collected reports in our EDW will continue to face the challenge of variable document structure and section heading expression.
The reference standard for this project may have introduced an element of bias into the analysis. Not all of the 3483 documents were read through by the annotator. Instead, texturally identical section headers were grouped and examples of section content were reviewed to make sure the topic assigned was consistent with section semantics. It is apparent that some section content in fact belonged to different semantic categories than the header would indicate. Indeed, this is frequently seen when, for instance, elements of the “Social History” or “Family History” are included in the “History of Present Illness” due to their apparent relevance to the patients presenting complaint. To the extent that this occurs, it may explain the reduced accuracy of section identifiers that include section content. Another contributor to this reduction in accuracy is the huge number of n-grams generated for the section content. The Bayesian algorithms could accommodate only a few thousand features, whereas accurate assignment of these sections to their semantic categories would have required tens of thousands or perhaps hundreds of thousands of N-gram-based features.
Conclusion
In this report, we have focused on tools to identify the semantic character of sections commonly found in medical documents. We describe this as assigning “topics” to these sections. The technology tested appears promising for this task and can be leveraged for other recognition tasks in natural language processing as well. We will continue to refine it and to study other approaches appropriate to semantic labeling tasks.
This work focuses on achieving accuracy in assigning topics to previously identified document components. In the future, we will embed this technology into a system designed to completely automate the identification of report components. This system will use both the text of medical documents and local document formatting characteristics to locate section headers and content. Subsequently, the tools described here will assign topics to these sections. Further processing of these labeled documents can then take advantage of an automatically generated document map to determine where relevant clinical information might be found. The ability to focus targeted natural language processing in those sections where relevant information is likely to be found should assist us in developing natural language processing systems that are both efficient and accurate.
The information extracted from medical documents has substantial value. It can contribute to research into the character and course of human illness and, in the future, will inform decision support systems capable of participate in clinical decision making at the bedside. We hope and expect that tools designed to identify sections in clinical text will help to realize the benefits of natural language processing systems.
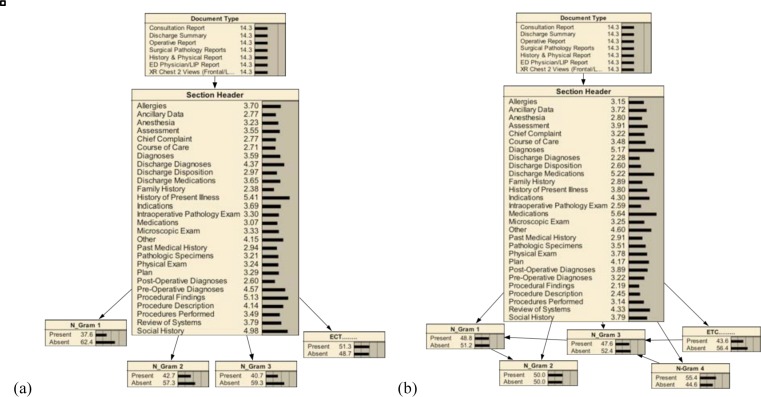
Figure 1:
Bayesian networks were used to recognize section topics. In three cases (a), the core of the network employed a naïve Bayesian model. N-grams generated from 1) the section headers, 2) the section content, and 3) a combination of header and content were used to detect the topics of the individual sections. In the three additional cases (b), the network used these same feature sources but applied a tree-augmented naïve (TAN) Bayesian model. In all cases, the network was extended by adding a node representing the document type being processed.
Acknowledgments
This research was made possible by funding from the Strategic Health IT Advanced Research Projects (SHARP) Program (90TR002) administered by the Office of the National Coordinator for Health Information Technology.
References
- 1.Nadkarni PM1, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. J Am Med Inform Assoc. 2011 Sep-Oct;18(5):544–51. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.HL7 Implementation Guide for CDA® Release 2: IHE Health Story Consolidation, DSTU Release 1.1. Draft Standard for Trial Use. Health Level 7. 2012 Jul; [Google Scholar]
- 3. http://loinc.org.
- 4.Denny J, Spickard A, Johnson K, Peterson N, Peterson J, Miller R. Evaluation of a method to identify and categorize section headers in clinical documents. Journal of the American Medical Informatics Association. 2009;16(6):806. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Ying, Gorman Sharon Lipsky, Elhadad Noémie. Section Classification in Clinical Notes Using a Supervised Hidden Markov Model. ACM International Health Informatics Symposium (IHI); Washington, DC: 2010. pp. 744–750. [Google Scholar]
- 6.Pearl J. Probabilistic reasoning in intelligent systems. San Francisco: Morgan-Kaufmann; 1988. [Google Scholar]
- 7.Friedman N, Geiger D, Goldszmidt M. Bayesian Network Classifiers. Machine Learning. 1997;29:131–63. [Google Scholar]
- 8.Chapman WW, Fiszman M, Chapman BE, Haug PJ. A comparison of classification algorithms to automatically identify chest X-ray reports that support pneumonia. J Biomed Inform. 2001 Feb;34(1):4–14. doi: 10.1006/jbin.2001.1000. [DOI] [PubMed] [Google Scholar]
- 9.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest X-ray reports. J Am Med Inform Assoc. 2000 Nov-Dec;7(6):593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiszman M, Haug PJ, Frederick PR. Automatic Extraction of PIOPED Interpretations from Ventilation/Perfusion (V/Q) Lung Scan Reports. Proceedings of the 1998 AMIA Annual Fall Symposium; pp. 860–864. [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman WW, Christensen LM, Wagner MM, Haug PJ, Ivanov O, Dowling JN, Olszewski RT. Classifying free-text triage chief complaints into syndromic categories with natural language processing. Artif Intell Med. 2005 Jan;33(1):31–40. doi: 10.1016/j.artmed.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Christensen L, Haug PJ, Fiszman M. MPLUS: a probabilistic medical language understanding system. Proceedings of the 40th Annual Meeting of the ACL (Association for Computational Linguistics); 2002. [Google Scholar]
- 13.Hand DJ, Till RJ. A simple generalization of the area under the ROC curve for multiple class classification problems. 2001 Machine Learning. 45:171–186. [Google Scholar]