Abstract
Adverse drug events (ADEs) are a critical factor for selecting cancer therapy options. The underlying molecular mechanisms of ADEs associated with cancer therapy drugs may overlap with their antineoplastic mechanisms; an aspect of toxicity. In the present study, we develop a novel knowledge-driven approach that provides an ADE-based stratification (ADEStrata) of tumor mutations. We demonstrate clinical utility of the ADEStrata approach through performing a case study of breast invasive carcinoma (BRCA) patients receiving aromatase inhibitors (AI) from The Cancer Genome Atlas (TCGA) (n=212), focusing on the musculoskeletal adverse events (MS-AEs). We prioritized somatic variants in a manner that is guided by MS-AEs codified as 6 Human Phenotype Ontology (HPO) terms. Pathway enrichment and hierarchical clustering of prioritized variants reveals clusters associated with overall survival. We demonstrated that the prediction of per-patient ADE propensity simultaneously identifies high-risk patients experiencing poor outcomes. In conclusion, the ADEStrata approach could produce clinically and biologically meaningful tumor subtypes that are potentially predictive of the drug response to the cancer therapy drugs.
1 Introduction
Adverse drug events (ADEs) have been well recognized as a cause of patient morbidity and increased health care costs in the United States. With rapid developments in genomics technology, the contribution of genetic factors to ADEs is being considered and has already influenced clinical recommendations for drug dosage and toxicity (1, 2), thus representing a major component of the movement to pharmacogenomics and individualized medicine (3, 4). Genetic susceptibility is an important feature of severe ADEs and there is considerable interest in developing genetic tests to identify at-risk patients prior to prescription (5). Preliminary studies also suggested that drug therapies based on an individual’s genetic makeup may result in a significant reduction in adverse outcomes (6).
To conduct a pharmacogenomics study of an ADE, ideally, multiple sources of evidence should be integrated to fully characterize the potential pharmacogenomics mechanism relevant to the ADE. For instance, a project known as PharmGKB (7, 8), initiated by the National Institute of Health (NIH), has a mission of collecting and disseminating human-curated information about the impact of human genetic variation on drug responses. In our previous studies, we proposed a knowledge-driven framework that aims to support pharmacogenomics-target prediction of ADEs (9). In the framework, we integrated a semantically annotated literature corpus, Semantic MEDLINE, with a semantically coded ADE knowledge base known as ADEpedia (10) using a Semantic Web-based framework. We developed a knowledge-discovery approach leveraging a network-based analysis of a protein-protein interaction (PPI) network to mine the knowledge of drug-ADE-gene interactions.
The recent advances in sequencing technology have underpinned the progress in several large-scale projects to systematically compile genomic informatics related to human cancer (11, 12). A notable example is The Cancer Genome Atlas (TCGA) (13) and projects that have focused on identifying links between cancer and genomic variation. More promisingly, TCGA Pan-Cancer Project (14) has been initiated to assemble coherent datasets across tumor types, analyze the data in a consistent fashion, and finally provide comprehensive interpretation. Tumor stratification has been regarded as one of the fundamental goals of cancer informatics, enabling Pan-Cancer studies in which the molecular profiles of tumors are used to determine subtypes (15), regardless of the organ in which it is manifest. In particular, the somatic mutation profile is emerging as a rich new source of data for uncovering tumor subtypes with different causes and clinical outcomes. A network-based stratification using the knowledge of molecular signaling could produce robust tumor subtypes that are biologically informative and have a strong association to clinical outcomes and emergence of drug resistance (15).
Preliminary studies have demonstrated that the underlying molecular mechanism of common ADEs known to cancer therapy drugs may overlap with that of the efficacy of the therapeutic drugs themselves. For example, breast cancer patients receiving aromatase inhibitors (AI) have a high incidence of musculoskeletal adverse events (MS-AEs); about half of patients treated with AIs have joint-related complaints (16, 17). Musculoskeletal complaints have been the most frequent reason given by patients on a clinical trial comparing the non-steroidal AI anastrozole with the steroidal AI exemestane as adjuvant therapy for early breast cancer (18). A case-control genome-wide association study (GWAS) from a Mayo Clinic group identified SNPs associated with MS-AEs in women treated with AIs, one of which created an estrogen response element (18). Another study in the same group at Mayo Clinic confirmed that single nucleotide polymorphisms (SNPs) in the aromatase CYP19 gene contribute to response to neoadjuvant AI therapy (19), two of which are significantly associated with both a greater change in aromatase activity after AI treatment and higher plasma estradiol levels pre- and post-AI treatment.
The objective of the present study is to develop a novel knowledge-driven approach that provides an ADE-based stratification of tumor mutations (ADEStrata). Our assumption here is that the ADE-based tumor stratification would potentially produce clinically and biologically meaningful tumor subtypes that are predictive of the drug response to the cancer therapy drugs. To test the assumption, we performed a case study of breast cancer patients receiving the AIs from TCGA. We utilized a variant prioritization tool upon the somatic mutation profiles of TCGA breast invasive carcinoma (BRCA) patients treated with three AI drugs. The phenotype input of the variant prioritization tools contains a set of MS-AEs represented by standard Human Phentoype Ontology (HPO) terms. We utilized the prioritized variants to cluster the target patients into subgroups and investigated their associations with clinical outcomes.
2 Materials and Methods
2.1 Materials
2.1.1 SIDER: A Side Effect Resource
The SIDER (SIDe Effect Resource) is a public, computer-readable side effect resource that contains reported adverse drug reactions(20). The information is extracted from public documents and package inserts; in particular, from the United States Food and Drug Administration (FDA) Structured Product Labels (SPLs). The standardized Coding Symbols for a Thesaurus of Adverse Reaction Terms (COSTART), which are part of the Unified Medical Language System (UMLS) Metathesaurus, were used as the basic lexicon of side effects. In the present study, we utilized the latest version SIDER 2 that was released on October 17, 2012.
2.1.2 HPO: Human Phenotype Ontology
The HPO project aims to provide a standardized vocabulary of phenotypic abnormalities encountered in human diseases (21). The HPO is being developed in collaboration with the OBO Foundry using information from Online Mendelian Inheritance in Man (OMIM) and medical literatures. The ontology contains more than 10,000 terms and equivalence mappings to other standard vocabularies such as MedDRA and UMLS. In the present study, we used the latest version of HPO-MedDRA mapping file that is publicly available from the HPO website (22).
2.1.3 eXtasy: A Variant Prioritization Tool
eXtasy is a pipeline developed at the University of Leuven, for ranking the likelihood that a given nonsynonymous single nucleotide variants (nSNVs) is related to given phenotype (23, 24). The pipeline utilizes a genomic data fusion methodology (25) that takes into account multiple strategies to detect the deleteriousness of mutations and prioritizes them in a phenotype-specific manner. The ultimate goal of the tool is to discriminate between putatively mildly deleterious rare variants and actual disease-causing variants. The eXtasy tool is open-source and publicly downloadable from its github site (26). The eXtasy pipeline takes a Variant Call File (VCF) and one or more gene prioritization files. Each prioritization file is pre-computed for a specific phenotype (HPO term). In the present study, we downloaded and installed the tool on a local Ubuntu server.
2.1.4 TCGA Data Portal
TCGA Data Portal provides a platform for researchers to search, download, and analyze data sets generated by TCGA (27). It contains clinical information, genomic characterization data, and high level sequence analysis of the tumor genomes. There are two data access tiers: Open Access data tier and Controlled Access. The data of more 30 tumor types are available from the data portal. As of February 2014, there are 1043 cases of breast invasive carcinoma (BRCA) with data. In the present study, we utilized the BRCA clinical data (including clinical drug data and follow-up data) and somatic mutation data through the Open Access data tier.
2.2 Methods
2.2.1 Identifying HPO ADE Terms Relevant to Aromatase Inhibitors
In the present study, we aim to conduct variant prioritization in a manner that can be guided by the ADEs relevant to AI drugs. In other words, the ADEs induced by AI drugs are treated as the phenotypes required by the eXtasy tool. In order to enable the use of the eXtasy tool for variant prioritization in a phenotype-specific manner, we need to identify the phenotypes that are represented in HPO terms.
We first mapped the ADE terms represented in MedDRA UMLS concept unique identifiers (CUIs) from the SIDER 2 database file to the HPO terms using an HPO-MedDRA mapping file produced by HPO development team. Second, we annotated those HPO terms with a flag using the eXtasy HPO term list to indicate whether a HPO-based ADE term can be processed by eXtasy or not. Third, we retrieved those entries (with drug-ADE pairs) using the drug names “anastrozole”, “exemestane” and “letrozole” which are the third-generation AI drugs. We reviewed all the ADE terms and identified those ADE terms belonging to musculoskeletal adverse events (MS-AEs) and their HPO term annotations.
2.2.2 Identifying Patient Cohorts by AI Drugs and Somatic Mutations from TCGA
We utilized the clinical drug file of the BRCA patients from TCGA data portal through its Open-Access HTTP Directory. The spelling corrections were taken for all variants of the three drugs to maximize the sample size of the patient cases. We then identified a set of patient cases (represented by patient barcodes) that were prescribed for the three AI drugs (AI cases).
We also downloaded the somatic mutation file of the BRCA patients from TCGA data portal in a Mutation Annotation Format (MAF). The format is a tab-delimited file containing somatic mutations for each patient. As eXtasy requires a VCF file as input, we converted the MAF file into a collection of VCF files. Each VCF file contains somatic mutations for a single patient tumor sample. We combined all VCF files for all AI cases into a single VCF file using the patient barcodes identified in the step above.
2.2.3 Variant Prioritization Using HPO ADE Terms
As mentioned above, we installed an instance of the eXtasy tool in a local server and ran the tool with a custom Ruby script. The input consists of a VCF file (produced in the Section 2.2.2) and a set of pre-computed gene prioritization files for those phenotypes represented by the HPO ADE terms of interest (identified in the Section 2.2.1). The output is a file with scores for the individual variants’ likelihood of impacting an individual HPO term. Order statistics (25) and aggregate scores are generated and range from 0 to 1, where 0 is likely to be disease-causing and 1 unlikely (in contrast with the normal eXtasy scores). This is a pseudo p-value that represents the probability that a variant is high-ranking in all different phenotypes given the null-hypothesis of random rankings. To understand how the variants affect function, we first classified the input variants into three functional impact categories, calling a variant “high” if it is a frameshift, nonsense, nonstop, or splice-site; and “medium” if it is a missense; and “silent” if it is a mutation not causing protein coding changes. And then we analyzed the function of those variants scored by eXtasy for AI-related HPO terms.
2.2.4 Tumor Mutation Stratification and Clinical Outcome Association Studies
We first selected statistically significant variants based on the eXtasy order statistics (pseudo p-value <0.05). Second, we aggregated genes affected by these prioritized variants across 1,320 canonical pathways collected from the Molecular Signature Database (MSigDB) (28, 29). In order to reduce false discoveries, multiple criteria were applied to further filter out less relevant pathways (binomial distribution p-value >0.05) or pathways containing too few genes (<10 genes). We excluded pathways with less than 10 genes, based on the consideration that small pathways are often subcomponents of larger pathways, and inclusion of them tends to introduce unnecessary redundancy. Third, we performed hierarchical clustering to highlight pathway-level patterns among AI-treated patients.
We used overall survival (OS) time (months) as a clinical endpoint to measure the outcome of TCGA patients in the identified cohort. We performed both univariate analysis and multivariate cox-regression to assess the association of clusters (produced by hierarchical clustering) with survival. In multivariate analysis, patient age and tumor stage were adjusted for to evaluate the independent contribution of each cluster. We also analyzed the distribution of patient age and tumor stage in the clusters identified.
3 Results
Out of 4,492 unique MedDRA terms represented by the UMLS CUIs from the SIDER database file, 2,827 (62.9%) MedDRA terms had mappings to 1,491 unique HPO terms. Out of the 1,491 HPO terms, 844 (56.6%) HPO terms are included in the eXtasy phenotype list. We identified 6 unique HPO terms representing the MS-AEs relevant to three AI drugs. The 6 HPO terms are HP:0003418|Back pain, HP:0002653|Bone pain, HP:0003011| Abnormality of musculature, HP:0001369|Arthritis, HP:0009763|Limb pain, and HP:0002758|Osteoarthritis. Table 1 shows the SIDER database entries with HPO terms identified for the musculoskeletal adverse events (MS-AEs) relevant to three AI drugs.
Table 1.
Entries with HPO terms identified for the musculoskeletal adverse events (MS-AEs) relevant to three AI drugs
| Drug label | Meddra umls cui | Meddra label | HPO id | HPO label | HPO Term in eXtasy |
|---|---|---|---|---|---|
| anastrozole | C0004604 | Back pain | HP:0003418 | Back pain | YES |
| anastrozole | C0151825 | Bone pain | HP:0002653 | Bone pain | YES |
| anastrozole | C0026857 | Musculoskeletal disorder | HP:0003011 | Abnormality of musculature | YES |
| anastrozole | C0003864 | Arthritis | HP:0001369 | Arthritis | YES |
| exemestane | C0004604 | Back pain | HP:0003418 | Back pain | YES |
| exemestane | C0151825 | Bone pain | HP:0002653 | Bone pain | YES |
| exemestane | C0030196 | Pain in extremity | HP:0009763 | Limb pain | YES |
| exemestane | C0026857 | Musculoskeletal disorder | HP:0003011 | Abnormality of musculature | YES |
| exemestane | C0029408 | Osteoarthritis | HP:0002758 | Osteoarthritis | YES |
| letrozole | C0004604 | Back pain | HP:0003418 | Back pain | YES |
| letrozole | C0026857 | Musculoskeletal disorder | HP:0003011 | Abnormality of musculature | YES |
| letrozole | C0151825 | Bone pain | HP:0002653 | Bone pain | YES |
| letrozole | C0030196 | Pain in extremity | HP:0009763 | Limb pain | YES |
| letrozole | C0003864 | Arthritis | HP:0001369 | Arthritis | YES |
Using the clinical drug file of TCGA BRCA patients, we identified a cohort of 212 patients who were prescribed with one of the three AI drugs (AI cases).
The algorithm eXtasy ranks coding variants according to their probability of being related to a given phenotype. We found that 23.8% of the input variants are silent and are ignored by eXtasy, while 11.6% are of high impact (see section 2.2.3) and almost assuredly affect the normal physiologic function of the affected gene. Of the variants scored by eXtasy for AI-related HPO terms, 43% are highly conserved among placental mammals. Variants were prioritized for each patient across the MS-AE phenotypes represented by 6 HPO terms (listed in Table 1), producing aggregate prioritization scores (max and order statistics). Table 2 lists the top 20 prioritized variants.
Table 2.
Top 20 variants prioritized for the MS-AE phenotypes using the eXtasy in the AI cases
| Chromosome | Ref base | Alt base | Position | Gene region | eXtasy combined max | eXtasy combined order statistics |
|---|---|---|---|---|---|---|
| X | G | C | 77289124 | ATP7A | 0.952 | 2.57E-13 |
| 10 | C | G | 89692883 | PTEN | 0.866 | 2.76E-13 |
| 12 | G | A | 115118782 | TBX3 | 0.88 | 3.47E-13 |
| 17 | G | A | 7577094 | TP53 | 0.852 | 1.28E-12 |
| 3 | A | T | 49455277 | AMT | 0.936 | 1.34E-12 |
| 9 | G | T | 132576329 | TOR1A | 0.958 | 1.91E-12 |
| 19 | C | G | 41838160 | TGFB1 | 0.874 | 2.26E-12 |
| 10 | A | T | 8115746 | GATA3 | 0.906 | 2.35E-12 |
| 5 | C | T | 174156285 | MSX2 | 0.98 | 2.97E-12 |
| 12 | A | G | 115120669 | TBX3 | 0.846 | 3.16E-12 |
| 17 | C | T | 7577547 | TP53 | 0.892 | 3.70E-12 |
| 12 | G | A | 121431482 | HNF1A | 0.942 | 5.98E-12 |
| 17 | A | C | 7577144 | TP53 | 0.834 | 6.21E-12 |
| 17 | G | A | 7577105 | TP53 | 0.83 | 7.33E-12 |
| 7 | C | G | 5567503 | ACTB | 0.876 | 9.62E-12 |
| 17 | A | G | 7577129 | TP53 | 0.832 | 1.07E-11 |
| 12 | C | T | 110781179 | ATP2A2 | 0.908 | 1.92E-11 |
| 12 | C | A | 121426687 | HNF1A | 0.928 | 2.23E-11 |
| 3 | G | A | 30729932 | TGFBR2 | 0.792 | 4.07E-11 |
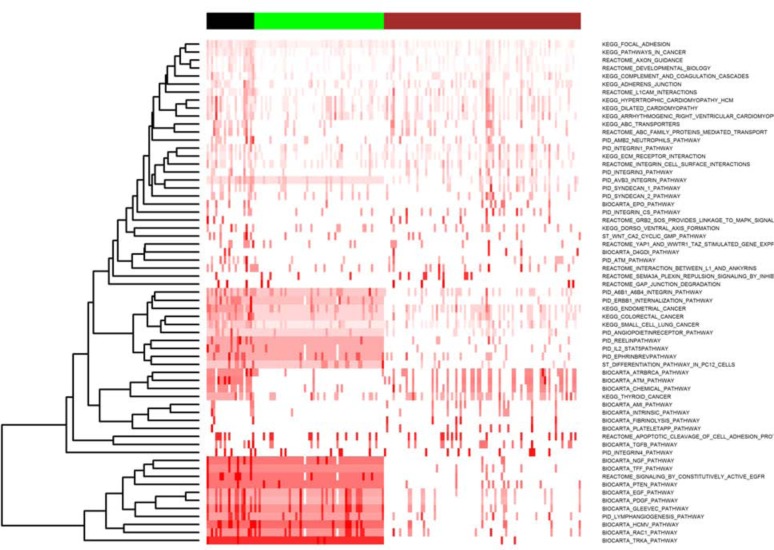
From the eXtasy output for the AI cases, 2,164 statistically significant variants were selected for pathway enrichment and clustering analysis. Among 1,320 canonical pathways, 63 of them passed the filtering criteria defined in section 2.2.4. By hierarchical clustering, 3 distinct patient clusters, organized by pathways (affected by prioritized variants), were identified and are displayed in Figure 1 containing 91, 60, and 22 patients each. Patients in Cluster 1 exhibit relatively silent pathway aberrations, while Cluster 2 and Cluster 3 have much stronger pathway activities. A summary of the 63 selected pathways, enriched in MS-AE relevant variants, can be found in Supplemental Table 1 posted at http://catargets.org.
Figure 1.
This ordered heatmap shows pathway-level clustering of 173 patients treated with AI across 63 pathways-enriched MS-AE relevant variants. The color of heatmap from white to red indicates low to high percentages (0% to 100%) of genes affected by MS-AE relevant variants. Column color-bar on top of the heatmap indicates three clusters of samples: Cluster 1 (brown), Cluster 2 (green) and Cluster 3 (black). Note that the number of the patients (n=173) with pathway enrichment is less than total number of the identified cohort (n=212) is because not all patients have prioritized variants listed.
Table 3 shows the results of the univariate and multivariate cox-regression analysis for the three clusters. We found that although Cluster 3 has a relatively small number of patients, the cluster is significantly association with poorer survival time in both univariate and multivariate analysis. Table 4 shows the distribution of age and stage in the 3 clusters identified. There is no significant association between the 3 clusters and age/stage, although we noticed that Cluster 3 is enriched with more Stage 2 patient cases.
Table 3.
The univariate and multivariate cox-regression analysis results of cluster labels. In multivariate analysis, patient age and tumor stage were adjusted for to determine the independent contribution of cluster membership.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| p-value | HR [95% CI] | p-value | HR [95% CI] | |
|
Cluster 2 (n=60) (Cluster 1 as ref) |
0.63 | 0.57 [0.06, 5.55] | 0.64 | 0.57 [0.06, 5.76] |
|
Cluster 3 (n=22) (Cluster 1 as ref) |
0.03* | 5.03 [1.13, 22.55] | 0.04* | 4.86 [1.07, 22.161] |
|
Overall model (log-rank test) |
0.02* | NA | 0.07 | NA |
denotes p<0.05.
Table 4.
The disribution of age and stage in the 3 clusters identified.
| Cluster 1 (n=91) | Cluster 2 (n=60) | Cluster 3 (n=22) | p-value* | |
|---|---|---|---|---|
|
Age mean [Q1, median, Q3] |
61 [55.2, 62.9, 66.1] | 63.4 [54.9, 62.8, 71.9] | 58.4 [54.1, 58.7, 62.8] | 0.16 |
|
Stage number (% per cluster) |
0.22 | |||
| s1 | 32 (35.2%) | 23 (38.3%) | 5 (22.7%) | |
| s2 | 48 (52.7%) | 27 (45%) | 15 (68.2%) | |
| s3 | 11 (12.1%) | 8 (13.3%) | 1 (4.5%) | |
| s4 | 0 (0%) | 2 (3.3%) | 1 (4.5%) |
p-value for age vs. cluster association was computed using ANOVA test; p-value for stage vs. cluster association was computed using Fisher’s exact test.
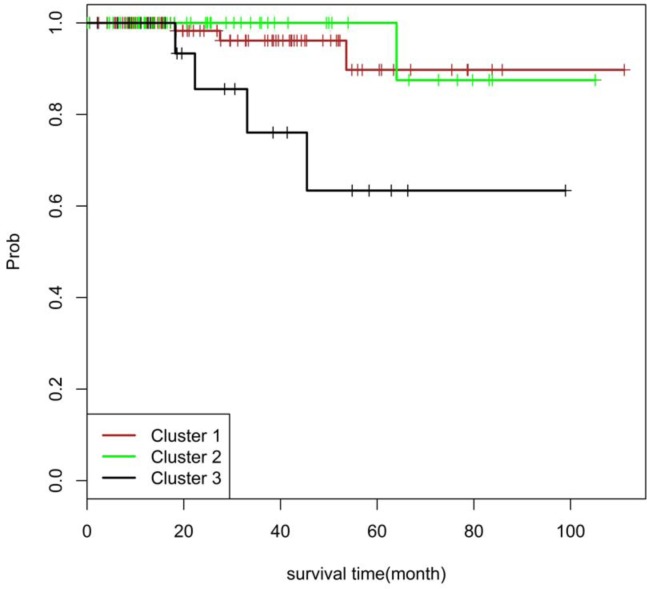
Figure 2 shows a Kaplan-Meier plot of survival time for the 3 clusters, derived from our pathway-level analysis. Interestingly, Cluster 2 does not have a significantly altered survival time, despite its similarity to Cluster 3. The exception is for a few activated pathways responsible for DNA damaging repair and apoptosis. In addition, we observed that many patients in Cluster 1 have somatic variants associated with ATM/thyroid pathways, while Cluster 2 has many other pathway features but lacks the ATM/thyroid pathway enrichment. Cluster 3, however, has the pathway features of both Cluster 1 and Cluster 2. This “two hits” pattern may account for the worse survival outcome associated with Cluster 3.
Figure 2.
Kaplan-Meier plot of survival time for patients within the 3 pathway-pattern clusters.
4 Discussion
In this study, we demonstrated that the ADE-based tumor stratification could produce clinically and biologically meaningful tumor subtypes that are potentially predictive of the drug response to the cancer therapy drugs. The preliminary results from the case study of TCGA breast cancer patients receiving AI drugs are very promising. We consider that our study approach and results have several important implications in terms of how to further understanding of disease, drug action or to improve treatment outcome. First, it would be possible that new patients can be assigned to different groups to inform clinical decision-making. Second, our approach could be potentially used in prediction of treatment outcomes or probability of ADEs based on tumor genome. Third, it would be possible that our study results could be used to gain a greater understanding of mechanism of action of targeted drugs or underlying causes of ADEs.
One of the key components of our approach is to leverage known ADE knowledge for tumor stratification. We found that a semantically coded ADE knowledge base is extremely useful for extracting known ADEs relevant to target drugs. We utilized the SIDER ADE dataset, in which the ADEs are annotated with the MedDRA codes represented by UMLS CUIs. In addition, the HPO team has produced a mapping file between MedDRA UMLS CUIs and HPO terms. These standard codes facilitated our ability to integrate and identify ADE terms across different datasets. For example, in the present study, eXtasy requires HPO terms as phenotype input and we were able to identify those MS-AE terms from the SIDER dataset represented in UMLS CUIs.
Use of standard phenotype/diagnosis codes enables the potential of an automatic mechanism to retrieve a set of ADE codes for a specific patient or disease domain. Although we manually identified and retrieved 6 ADE codes related to MS-AEs in this study, it would be valuable to build a valueset definition mechanism based on the semantic hierarchical relationship (eg, parent-child relation) asserted in standard terminologies, ideally through standard terminology services such as the recent development of the OMG/HL7 standard – common terminology services 2 (CTS2) (30, 31). Drug data normalization is another typical use case for utilizing standard terminologies and an important prerequisite for accurate ADE association. We noticed that the scope of drug name normalization must move beyond synonym mapping (itself a difficult task) as commonly used public resources also contain spelling errors and other types of irregularities. For example, from TCGA clinical drug data, the names “CYOTXAN”, “CYTOXAN”, “CYTOXEN”, “CYCLOPHASPHAMIDE”, “CYCLOPHOSPHAMID” are used to record the drug “Cyclophosphamide”. A more detailed investigation on drug data normalization is beyond the scope of the present study and is addressed in a separate paper (32).
We also found that the severity and frequency of the ADEs are important factors for enabling our tumor stratification approach. As we mentioned in the Introduction section, breast cancer patients receiving AIs have a high incidence of MS-AEs; about half of patients treated with AIs have joint-related complaints (16, 17). Musculoskeletal complaints have been the most frequent reason given by patients on a clinical trial comparing the non-steroidal AI anastrozole with the steroidal AI exemestane as adjuvant therapy for early breast cancer (18). We believe that an ADE knowledge base with such severity and frequency information would be greatly useful in selecting the ADE phenotypes for tumor stratification, which is one of the goals of our ongoing ADEpedia project. In a previous study, we have developed an approach to build a severe ADE knowledge base based on the FDA Adverse Event Reporting System (AERS) reporting data (33). In this work, we propose a computational approach to screen genomic variants from individual patients and relate them to the probability of that patient experiencing an ADE while on a particular treatment.
In this study, we utilized the variant prioritization tool eXtasy for two primary reasons. First, eXtasy uses summary statistics of multiple criteria to evaluate the importance and contextual relevance of nonsynonymous SNVs according to conservation, interaction networks derived from experimental and knowledge databases, gene ontology, etc. This provides a succinct computable mechanism for aggregating knowledge associated with the nonsynonymous SNVs. Second, variant prioritization is guided by specific phenotypes, which in particular are organized using standard HPO terms. This provides a standard interface, allowing us to use the ADEs in HPO terms as input to the tool. In the future, we plan to evaluate the eXtasy tool in comparison with other variant prioritization algorithms. Pre-filtering the variants to only those we believe likely to affect function may be worth exploring, but may suffer from eXtasy having less power to make an appropriate association. The eXtasy algorithm only utilizes non-synonymous variants; all silent mutations (about 23.8%) are filtered out. It is intuitive that those affecting splicing, frameshift, or truncating would be more impactful than missense variants, but they are also more rare.
The rich cancer genomic data produced by TCGA Research Network provides a major opportunity to develop an integrated picture of commonalities, differences and emergent themes across tumor lineages. TCGA Pan-Cancer project (14) envisions that there are six types of omics characterization that create a data stack for maximizing the potential of integrative analysis. The six types comprise mutation, copy number, gene expression, DNA methylation, MicroRNA, reverse-phase proteomic arrays (RPPA) and clinical data. Hofree, et al, introduced a network-based stratification (NBS) method to integrate somatic tumor genomes with gene networks, which could identify subtypes in ovarian, uterine and lung cancer cohorts from TCGA (15). They demonstrated that the subtypes are predictive of clinical outcomes such as patient survival, response to therapy or tumor histology. By integrating mRNA, microRNA (miRNA), and DNA methylation next-generation sequencing data from TCGA, Volinia, et al. performed survival analysis on a cohort of 466 patients with primary invasive ductal carcinoma (IDC), and produced an integrated RNA signature that has been demonstrated prognostic to the IDC patients (34). The novelty of the present study is to build a tumor stratification method that utilizes the ADE-based variant prioritization, with the assumption that the underlying molecular mechanism of common ADEs known to cancer therapy drugs may overlap with that of the efficacy of the therapeutic drugs themselves, or have common indications. Although not presently utilizing the full data stack, our approach did identify subtypes predictive of patient survival time. We believe that an integrative analysis with more omics data types would provide greater insights into the underlying biological mechanisms of the identified subtypes.
Since the impact of an individual variant is often difficult to interpret, in particular those only mutated in one (or a few) patient(s), directly comparing the landscape of genomic differences across patients is of great difficulty. To address the challenge, we performed a pathway enrichment analysis with multiple criteria and identified 63 canonical pathways that are highly related to the prioritized variants selected using the MS-AE phenotypes. In general, the definition of a pathway is a convenient abstraction for underlying molecular regulations, but cross-talk between pathways is often observed. Capitalizing upon cross-talk, we were able to perform a hierarchical clustering analysis to highlight the pathway-level patterns for the somatic variants among AI treated patients. Three clusters were identified, in which we found that the pathway pattern in Cluster 1 demonstrated its own characteristics in terms of pathway aberrations in comparison with the pathway patterns in Cluster 2 and 3. More promisingly, clinical outcome association analysis demonstrated that the survival time among the three clusters is significantly different, with Cluster 3 having the worst survival time (see Figure 2). We find that Clusters 2 and 3 have a similar pathway pattern in general, except for a few activated pathways responsible for DNA damaging repair and apoptosis. This perhaps represents a “two hits” pattern for Cluster 3 that may be responsible for the poorer survival outcome. We consider that our pathway-based clustering approach would make the findings from clinical outcome association studies more interpretable.
In summary, we developed a novel knowledge-driven approach that provides an ADE-based stratification of tumor mutations. We demonstrated that the prediction of per-patient ADE propensity simultaneously identifies high-risk patients experiencing poor outcomes. We plan to evaluate and validate our approach by incorporating more other data types (eg, germline variants) and other tumor types, and explore its potential in enabling pan-cancer studies in the future. The datasets and supplementary results produced by the study are publicly available at http://catargets.org.
References
- 1.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clinical pharmacology and therapeutics. 2011 Mar;89(3):464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genetics in medicine : official journal of the American College of Medical Genetics. 2013 Jul;15(7):565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L. Pharmacogenomics: a systems approach. Wiley interdisciplinary reviews Systems biology and medicine. 2010 Jan-Feb;2(1):3–22. doi: 10.1002/wsbm.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karczewski KJ, Daneshjou R, Altman RB. Chapter 7: Pharmacogenomics. PLoS computational biology. 2012;8(12):e1002817. doi: 10.1371/journal.pcbi.1002817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly AK. Pharmacogenomics of adverse drug reactions. Genome medicine. 2013 Jan 29;5(1):5. doi: 10.1186/gm409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. JAMA : the journal of the American Medical Association. 2001 Nov 14;286(18):2270–9. doi: 10.1001/jama.286.18.2270. [DOI] [PubMed] [Google Scholar]
- 7.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics. PharmGKB Pharmacogenomics. 2010 Apr;11(4):501–5. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clinical pharmacology and therapeutics. 2012 Oct;92(4):414–7. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang G, Wang C, Zhu Q, Chute CG. A Framework of Knowledge Integration and Discovery for Supporting Pharmacogenomics Target Predication of Adverse Drug Events: A Case Study of Drug-Induced Long QT Syndrome. AMIA Summits on Translational Science proceedings AMIA Summit on Translational Science; 2013. pp. 88–92. [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang G, Solbrig HR, Chute CG. ADEpedia: a scalable and standardized knowledge base of Adverse Drug Events using semantic web technology. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2011. pp. 607–16. [PMC free article] [PubMed] [Google Scholar]
- 11.Vazquez M, de la Torre V, Valencia A. Chapter 14: Cancer genome analysis. PLoS computational biology. 2012;8(12):e1002824. doi: 10.1371/journal.pcbi.1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013 Mar 29;339(6127):1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Cancer Genome Atlas [cited February 17, 2014]; Available from: http://cancergenome.nih.gov/
- 14.Weinstein JN, Collisson EA, Mills GB, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nature genetics. 2013 Oct;45(10):1113–20. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofree M, Shen JP, Carter H, Gross A, Ideker T. Network-based stratification of tumor mutations. Nature methods. 2013 Nov;10(11):1108–15. doi: 10.1038/nmeth.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crew KD, Greenlee H, Capodice J, et al. Prevalence of joint symptoms in postmenopausal women taking aromatase inhibitors for early-stage breast cancer. Journal of clinical oncology official journal of the American Society of Clinical Oncology. 2007 Sep 1;25(25):3877–83. doi: 10.1200/JCO.2007.10.7573. [DOI] [PubMed] [Google Scholar]
- 17.Henry NL, Giles JT, Ang D, et al. Prospective characterization of musculoskeletal symptoms in early stage breast cancer patients treated with aromatase inhibitors. Breast cancer research and treatment. 2008 Sep;111(2):365–72. doi: 10.1007/s10549-007-9774-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingle JN, Schaid DJ, Goss PE, et al. Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010 Nov 1;28(31):4674–82. doi: 10.1200/JCO.2010.28.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Ellsworth KA, Moon I, et al. Functional genetic polymorphisms in the aromatase gene CYP19 vary the response of breast cancer patients to neoadjuvant therapy with aromatase inhibitors. Cancer research. 2010 Jan 1;70(1):319–28. doi: 10.1158/0008-5472.CAN-09-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhn M, Campillos M, Letunic I, Jensen LJ, Bork P. A side effect resource to capture phenotypic effects of drugs. Molecular systems biology. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohler S, Doelken SC, Mungall CJ, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic acids research. 2014 Jan 1;42(1):D966–74. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The Human Phenotype Ontology URL. [cited February 17, 2014]; Available from: http://www.human-phenotype-ontology.org/
- 23.Sifrim A, Popovic D, Tranchevent LC, et al. eXtasy: variant prioritization by genomic data fusion. Nature methods. 2013 Nov;10(11):1083–4. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- 24.eXtasy URL. [cited February 15, 2014]; Available from: http://homes.esat.kuleuven.be/~bioiuser/eXtasy/
- 25.Aerts S, Lambrechts D, Maity S, et al. Gene prioritization through genomic data fusion. Nature biotechnology. 2006 May;24(5):537–44. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 26.eXtasy GitHub URL. [cited February 15, 2014]; Available from: https://github.com/asifrim/eXtasy.
- 27.TCGA Data Portal [cited February 17, 2014]; Available from: https://tcga-data.nci.nih.gov/tcga/
- 28.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011 Jun 15;27(12):1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America; 2005. Oct 25, pp. 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathak J, Bailey KR, Beebe CE, et al. Normalization and standardization of electronic health records for high-throughput phenotyping: the SHARPn consortium. Journal of the American Medical Informatics Association : JAMIA. 2013 Dec;20(e2):e341–8. doi: 10.1136/amiajnl-2013-001939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.CTS2 Wiki. [cited March 12, 2014]; Available from: http://informatics.mayo.edu/cts2/index.php/Main_Page.
- 32.Jiang G, Sohn S, Zimmermann MT, Liu H, Chute CG. Drug Normalization for Cancer Therapeutic and Druggable Genome Target Discovery. Proceedings of ICBO 2014 – International VDOS Workshop (in submission); Houston, TX. 2014. [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang G, Wang L, Liu H, Solbrig HR, Chute CG. Building a knowledge base of severe adverse drug events based on AERS reporting data using semantic web technologies. Studies in health technology and informatics. 2013;192:496–500. [PubMed] [Google Scholar]
- 34.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proceedings of the National Academy of Sciences of the United States of America; 2013. Apr 30, pp. 7413–7. [DOI] [PMC free article] [PubMed] [Google Scholar]