Abstract
Competence is essential for health care professionals. Current methods to assess competency, however, do not efficiently capture medical students’ experience. In this preliminary study, we used machine learning and natural language processing (NLP) to identify geriatric competency exposures from students’ clinical notes. The system applied NLP to generate the concepts and related features from notes. We extracted a refined list of concepts associated with corresponding competencies. This system was evaluated through 10-fold cross validation for six geriatric competency domains: “medication management (MedMgmt)”, “cognitive and behavioral disorders (CBD)”, “falls, balance, gait disorders (Falls)”, “self-care capacity (SCC)”, “palliative care (PC)”, “hospital care for elders (HCE)” – each an American Association of Medical Colleges competency for medical students. The systems could accurately assess MedMgmt, SCC, HCE, and Falls competencies with F-measures of 0.94, 0.86, 0.85, and 0.84, respectively, but did not attain good performance for PC and CBD (0.69 and 0.62 in F-measure, respectively).
1. Introduction
National accreditation bodies including the Accreditation Council for Graduate Medical Education (ACGME) and the American Association of Medical Colleges (AAMC) have called for competency based curriculum and assessment models for training programs. Many medical schools have responded with education portfolios to capture student experiences in real and simulated environments matched to shared competency goals. Education portfolios, however, have not achieved widespread adoption, partially because current methods require significant manual entry of a limited amount of clinical data. Automatic and valid methods of capturing the richness of students’ clinical experiences are needed.
In this project, we developed and validated the machine learning based methods used to identify student experiences in six AAMC geriatrics competency domains: medication management (MedMgmt), cognitive and behavioral disorders (CBD), falls, balance, gait disorders (Falls), self-care capacity (SCC), palliative care (PC), and hospital care for elders (HCE). We tested different feature sets such as bag of words, use of biomedical concepts identified through natural language processing (NLP), and a refined list of physician-identified concepts corresponding to a particular competency. This work also highlighted the significant challenges for identifying medical student competency from clinical notes.
2. Background
Competency-based assessment methods combine a variety of modalities to provide a comprehensive evaluation of a learner’s knowledge and proficiency. 1,2 Medical schools using competency-based assessments typically rely on education portfolios to track students’ progress.3–5 Portfolio components can include personal reflections, examinations, individual and small group projects, simulation encounter reports such as observed structured clinical examinations (OSCEs), mentoring experiences, and clinical exposures. Handwritten log books or score sheets of clinical data,6,7 replaced now by portable electronic solutions,8–11 allow students to enter patient information including demographics, diagnosis, procedures performed, and/or severity of illness. Use of these systems is limited for various reasons, including lack of time. Furthermore, teachers often disagree with students on primary diagnoses. We propose a system that automatically captures all concepts in a student’s notes and organizes the data automatically to reflect the student’s full experience and proficiency along important clinical outcomes.
The AAMC and the John A. Hartford Foundation developed a minimum set of graduating medical student competencies to ensure competent care of older patients by new interns. 12 With the help of leading geriatric educators and survey responses from educators in a number of clinical domains, the consensus panel established eight core geriatric competency domains. Each competency domain contains 2–5 competencies, outlining detailed goals for medical students in each domain. For example, the “medical management” domain includes 3 subtopics: 1) age related sensitivity to drug selection and dosing based on patient factors (e.g., renal and hepatic dysfunction); 2) identifying medications (e.g., anticholinergic and analgesics) that should be avoided or used with caution in the elderly; and 3) documenting the patient’s complete medication list (including herbal and over-the-counter medications) and recognizing possible side effects. These competency domains represent an agreed-upon framework to guide educational curricula and assessment of medical students.
Competency-based assessment may be amenable to clinical NLP and machine learning (ML) methods. Prior work has shown that the logistic regression and concept-based queries can identify student exposure to some common presentations, such as chest pain or fever.13 Such systems are currently being used at Vanderbilt to track student exposure to 16 common presenting problems through the Learning Portfolio website, which captures all student-authored clinical notes.14 ML-based methods have not been studied extensively in the medical education domain. ML-based models are trained on available annotated datasets, and then applied to new samples to identify their labels. As a preliminary study, we focused on developing ML-based methods to identify six of the geriatric competency domains from clinical notes written by third and fourth year medical students in Vanderbilt.
3. Methods
The automatic competency detection tool consists of three components. The first is Learning Portfolio, a web-based system that gathers all students’ clinical notes from patient encounters in the electronic medical record (EMR). 15 Portfolio employs NLP, using KnowledgeMap Concept Indexer (KMCI) and SecTag to identify biomedical concepts from these notes mapped to Unified Medical Language System (UMLS) with their local context (negation and section information). We then applied ML based competency detectors to the NLP output to determine the relevance of a note with respect to different geriatric competency domains based on identified biomedical concepts. Figure 1 illustrates the components used in this project.
Figure 1.
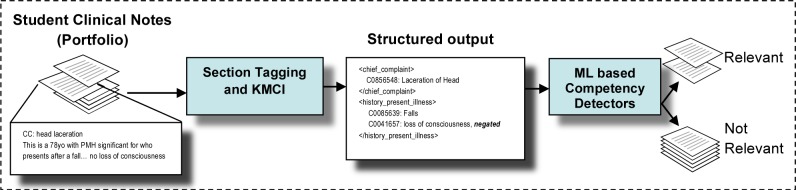
Overview of project: Machine learning based competency detectors assess a clinical note’s relevance for each competency based on structured output of the KMCI.
3.1 Section Tagging
We used the SecTag section tagger to recognize clinical note sections and their boundaries.16 The SecTag algorithm uses a concept-oriented, hierarchical section header terminology.17 In this study, we only used the top sections and some of the first subsections shown in Table 1 as clinician-identified relevant sections. For example, “Family Medical History” under top section “Patient History” is kept as “Family Medical History.” Key/first subsections listed in Table 1 replaced their subsections. For example, we have a hierarchical structure of the following sections: Objective Data -> Physical Examination -> Cardiovascular exam. “Objective Data” is the top section, “Physical Examination” is the first or key subsection, and “Cardiovascular exam” is the subsection of the key subsection. Then “Cardiovascular exam” was replaced by “Physical Examination”. We also added a blank section “None” to indicate the content outside of these sections.
Table 1.
Note sections and subsections for use
|
Note: “None” represents the location of words or concepts outside section boundaries.
3.2 KnowledgeMap Concept Indexer (KMCI)
KMCI is a tool for analyzing unstructured text. For each clinical note, KMCI utilizes UMLS knowledge resources to encode concepts with Concept Unique Identifiers (CUIs). Once assigned a CUI, the UMLS provides, for each concept, semantic type information (e.g., “congestive heart failure” is a “disease or syndrome”) and relationships to other concepts (e.g., “anterior myocardial infarction” is a type of “heart disease). Locating concepts in the appropriate section of a note can allow an educator or an algorithm to rate a student’s mastery of a concept. For example, a search for key concepts related to back pain effectively identifies a note with back pain as a chief complaint, the extent of an appropriate back exam, and the presence or absence of differential diagnoses of back pain found in the student’s written assessment.
3.3 Gold Standard of Competency Relevance
Our gold standard note corpus consists of 399 clinical notes from randomly selected inpatients older than 65 years or admitted to the geriatric service and followed by medical students. Geriatric educators rated a student’s notes for each admission as containing high, medium, low, or no relevance to each geriatric competency domain. Each reviewer was a board-certified internal medicine physician who was either a geriatrician or has significant experience with geriatrics, including covering the inpatient geriatric service. A total of five reviewers scored each admission. Before scoring, physician reviewers reviewed test sets of admissions and scored them, developing a formal rubric for high, medium, and low/no for each competency domain. Then admissions were scored, disagreements discussed, and consensus achieved. The rubric was developed over the course of several 1-hour meetings of the physicians. The relevance scores of 3, 2, 1, and 0 represent the relevance level from “high”, “medium”, “low”, and “no”, respectively. A “high relevance” document contains primary discussion of the components that indicate an experience in that competency domain. In total, 119 admissions were scored by between one and five geriatric educators. For disagreement on scores, we applied the majority vote strategy and finalized the score using the score with the most votes for each student. If scores remain tied, we assign the mean of the tied scores as the final score. We considered students’ notes with ‘high’ or ‘medium’ relevance (with final score of higher than 1) to have positive relevance and those with ‘low’ or ‘no (with final score of 1 or smaller) to have negative relevance. Table 3 shows the class distribution for each competency domain.
Table 3.
Class distribution for six competency domains
| Competency Domains | Number of Positive Samples | Number of Negative Samples | Total Samples |
|---|---|---|---|
| Medication Management (MedMgmt) | 94 (88%) | 13 (12%) | 107 |
| Cognitive and Behavioral Disorders (CBD) | 46 (43%) | 61 (57%) | 107 |
| Falls, balance, gait disorders (Falls) | 77 (72%) | 28 (28%) | 107 |
| Self-care capacity (SCC) | 78 (74%) | 28 (26%) | 106 |
| Palliative care (PC) | 44 (45%) | 54 (55%) | 98 |
| Hospital care for elders (HCE) | 79 (74%) | 28 (26%) | 107 |
3.4 Machine Learning Based Competency Detectors
We applied supervised machine learning techniques to determine a student’s experience with geriatric competencies by identifying relevant concepts or other features in the corpus of notes. Experts labeled each case relevant or irrelevant to competency domains. We tested several different feature sets including “Bag of Words” as the baseline feature set, CUIs identified by KMCI, CUIs coupled with Section (SEC), Negation (NEG), and semantic type (STY). We tested other features such as the counts of CUIs in each note as well as the normalized values of CUIs based on term frequency-inverse document frequency (TFIDF). In addition, we added the number of notes in each admission and the age of patient as two basic features in all experiments. Table 2 shows detailed descriptions of all feature sets in our experiments.
Table 2.
Description of feature sets used in machine learning experiments
| Name of Feature Set | Type of Feature | Description |
|---|---|---|
| Note_count | Integer | The number of clinical notes by a medical student for each admission |
| Age | Integer | Age of patient |
| Words (baseline) | Binary | Bag of Words features; “1” if word present; “0” if word absent |
| CUI | Binary | Concept code features; “1” if CUI present; “0” if CUI absent |
| CUI_NEG | Binary | Concept code with negation: If a CUI is negated in the note, the value of CUI is “0” and the value of CUI_NEG is “1” |
| CUI_SEC | Binary | Dyad of concept code (CUI) and section: “1” if CUI present in the section; “0” if CUI absent in the section |
| CUI_count | Integer | The count of each CUI in the notes for one admission |
| CUI_count_tfidf | Numeric | The TFIDF value of each CUI in the notes for one admission |
| STY | Binary | Semantic type features (as defined in the UMLS): “1” if semantic type present; “0” if semantic type absent |
We tried to mimic the way geriatric educators identified the relevance of competency from students’ clinical notes. Instead of using all CUIs from the notes, we also tested the feature set with a refined list of CUIs associated with corresponding competency domains, just as geriatric educators use searches for key concepts to assess students’ notes. These lists of CUIs were developed by clinicians using web-based tools part of the KnowledgeMap curriculum website,18 in which complex concept queries are used to track themes in the medical school curriculum. We have previously shown good performance using concept queries of this sort to identify broad themes in the curriculum (e.g., “genetics”, “radiology”).18 This reduced the size of features dramatically.
Naïve Bayes (NB, the baseline classifier, implemented in CLOP19), logistic regression (LR), and support vector machines (SVM) with linear kernel are three efficient supervised learning tools for the data in high dimensional space. We applied both LR and linear SVM in the package Liblinear. 20 We constructed a classifier for each competency domain using all feature sets including refined lists of CUIs.
3.5 Validation
For each competency domain, we ran experiments using 10-fold cross validation for three machine learning algorithms and all feature set candidates. Each fold was stratified so that the distribution of classes in each fold was similar to the original distribution over all samples. These algorithms produced a numeric “relevance score” for the test samples on each fold. We compared the performances of each method with each feature set by computing the average area under receiver operator characteristic curve (AUC) over the cross validation. In addition, using different thresholds for “relevance scores” from the scores of the ML algorithms, we generated different sets of binary predictions and analyzed the precision (i.e., positive predictive values), recall (i.e., sensitivity), and F-measure (the harmonic mean of recall and precision) for all test samples.
4. Results
We evaluated the competencies at the admission level, and a student could write multiple notes for each admission. Totally we had 119 admissions that consisted of 399 clinical notes. There were total 11,249 unique CUIs for all these notes with at least one grade for relevance. Refining this list using just the physician-identified relevant CUIs dramatically reduced the number of features. Given hundreds of refined CUIs for each competency by a geriatric and NLP expert, only 24 CUIs occurred in the notes related to competency domain of medication management, 33 related to CBD, 52 related to Falls, 35 related to PC, 93 related to HCE, and 61 related to SCC. Tables 4 to 9 show the AUC scores for different feature sets and machine learning algorithms for Medication Management, CBD, Falls, Self-care capacity, Palliative Care, and Hospital care for elders competencies, respectively. The best AUC score for each competency was highlighted in bold.
Table 4.
AUC results for competency MedMgmt over different feature sets
| Feature Sets | Num of Features | Naïve | LR | SVM |
|---|---|---|---|---|
| Words | 27406 | 0.72 | 0.73 | 0.75 |
| CUI | 10258 | 0.84 | 0.82 | 0.78 |
| CUI_SEC | 25947 | 0.72 | 0.77 | 0.80 |
| CUI_SEC + CUI_NEG | 27978 | 0.67 | 0.77 | 0.79 |
| CUI_SEC + CUI_NEG + STY | 28237 | 0.68 | 0.78 | 0.81 |
| CUI_count | 10576 | 0.82 | 0.89 | 0.91 |
| CUI_count_tfidf | 10572 | 0.77 | 0.68 | 0.86 |
| Refined CUI_count | 24 | 0.76 | 0.61 | 0.74 |
| Refined CUI_count+CUI_SEC+CUI_NEG +STY | 318 | 0.68 | 0.76 | 0.84 |
Table 9.
AUC results for competency SCC over different feature sets
| Feature Sets | Num of Features | Naïve | LR | SVM |
|---|---|---|---|---|
| Words | 27261 | 0.75 | 0.71 | 0.70 |
| CUI | 10227 | 0.78 | 0.70 | 0.69 |
| CUI_SEC | 25850 | 0.80 | 0.78 | 0.77 |
| CUI_SEC + CUI_NEG | 27867 | 0.79 | 0.75 | 0.75 |
| CUI_SEC + CUI_NEG + STY | 28126 | 0.78 | 0.78 | 0.77 |
| CUI_count | 10540 | 0.71 | 0.69 | 0.69 |
| CUI_count_tfidf | 10536 | 0.68 | 0.54 | 0.58 |
| Refined CUI_count | 61 | 0.58 | 0.70 | 0.68 |
| Refined CUI_count+CUI_SEC+CUI_NEG +STY | 290 | 0.64 | 0.65 | 0.64 |
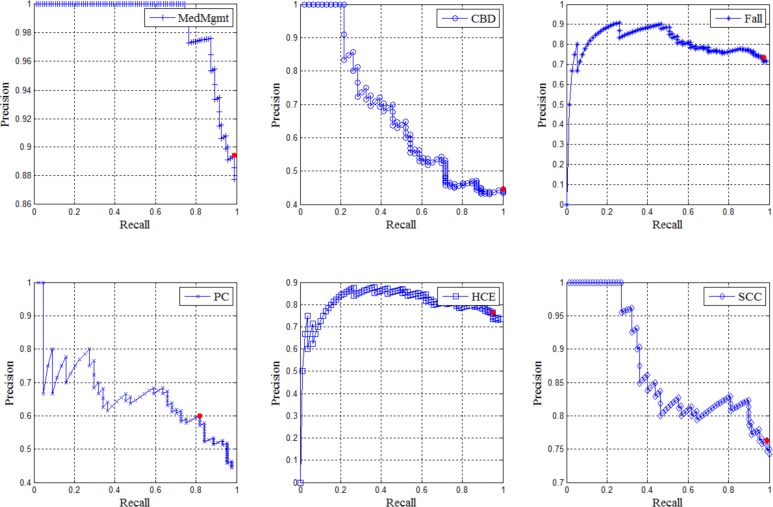
The SVM classifier generated the models with the best AUC scores of 0.91, 0.76, and 0.69 for competencies MedMgmt, PC, and HCE, respectively. The Naïve Bayes classifier achieved the best models with AUC scores of 0.73, 0.75, and 0.80 for competencies CBD, Falls, and SCC, respectively. For these six best models, we performed a precision-recall analysis based on different thresholds of numeric outputs, or “relevance score”, by the classifiers (see Figure 2). The red dot in each graph in Figure 2 represents the precision and recall using the best threshold.
Figure 2.
Precision-Recall graphs for the best-performing models for each competency domain. The red dot represents the point with the maximum F-measure (the harmonic mean of recall and precision).
By using the best threshold that can maximize the F-measure (the harmonic mean of recall and precision), we generated the table of precision, recall, and F-measure for each competency in Table 10. ML-model for MedMgmt competency achieved the best result with 0.89 in precision, 0.99 in recall, and 0.94 in F-measure. The next best competencies are SCC, HCE, and Falls with 0.86, 0.85, and 0.84 in F-measure, respectively. PC and CBD are two hard competency identification tasks because the best ML models can only achieve 0.69 and 0.62 in F-measure, respectively.
Table 10.
Results of precision, recall, and F-measure for the best model for each competency
| Precision | Recall | Fmeasure | |
|---|---|---|---|
| MedMgmt | 0.89 | 0.99 | 0.94 |
| CBD | 0.45 | 1.00 | 0.62 |
| Falls | 0.74 | 0.97 | 0.84 |
| PC | 0.60 | 0.82 | 0.69 |
| HCE | 0.77 | 0.95 | 0.85 |
| SCC | 0.76 | 0.99 | 0.86 |
5. Discussion
Medical student competency assessment, an ultimate goal of medical education, has typically been largely performed through standardized tests and the subjective assessments of clinical preceptors. This research represents an attempt to implement an objective assessment based on a complete capture of all clinical notes that students write. We chose geriatric competencies due to local interest and 2008 Institute of Medicine report on the importance of geriatrics for the aging US population.21 We applied NLP and ML methods to 6 of the 8 agreed-upon geriatric competencies as a novel, automated assessment of medical student’s competency that are poorly assessed currently, and when they are assessed, it is usually either via a survey or via manual effort by educators. We did not pursue the other two competencies (HCPP: Health care planning and promotion and APD: Atypical presentation of disease) for machine learning. Regarding APD, the physicians failed to resolve an operational definition and consistent rating between them – some Kappas between reviewers for these categories were <0, indicating the difficulty in defining educational competency even despite multiple face-to-face meetings. HCPP was highly unbalanced with respect to the ratio between positive and negative samples, with only 7% of the notes marked as irrelevant to the competency.
This is one of the first applications of NLP and machine learning methods to competency assessment. Based on this preliminary study, identifying the competency domain from the students’ clinical notes is a hard problem and requires a variety of features to achieve effective results. The best AUC score of 0.91 in assessing MedMgmt is a desired performance using KMCI CUI counts as feature set. Moreover, methods using NLP methods significantly outperformed “Bag of Words” approaches, whose performance often did not differ significantly from random chance. It is important to note, however, that all of these algorithms had portions of the precision curves in which the precision exceeded 0.8 and 0.9. Such algorithms, implemented with high thresholds, could still significantly enhance upon current methods since they could automatically scan the hundreds to thousands of notes written by clinical students.
We expected that using an expert-refined list of concepts that are highly associated with the competency would improve results, as has performed well in the past.13,18 In our experiments, the performances on the CBD, Falls, and HCE competencies improved with the refined list of concepts related to the corresponding competency. They are significantly better than the results generated by baseline “Bag of Words”. These results told a similar story to the recent I2B2/VA challenge, where most of the research teams used concepts and their related features to improve the text classification performance.22–26 For the MedMgmt, PC, and SCC competencies, however, the performance decreased with the refined list. This implied that our refined list of concepts for medication management might not be sufficient to cover the entire range of medication management, self-care capacity, and palliative care concepts, or we missed hidden relationships among these concepts that could help the detection. In addition, the best model for each competency domain could generate high recall/precision outcome if we adjust the threshold in figure 2.
Regarding the classifiers we used, Naïve Bayes, Logistic Regression, and linear SVM are all linear classifiers and could run very fast for training. We will try other complex models such as SVM with polynomial or Gaussian kernel27 and Random Forest28 to find better models in the future.
Our study has limitation in the following aspects. First, the sample sizes for machine learning are relatively small comparing to other similar NLP tasks. We found the annotation extremely challenging; annotators were spending 30 minutes or more per admission reviewing content for the 8 domains. With the size of training samples less than 150, building a model with high performance when considering multiple documents is extremely hard. Secondly, the quality of annotation result or gold standard in our study is less than perfect. Developing a rubric for relevancy of content was a source of considerable discussion between the physicians. Often, physicians would pick up on subtle hints of disease progression under different situations that would lend them to infer relevance of a given competency – such “hints” of relevance may be difficult for machine learning algorithms to assess. We implemented a majority vote scheme to decide the final label for each admission. There were several cases where the disagreement among raters is high (3 raters voted for high relevance, and another 3 raters voted for low relevance). We have not resolved these cases yet.
In the future, we may try competency detection models at an individual document level instead of a complete admission, as well as a larger annotated set. Machine learning approaches could be more powerful with a refined set of concept related features as well as non-controversial gold standard. Automatic feature section methods could help reduce the dimension of data and extract the most important concept codes with respect to the competency. Expert systems, coupling with machine learning approach, may improve the performance by incorporating the domain knowledge in medical education. Finally, we intend to extend our study to refine the models for all geriatric competency domains by constructing more reliable labels for the training data and test more types of NLP feature sets.
6. Conclusion
In this study, we used machine learning approaches to automate the assessment of geriatric competency for medical students using their clinical portfolios. Use of NLP to generate concept related feature sets as the input of machine learning based competency detectors improved performance. We found use of a physician-generated list of concepts to be our best performing feature set for 3 out of 6 competency assessment tasks. Our model could achieve optimal performance for MedMgmt, SCC, HCE, and Falls with high F-measures, and achieved high precision at some score threshold for all tested competencies.
Table 5.
AUC results for competency CBD over different feature sets
| Feature Sets | Num of Features | Naïve | LR | SVM |
|---|---|---|---|---|
| Words | 27406 | 0.61 | 0.62 | 0.62 |
| CUI | 10258 | 0.68 | 0.68 | 0.69 |
| CUI_SEC | 25947 | 0.65 | 0.60 | 0.61 |
| CUI_SEC + CUI_NEG | 27978 | 0.63 | 0.61 | 0.62 |
| CUI_SEC + CUI_NEG + STY | 28237 | 0.65 | 0.60 | 0.60 |
| CUI_count | 10576 | 0.66 | 0.68 | 0.68 |
| CUI_count_tfidf | 10572 | 0.64 | 0.57 | 0.66 |
| Refined CUI_count | 33 | 0.73 | 0.68 | 0.68 |
| Refined CUI_count+CUI_SEC+CUI_NEG +STY | 387 | 0.70 | 0.66 | 0.64 |
Table 6.
AUC results for competency Falls over different feature sets
| Feature Sets | Num of Features | Naïve | LR | SVM |
|---|---|---|---|---|
| Words | 27406 | 0.60 | 0.60 | 0.65 |
| CUI | 10258 | 0.64 | 0.59 | 0.63 |
| CUI_SEC | 25947 | 0.61 | 0.51 | 0.59 |
| CUI_SEC + CUI_NEG | 27978 | 0.59 | 0.50 | 0.61 |
| CUI_SEC + CUI_NEG + STY | 28237 | 0.60 | 0.52 | 0.61 |
| CUI_count | 10576 | 0.62 | 0.54 | 0.62 |
| CUI_count_tfidf | 10572 | 0.62 | 0.66 | 0.67 |
| Refined CUI_count | 52 | 0.65 | 0.50 | 0.57 |
| Refined CUI_count+CUI_SEC+CUI_NEG +STY | 469 | 0.75 | 0.56 | 0.64 |
Table 7.
AUC results for competency PC over different feature sets
| Feature Sets | Num of Features | Naïve | LR | SVM |
|---|---|---|---|---|
| Words | 25904 | 0.59 | 0.60 | 0.62 |
| CUI | 9924 | 0.65 | 0.75 | 0.76 |
| CUI_SEC | 24767 | 0.63 | 0.67 | 0.68 |
| CUI_SEC + CUI_NEG | 26707 | 0.60 | 0.65 | 0.66 |
| CUI_SEC + CUI_NEG + STY | 26958 | 0.59 | 0.64 | 0.65 |
| CUI_count | 10245 | 0.68 | 0.68 | 0.67 |
| CUI_count_tfidf | 10241 | 0.72 | 0.50 | 0.59 |
| Refined CUI_count | 35 | 0.58 | 0.58 | 0.60 |
| Refined CUI_count+CUI_SEC+CUI_NEG +STY | 174 | 0.60 | 0.51 | 0.53 |
Table 8.
AUC results for competency HCE over different feature sets
| Feature Sets | Num of Features | Naïve | LR | SVM |
|---|---|---|---|---|
| Words | 27406 | 0.58 | 0.62 | 0.64 |
| CUI | 10258 | 0.57 | 0.67 | 0.66 |
| CUI_SEC | 25947 | 0.65 | 0.66 | 0.69 |
| CUI_SEC + CUI_NEG | 27978 | 0.64 | 0.67 | 0.66 |
| CUI_SEC + CUI_NEG + STY | 28237 | 0.64 | 0.65 | 0.68 |
| CUI_count | 10576 | 0.62 | 0.55 | 0.55 |
| CUI_count_tfidf | 10572 | 0.61 | 0.65 | 0.64 |
| Refined CUI_count | 93 | 0.66 | 0.54 | 0.51 |
| Refined CUI_count+CUI_SEC+CUI_NEG +STY | 670 | 0.66 | 0.64 | 0.69 |
Acknowledgments
This study was funded under a grant from the Stemmler Foundation of the National Board of Medical Examiners.
Reference
- 1.Davis MH HR. Competency-based assessment: making it a reality. Medical teacher. 2003;25(6):565–568. doi: 10.1080/0142159032000153842. [DOI] [PubMed] [Google Scholar]
- 2.Whitcomb M. Redirecting the assessment of clinical competence. Acad Med. 2007;82(6):527–528. doi: 10.1097/ACM.0b013e31805556f8. [DOI] [PubMed] [Google Scholar]
- 3.Smith SR, Dollase RH, Boss JA. Assessing students’ performances in a competency-based curriculum. Academic Medicine. 2003 Jan;78(1):97–107. doi: 10.1097/00001888-200301000-00019. [DOI] [PubMed] [Google Scholar]
- 4.Dannefer EF, Henson LC. The portfolio approach to competency-based assessment at the Cleveland Clinic Lerner College of Medicine. Academic Medicine. 2007 May;82(5):493–502. doi: 10.1097/ACM.0b013e31803ead30. [DOI] [PubMed] [Google Scholar]
- 5.Litzelman DK, Cottingham AH. The new formal competency-based curriculum and informal curriculum at Indiana University School of Medicine: Overview and five-year analysis. Academic Medicine. 2007 Apr;82(4):410–421. doi: 10.1097/ACM.0b013e31803327f3. [DOI] [PubMed] [Google Scholar]
- 6.Langdorf MI, Montague BJ, Bearie B, Sobel CS. Quantification of procedures and resuscitations in an emergency medicine residency. J Emerg Med. 1998 Jan-Feb;16(1):121–127. doi: 10.1016/s0736-4679(97)00252-7. [DOI] [PubMed] [Google Scholar]
- 7.Rattner SL, Louis DZ, Rabinowitz C, et al. Documenting and medical students’ comparing clinical experiences. Jama-J Am Med Assoc. 2001 Sep 5;286(9):1035–1040. doi: 10.1001/jama.286.9.1035. [DOI] [PubMed] [Google Scholar]
- 8.Alderson TS, Oswald NT. Clinical experience of medical students in primary care: use of an electronic log in monitoring experience and in guiding education in the Cambridge Community Based Clinical Course. Med Educ. 1999 Jun;33(6):429–433. doi: 10.1046/j.1365-2923.1999.00336.x. [DOI] [PubMed] [Google Scholar]
- 9.Bird SB, Zarum RS, Renzi FP. Emergency medicine resident patient care documentation using a hand-held computerized device. Acad Emerg Med. 2001 Dec;8(12):1200–1203. doi: 10.1111/j.1553-2712.2001.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 10.Gordon JS, McNew R, Trangenstein P. The development of an online clinical log for advanced practice nursing students: a case study. Stud Health Technol Inform. 2007;129(Pt 2):1432–1436. [PubMed] [Google Scholar]
- 11.Sumner W, 2nd, Campbell J, Irving SC. Developing an educational reminder system for a handheld encounter log. Fam Med. 2006 Nov-Dec;38(10):736–741. [PubMed] [Google Scholar]
- 12.The Medical Student Competencies in Geriatric Medicine. [Accessed October 10, 2007]. at http://www.pogoe.org.
- 13.Denny JC, Bastarache L, Sastre EA, Spickard A. Tracking medical students’ clinical experiences using natural language processing. J Biomed Inform. 2009 Oct;42(5):781–789. doi: 10.1016/j.jbi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spickard A, 3rd, Ridinger H, Wrenn J, et al. Automatic scoring of medical students’ clinical notes to monitor learning in the workplace. Med Teach. 2014 Jan;36(1):68–72. doi: 10.3109/0142159X.2013.849801. [DOI] [PubMed] [Google Scholar]
- 15.Spickard A, 3rd, Gigante J, Stein G, Denny JC. Automatic capture of student notes to augment mentor feedback and student performance on patient write-ups. J Gen Intern Med. 2008 Jul;23(7):979–984. doi: 10.1007/s11606-008-0608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JC. Evaluation of a novel terminology to categorize clinical document section headers and a related clinical note section tagger. Nashville, TN: Vanderbilt University; 2007. [Google Scholar]
- 17.Denny JC, Miller RA, Johnson KB, Spickard A., 3rd Development and evaluation of a clinical note section header terminology. AMIA Annu Symp Proc. 2008:156–160. [PMC free article] [PubMed] [Google Scholar]
- 18.Denny JC, Smithers JD, Armstrong B, Spickard A. BRIEF REPORT: “Where do we teach what?” - Finding broad concepts in the medical school curriculum. J Gen Intern Med. 2005 Oct;20(10):943–946. doi: 10.1111/j.1525-1497.2005.0203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. http://clopinet.com/CLOP/
- 20.Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A Library for Large Linear Classification. J Mach Learn Res. 2008 Aug;9:1871–1874. [Google Scholar]
- 21.Report, Institute of Medicine of the National Academies. Apr 11, 2008. Retooling for an Aging America: Building the Health Care Workforce. [PubMed] [Google Scholar]
- 22.Torii M, Wagholikar K, Liu HF. Using machine learning for concept extraction on clinical documents from multiple data sources. J Am Med Inform Assn. 2011 Sep;18(5):580–587. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Avolio LW, Nguyen TM, Goryachev S, Fiore LD. Automated concept-level information extraction to reduce the need for custom software and rules development. J Am Med Inform Assn. 2011 Sep;18(5):607–613. doi: 10.1136/amiajnl-2011-000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minard AL, Ligozat AL, Ben Abacha A, et al. Hybrid methods for improving information access in clinical documents: concept, assertion, and relation identification. J Am Med Inform Assn. 2011 Sep;18(5):588–593. doi: 10.1136/amiajnl-2011-000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu XD. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. J Am Med Inform Assn. 2011 Sep;18(5):557–562. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang M, Chen YK, Liu M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assn. 2011 Sep;18(5):601–606. doi: 10.1136/amiajnl-2011-000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang C-CaL Chih-Jen. LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology. 2011;2(3):27:21–27:27. [Google Scholar]
- 28.Breiman L. RANDOM FORESTS. Machine Learning. 2001;45(1):5–32. [Google Scholar]