Abstract
Patient-generated health data (PGHD) offers a promising resource for shaping patient care, self-management, population health, and health policy. Although emerging technologies bolster opportunities to extract PGHD and profile the needs and experiences of patients, few efforts examine the validity and use of such profiles from the patient’s perspective. To address this gap, we explore health interest profiles built automatically from online community posts. Through a user evaluation with community members, we found that extracted profiles not only align with members’ stated health interests, but also expand upon those manually entered interests with little user effort. Community members express positive attitudes toward the use and expansion of profiles to connect with peers for support. Despite this promising approach, findings also point to improvements required of biomedical text processing tools to effectively process PGHD. Findings demonstrate opportunities to leverage the wealth of unstructured PGHD available in emerging technologies that patients regularly use.
Introduction
One of the most promising trends in health informatics is the rise of patient-generated health data (PGHD), ranging from patient-reported outcomes (PRO)1–2 and observations of daily living3 to quantified self4 and qualitative illness narratives5 collected outside of clinical care. Whereas traditional consumer health technologies focus largely on pushing resources to patients, emerging opportunities leverage existing PGHD for better care. Health information technology, including social media, provides a vital source of PGHD used as the basis of automated health profiling for targeted prevention6 and treatment.7–8 To promote social support in the context of online health communities, we leverage PGHD to automatically profile the health interests of online community members and then use those profiles to facilitate peer connections and support for cancer.9
Although PGHD has long been the prized treasure of online health communities,10 it has become recognized as a critical tool for improving clinical care and population health by complementing traditional forms of data collected in the clinic.1,11–12 For example, social support provided through narrative posts on online health communities promotes empowerment by improving psychological adjustment to cancer,13 increasing social wellbeing, and helping patients feel better informed.14 Electronic self-reported quality of life collected through structured PRO tools can reduce symptom distress and improve patient-provider communication.15–16 Patient illness collected through a self-report tool was found to identify respiratory illnesses with greater sensitivity than chief complaint data used by traditional disease surveillance systems.17 Growing patient engagement in health care highlights the important role that patient experience plays in policy, such as meaningful use criteria.11,18 In particular, the Office of the National Coordinator for Health Information Technology initiated several activities to advance the application of PGHD in clinical workflows, research and development, and policy.12
Emerging technology (e.g., social media, mobile devices, sensors) bolsters the opportunity for using PGHD to profile the needs and experiences of patients, making this an important area for research and policy.19 Thus, examining the validity of inferences extracted from PGHD is critical. Whereas progress has been made processing structured PGHD, such as PROs,2 opportunities remain to leverage the wealth of unstructured PGHD in social media and other technologies that patients regularly use. Several studies benchmark the validity of inferences drawn from PGHD against clinical comparisons.17,20 Yet few efforts examine the validity and use of health profiles extracted from PGHD from the patient’s perspective.
Using automated text processing, we extract health-related terms from online community posts to summarize individual members’ health-related interests.21 Our long-term goal is to use the resulting health interest profiles to connect members with shared interests for peer support.9 Our partnership with CancerConnect.com provides a unique opportunity to evaluate this approach with online community members. As a first step, we conducted a user study to evaluate individualized health interest profiles with users based on their posts. In this work, we address two key research questions:
RQ1. How closely do health interest profiles extracted from PGHD align with members’ stated interests?
RQ2. What are members’ preferences for using health interest profiles to connect with peers for support?
Profiling users from PGHD in online health communities
Growth in health-related use of social media,22 including online health communities, helps patients share experience and advice with peers (i.e., patient expertise).23 Many individuals now use these tools more often to exchange information and advice than to obtain emotional support.24 Yet, reading numerous posts to identify peers with shared interests takes time and effort. It can be difficult for community members to relate to the health experiences of other users25 and build relationships that support rich exchange of patient expertise.13
Profiles about users and their interests provide a key means for exploring potential relationships in online communities. Most online communities encourage users to create a profile by manually entering a few key details, such as diagnosis or treatment. Detailed user profiles are invaluable for summarizing the experience and expertise of available from peers,26 yet manual upkeep of detailed profiles takes time and energy away from managing a serious illness. We explore one possible solution that augments user profiles with details extracted automatically from community posts contributed by each user, such as treatments, tests, or other topics of interests.
Members of early online communities without user profiles were often limited to communicating their personal characteristics and interests through “signature line” descriptions at the end of message board posts. Today, user profiles are a fundamental component of modern social media that represent individual community members.27 By aggregating distinctive features that characterize a user, a user profile provides a synopsis, typically through a combination of manually entered elements (e.g., personal interests) and semi-automatically generated elements (e.g., number of followers or friends). From these elements, users form “thin slice” impressions when establishing online connections.28 Thus, user profiles help establish social context as conversation starters.29
In the health domain, some researchers enrich user profiles by dynamically leveraging PGHD. For example, Nuschke and colleagues30 designed a community-based diet and exercise journal that dynamically illustrates progress towards health goals and community participation on user profiles. Similarly, profiles on PatientLikeMe.com summarize historical trends in PGHD that users post about their experience with treatments, symptoms, outcomes, and community participation with dynamic icons.31 Temporal charts and graphs extend profiles to illustrate trends in these metrics over time. Although automatic extraction of PGHD can help users to build detailed profiles, this approach raises a number of questions about how machines might assist users–does automatic extraction produce more content than what users manually enter? How accurate is extracted content? Does automatic extraction capture interests and experiences that users do not otherwise enter?
In our work, we are enriching user profiles with health-related interests automatically extracted from a user’s community posts.21 The resulting health interest profile could efficiently summarize a user’s experience through the health terms they discuss in posts as their community participation evolves over time. One could imagine extending such profiles with additional personal characteristics that members wish to share when forging connections with community members, such as demographics, education, livelihood, or connection to cancer as a patient, survivor, caregiver, or other role.26 Despite the potential value of reducing the effort required for profile creation and maintenance, user perceptions about the accuracy and value of automated profile generation remain unknown.
Extracting Health Interest Profiles in CancerConnect Online Community
Within the context of CancerConnect (http://cancerconnect.com/), our partnering online health community, we examined the automatic extraction of individualized health interest profiles by processing the text of community members’ posts. CancerConnect is an award winning resource for web-based cancer resources that facilitates peer support for cancer through forum-style community posts. Health interests profiles provide the basis for our broader effort aimed at peer matching to recommend “mentors” with shared interests.9
We developed a text extraction approach to automatically generate health interest profiles from online community text.21 The profile is a vector of terms representing the health interests of an individual member based on all posts that user has contributed to the community. Our profile extraction pipeline includes MetaMap to support automatic extraction of health-related terms and sematic concepts that populate health interest profiles. MetaMap31 is a natural language processing tool designed to extract health-related terms from biomedical text that map to concepts in the Uniform Medical Language System (UMLS).33 The UMLS consists of more than 1.3 million concepts from over 100 biomedical vocabularies.34 Each concept in the UMLS is classified into one or more semantic types.35 Together, semantic types make up the UMLS Semantic Network that unifies the vocabularies within the UMLS, and thus, provide a means for grouping semantically similar terms. Because health terminology used by biomedical professionals can differ from the ways many patients express and think about health topics, researchers map patient-friendly terms to UMLS concepts in an effort to develop consumer health vocabularies (CHV).36 Recent efforts include computer assisted updates that leverage social network data from PatientsLikeMe.com to identify new terms for inclusion in CHV.37
To generate a health interest profile for an individual community member, we collect all of the posts that member contributes to the community. We then process those posts to automatically extract health-related terms, which we refer to as “health interests.” Since we wish to present those health interests to users, we were faced with a small dilemma. Do we present the user with an uncategorized set of extracted health interests? Or do we attempt to group health interests in some coherent way? We chose to group similar health interests using UMLS semantic types.35 There are a number of strategies for grouping UMLS semantic types, such as maximizing semantic coherence.38 Our aim was to present groups of terms relevant to the cancer experience that would be sensible to users who view health interest profiles. To do so, we considered a sample set of terms with their associated UMLS semantic types and created our own five categories that group similar semantic types (Table 1). Through this process, we populate a user’s health interest profile with the health interests we extracted across the five categories (see example in Figure 1). This categorization enabled us to generate individualzed profiles that present information about uers in logical chunks, similar to user profiles they might find in any other online community.
Table 1.
Health interest profile categories and associated UMLS semantic types
| Category | UMLS Semantic type |
|---|---|
| Health problems | Neoplastic Process Disease or Syndrome Acquired Abnormality Injury or Poisoning Anatomical Abnormality Finding Sign or Symptom Pathologic Function Clinical Attribute Laboratory or Test Result |
| Treatments | Antibiotic Biomedical or Dental Material Medical Device Pharmacologic Substance Therapeutic or Preventive Procedure Hormone Vitamin |
| Diagnostics & tests | Diagnostic Procedure Research Activity Laboratory Procedure |
| Provider care | Health Care Activity |
| Genetics | Gene or Genome |
Figure 1.
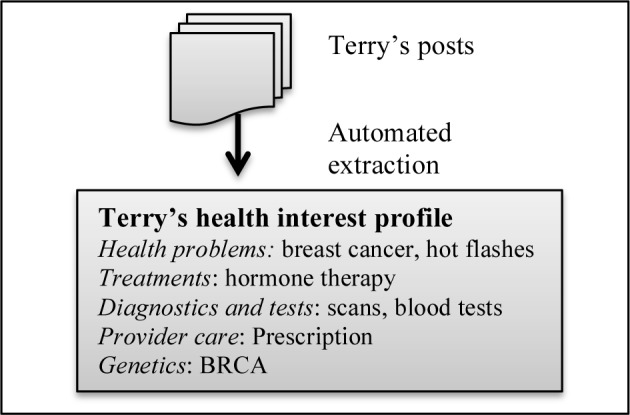
Example health interest profile populated with health interests extracted from community posts contributed by fictitious member “Terry”
We evaluate these individualized health interest profiles through a user study with community members. Our CancerConnect partnership enabled us to examine both the accuracy and perceived value of health interest profiles. We could generate health interest profiles for individual community members, and then ask those members to evaluate their own individualized profile. We were particularly interested in evaluating the accuracy of our automatically extracted health interest profiles compared to the stated health interests that community members could provide directly through manual entry (RQ1), as well as examining community members’ perceived value of using those profiles to connect with other members for peer support (RQ2).
Methods
We conducted a web-based user study by recruiting members of CancerConnect to answer our two key research questions about the accuracy and perceived value of extracting health interest profiles from PGHD in online health communities. Before the study, we processed the text from all posts each community member contributed to the CancerConnect community. Using the extracted terms, we constructed individualized health interest profiles that summarize health-related issues each member discussed in their posts across our five categories (Table 1). To be eligible for participation, members were required to have posted sufficient text to the CancerConnect online community over the past six months to extract a health interest profile with at least ten unique terms.
We used the individualized health interest profiles with participants during the two-part user study. To examine the alignment of health interests extracted from members’ posts with members’ stated health interests (RQ1), participants first completed a set of recall and recognition tasks during which they manually entered health issues of personal interest. Then, participants completed a set of preference ratings to describe their perceptions about using health profiles to disclose their personal interests and characteristics to other community members (i.e., “peers”), to learn about the interests and characteristics of peers in the community, and to be matched and interact with peers with shared interests and characteristics (RQ2). At the end of the study we collected participant demographics. IRB approval was granted from the University of Washington Human Subjects Division. We configured and administered the user study using Lime Survey (http://www.limesurvey.org/en/).
Part 1. Recall and recognition tasks
Each participant completed a sequential set of recall and recognition tasks to assess the accuracy of their individualized health interest profile. The participant first completed a free recall task in which they were asked to enter terms or phrases that describe the health issues they discuss within the community for as many of the five categories as they wished (i.e., health problems, treatments, diagnostics and tests, provider care, and genetics). Next in the recognition task, we primed the participant by showing the extracted health interests from their profile across the five categories. We asked the participant to add or remove terms and phrases until they were satisfied. Terms and phrases were shown as tags that could be easily added or clicked on to remove (Figure 2).
Figure 2.

Example of recognition task to add or remove health interests for category “health problems”
At the conclusion of the recall and recognition tasks, we obtained three lists of health interests for each participant: (1) extracted terms making up the participant’s health interest profile that we automatically generated, (2) recalled terms that the participant entered from memory during the recall task, and (3) recognized terms that resulted after the participant added and removed terms from their automatically generated health interest profile during the recognition task. We assessed the alignment of those lists of health interests using Wilcoxon sign rank to compare the number of terms between lists, as well as Jaccard index, precision, and recall to compare term similarity between lists. In particular, we examined the alignment between extracted terms from the health interest profile with the member’s stated health interests through recall and recognition tasks (RQ1). This analysis enables us to examine how close a set of extracted terms can come to an acceptable set of profile terms. It also allows us to consider the similarity/differences between the health interests we can extract and the health interests users choose to enter.
Part 2. Preference ratings
Following the recall and recognition tasks, the participant provided structured feedback by rating their preference across a range of potential uses of their personal health profile (RQ2). We were particularly interested the perceived value of using profiles to share health interests with community members, as well as expanding profiles with other personal characteristics. Guided by our prior work,25 we chose 10 personal characteristics which that could be used as the basis for peer matching and interaction including: all health interests, only common health interests, personality, participation level, demographics, education and livelihood, geographic location, common users followed, common groups followed, and common posts responded to. Preference ratings were made on a 5-point Likert scale (1=“not at all” to 5=“very”) and covered four main areas: (1) comfort in disclosing personal interests and characteristics to other members of the online community on the profile, (2) interest in viewing personal interests and characteristics of other community members on their profiles, (3) importance of matching characteristics sought when connecting with online community members (e.g., someone with the same diagnosis or similar age), and (4) interest in interaction styles for connecting with online community members. Interaction styles included four types: (1) “one shot” anonymous interaction that is one-to-one, short-term, and impersonal, (2) one-to-one interaction that is personal, confidential, and sustained over time with a “buddy,” (3) group interaction that occurs regularly among individuals with a shared issue modeled after a “support group,” and (4) group interaction that occurs as needed and focused on a topic of interest in a “group campaign.”
We report preferences using descriptive statistics and comparisons based on Friedman chi square (X2) and Wilcoxon signed rank (V) tests. We also offered participants the option to provide open-ended responses for concerns and suggestions regarding the use of health profiles to connect with community members, which we grouped qualitatively for emergent themes. This analysis enabled us to examine participants’ attitudes regarding a range of potential uses of health profiles for sharing their health interests as well as broader personal characteristics for connecting with peers in the online community.
Results
Participants
A total of 34 CancerConnect members participated in the study with one participant completing only the recall and recognition tasks in part 1. The remaining participants ranged in age from 31 to 76 and are mostly female and white (Table 2). Table 3 shows participants’ online community participation including average weeks worth of posts, posts per week, words per post, and extracted health interests per week.
Table 2.
Demographics of participants
| Age mean(sd) range | 55(11) 31–76 |
| Sex | 85% Female 9% Male 6% na |
| Education | 9% High school graduate 31% Some college 33% College graduate 24% Post graduate 3% na |
| Race/ethnicity | 94% white 6% na |
| Social network size | 24% Extensive 49% Moderate 18% Small 9% na |
| Geographic location | 22% Western United States 15% Midwestern United States 27% South United States 27% Northeastern United States 9% na |
| Top personal interests/hobbies | Reading, exercise, cooking, gardening, television/movies |
Table 3.
Online community participation
| Mean (sd) | Range | |
|---|---|---|
| Weeks worth of posts | 32 (38.4) | 0.1 – 129 |
| Posts/week % Initiating posts % Replies |
3 (4.5) 20% 80% |
0.1 – 14 0 – 100% 0 – 100% |
| Mean words/post | 105(55.0) | 36 – 227 |
| Mean extracted terms/week | 13 (137) | 0.1 – 49 |
Recall and recognition: Profile validation
We report on alignment in the number and similarly of health interest terms we automatically extracted, terms participants freely recalled, and terms resulting after the participant completed the recognition task.
The number of terms that resulted following automated extraction, the recall task, and the recognition task are shown in Table 4. Compared to recalled terms, we extracted significantly more health problems (V=450, p=0.001) and treatments (V=444, p=0.004). Participants entered more terms than we extracted for diagnostics and tests, but that difference was not significant. Compared to what we automatically extracted, participants entered significantly more genetic terms and provider care terms (V=109, p=0.004). Automated extraction was limited for those two categories–we extracted provider care terms for 19 participants and extracted genetics terms for only three.
Table 4.
Number of terms that resulted following the text extraction, recall task, and recognition task
| Extracted terms | Recalled terms | Recognized terms | ||||
|---|---|---|---|---|---|---|
| mean (sd) | range | mean (sd) | range | mean (sd) | range | |
| Health problems | 13.15 (7.16) | 3 – 26 | 6.94 (5.23) | 1 – 23 | 11.74 (6.18) | 2 – 26 |
|
| ||||||
| Treatments | 9.79 (7.80) | 0 – 25 | 5.09 (3.82) | 0 – 20 | 9.88 (7.41) | 0 – 24 |
|
| ||||||
| Diagnostics and tests | 3.38 (3.23) | 0 – 13 | 4.65 (4.85) | 0 – 25 | 4.38 (3.61) | 0 – 17 |
|
| ||||||
| Provider care | 1.82 (2.96) | 0 – 13 | 4.15 (3.86) | 0 – 17 | 3.00 (2.83) | 0 – 12 |
|
| ||||||
| Genetics | 0.09 (0.29) | 0 – 1 | 1.71 (1.73) | 0 – 6 | 0.82 (1.14) | 0 – 4 |
In contrast, the difference in number of terms that resulted following automated extraction and the recognition task was much less striking. Participants removed an average of 2.4 health problems from the health interest profile, resulting in significantly fewer terms on the resulting recognized term list than we extracted (V=205, p=0.05). With the exception of treatments, participants added terms for remaining categories, resulting in more terms on the resulting recognized term list than we extracted for diagnostics and tests (V=21, p=0.02), provider care (V=34, p=0.008), and genetics (V=0, p=0.001). Table 5 shows numbers and examples of added and removed terms.
Table 5.
Terms added and removed during recognition task
| Terms added | Examples of added terms | Terms Removed | Examples of removed terms | |||
|---|---|---|---|---|---|---|
| mean (+sd) | range | mean (sd) | range | |||
| Health Problems | 1.0 (1.8) | 0 – 7 | no side effects | 2.4 (2.8) | 0 – 10 | alone, pain, HIV |
|
| ||||||
| Treatments | 0.9 (2.0) | 0 – 9 | folfiri 5fu, nutrition | 0.9 (1.2) | 0 – 4 | oxygen, procedure |
|
| ||||||
| Diagnostics and tests | 1.4 (2.1) | 0 – 7 | ct scan, mri, blood test | 0.4 (0.8) | 0 – 3 | hgb, research, color |
|
| ||||||
| Provider care | 1.4 (2.1) | 0 – 7 | 2nd opinion, exams | 0.3 (0.6) | 0 – 3 | report, documented |
|
| ||||||
| Genetics | 0.7 (1.1) | 0 – 4 | brca, family history | 0.0 (0.0) | 0 – 0 | (none) |
Findings on alignment of the number of extracted, recalled, and recognized terms demonstrate that automatic extraction can help users populate their profiles. This machine assistance was most effective for categories in which we extracted more terms (i.e., health problems and treatments). When given the opportunity, participants removed extracted terms they found inappropriate. Some removed health problems, such as “alone” and “clarity”, point to limitations of extracting terms with MetaMap that do not seem health-related from the perspective of participants. Other removed terms were more clearly health-related, such as “pain” and “HIV,” but participants chose not to keep them on their health interest profile. Categories with fewer extracted terms (i.e., provider care, genetics) required participants to add terms during the recognition task because extraction alone did not sufficiently populate their profile. Health problems that participants added, such as “no side effects”, might be impossible to extract unless the user explicitly stated in a post. Thus the size of our categories varied and this had impact on our approach. These findings suggest that machines can help augment profiles through automated extraction, but that users should be provided the opportunity to edit extracted data. These findings also illustrate limitations of applying biomedical text processing tools to unstructured PGHD.
Term similarity between extracted terms and recognized terms was substantially higher than between extracted terms and recalled terms across all 5 categories of health interests (Table 6). With the exception of genetics, where terms were extracted for only 3 participants, overlap between extracted terms was recognized terms was substantial with Jiccard indices ranging from 0.48 to 0.79. When recognized terms served as the gold standard, the precision and recall of extracted terms was also high with worsening performance as categories become sparser moving from provider care to genetics. When we used recalled terms as the gold standard, overlap with extracted terms was much lower, including low precision and recall across all categories.
Table 6.
Similarity between extracted terms and recognized terms, recalled terms
| Extracted (test) vs. Recalled (gold standard) | Extracted (test) vs. Recognized (gold standard) | |||
|---|---|---|---|---|
| mean (sd) | range | mean (sd) | range | |
| Health problems | ||||
| Jaccard index | 0.04 (0.04) | 0.00 – 0.17 | 0.75 (0.20) | 0.36 – 1.00 |
| Precision | 0.05 (0.06) | 0.00 – 0.20 | 0.82 (0.19) | 0.36 – 1.00 |
| Recall | 0.16 (0.22) | 0.00 – 1.00 | 0.91 (0.16) | 0.44 – 1.00 |
|
| ||||
| Treatments | ||||
| Jaccard index | 0.08 (0.13) | 0.00 – 0.60 | 0.79 (0.25) | 0.00 – 1.00 |
| Precision | 0.13 (0.17) | 0.00 – 0.75 | 0.89 (0.18) | 0.40 – 1.00 |
| Recall | 0.26 (0.31) | 0.00 – 1.00 | 0.89 (0.23) | 0.00 – 1.00 |
|
| ||||
| Diagnostics & tests | ||||
| Jaccard index | 0.03 (0.06) | 0.00 – 0.20 | 0.61 (0.42) | 0.00 – 1.00 |
| Precision | 0.08 (0.20) | 0.00 – 1.00 | 0.80 (0.35) | 0.00 – 1.00 |
| Recall | 0.04 (0.09) | 0.00 – 0.33 | 0.65 (0.42) | 0.00 – 1.00 |
|
| ||||
| Provider care | ||||
| Jaccard index | 0.01 (0.03) | 0.00 – 0.14 | 0.48 (0.44) | 0.00 – 1.00 |
| Precision | 0.04 (0.13) | 0.00 – 0.50 | 0.83 (0.28) | 0.00 – 1.00 |
| Recall | 0.01 (0.04) | 0.00 – 0.20 | 0.55 (0.48) | 0.00 – 1.00 |
|
| ||||
| Genetics | ||||
| Jaccard index | 0.00 (0.0) | 0.00 – 0.00 | 0.17 (0.36) | 0.00 – 1.00 |
| Precision | 0.00 (0.00) | 0.00 – 0.00 | 1.00 (0.00) | 0.00 – 1.00 |
| Recall | 0.00 (0.00) | 0.00 – 0.00 | 0.17 (0.36) | 0.00 – 1.00 |
Findings on term similarity further support our claim that participants appear largely satisfied with the accuracy of individualized health interest profiles we extracted and thus made few changes. Users are beginning to discuss emergent topics, such as genetics, which may be challenging to extract using tools like MetaMap. Such categories may be small with few terms, but that does not mean they are unimportant and users should be solicited to input terms. Further, lack of overlap between extracted terms and recalled terms suggests that users and machines might contribute different kinds of health interests to profiles. To further investigate this possibility, we compared the similarity between recalled terms and the terms participants added during the recognition task (Table 7). The small term overlap and low precision and recall suggest that when participants are prompted with extracted terms, they add different kinds of terms than those they freely recall. This finding holds despite the lack of a washout period between sequential recall and recognition tasks. Our findings suggest a valuable role for machines to assist users not only in augmenting profiles with extracted terms that they are unlikely to recall and enter manually—by showing users the extracted terms, machines can also remind users of additional terms they are unlikely to recall on their own.
Table 7.
Similarity between recalled terms & added terms
| Added terms (test) vs. Recalled terms (gold stand.) | ||
|---|---|---|
| mean (sd) | range | |
| Health problems | ||
| Jaccard index | 0.03 (0.13) | 0.00–0.67 |
| Precision | 0.03 (0.13) | 0.00–0.67 |
| Recall | 0.15 (0.33) | 0.00–1.00 |
|
| ||
| Treatments | ||
| Jaccard index | 0.05 (0.16) | 0.00–0.78 |
| Precision | 0.07 (0.24) | 0.00–1.00 |
| Recall | 0.27 (0.38) | 0.00–1.00 |
|
| ||
| Diagnostics & tests | ||
| Jaccard index | 0.15 (0.29) | 0.00–1.00 |
| Precision | 0.17 (0.31) | 0.00–1.00 |
| Recall | 0.48 (0.45) | 0.00–1.00 |
|
| ||
| Provider care | ||
| Jaccard index | 0.03 (0.07) | 0.00–0.33 |
| Precision | 0.07 (0.19) | 0.00–1.00 |
| Recall | 0.15 (0.17) | 0.00–0.50 |
|
| ||
| Genetics | ||
| Jaccard index | 0.21 (0.36) | 0.00–1.00 |
| Precision | 0.22 (0.36) | 0.00–1.00 |
| Recall | 0.47 (0.49) | 0.00–1.00 |
Preference ratings: Attitudes toward profile use
Self-disclosure
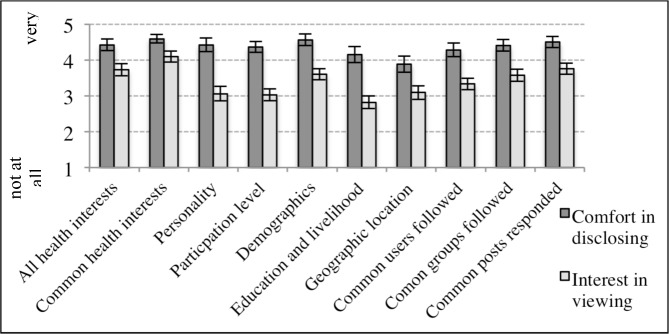
On average, participants expressed comfort in disclosing 10 types of personal interests and characteristics by publicly displaying them on their profile (Figure 3), with the greatest mean comfort expressed for disclosing common health interests and the least mean comfort disclosing geographic location. For participants without missing data (n=30), the difference in comfort level among types of personal characteristics was significant (X2 = 19.7, p=0.02). Pairwise comparison between the highest level of comfort disclosing common health interests and lowest level of comfort disclosing geographic location shows a significant difference (V=93, p=0.005). Most participants (25/33) expressed no concerns about disclosing personal characteristics to other community members. When asked about concerns, 11 left their open-ended response blank and 14 responded with an explicit statement (e.g., no concerns). Concerns expressed by the remaining 8 participants included revealing personal information that could be used in unapproved ways (e.g., targeted advertising) (4/33), desire to choose with whom to share personal information (1/33), desire to keep personal information private (1/33), and creating stress (1/33) or embarrassment (1/33).
Figure 3.
Comfort in disclosing vs. Interest in viewing personal characteristics
Viewing preferences
On average, participants expressed interest in viewing the 10 personal characteristics of other members on user profiles (Figure 3), but this interest level varied significantly among the 10 types (X2=62 p<0.001). Participants expressed the greatest interest in viewing common health interests and the least interest in viewing education and livelihood. Pairwise comparison between the highest interest in viewing common health interests and lowest interest in viewing education and livelihood location shows a significant difference (V=210 p<0.001). Interest was significantly higher for viewing common health interests than any other personal characteristic except common posts responded to. Interest in viewing education and livelihood was significantly lower than other characteristics except personality, participation level, and geographic location. When compared to ratings for comfort disclosing personal characteristics, participants expressed greater comfort disclosing than interest in viewing all personal characteristics (Figure 3).
Matching preferences
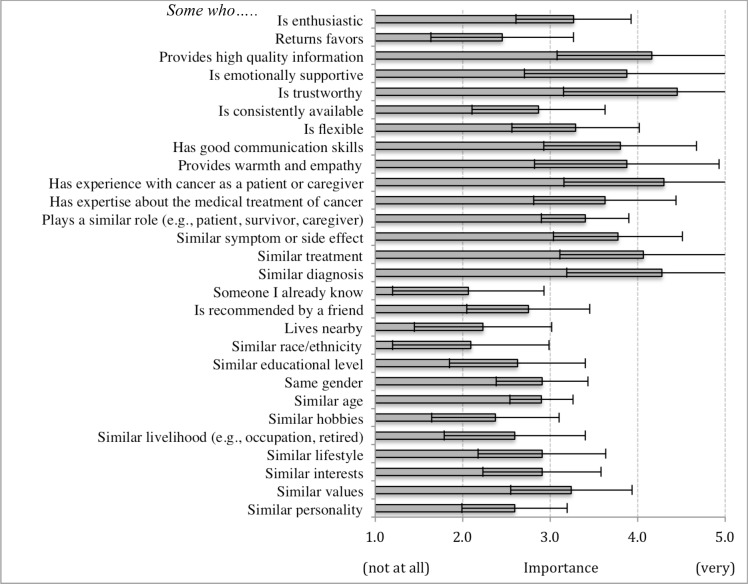
When considering connecting with an online peer, participants rated the importance of a range of matching characteristics (e.g., someone who…). Figure 4 shows that on average participants rated some matching characteristics more important than others. For participants without missing data (n=19), this difference was significant (X2=27, p<0.001). Matching characteristics rated highest include someone who is trustworthy, has experience with cancer as a patient or caregiver, and has a similar diagnosis. Characteristics rated least important include someone who lives nearby, who I already know, and who is of similar race/ethnicity. Pairwise comparisons between highest and lowest rated characteristics show significant differences at or below the 0.001 level.
Figure 4.
Mean importance of matching characteristics
Interaction preferences
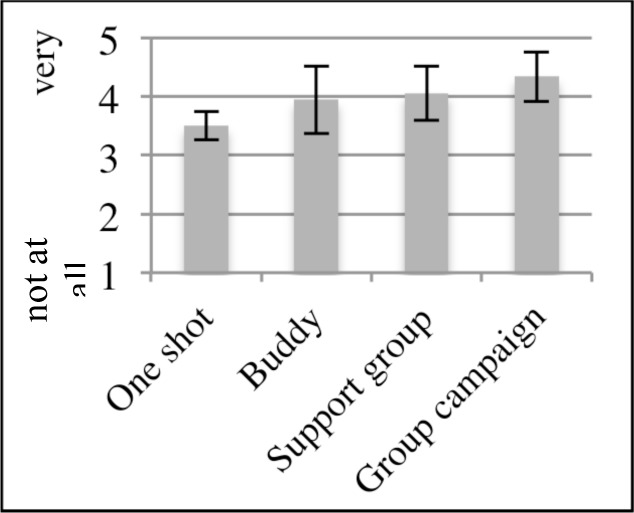
On average, participants expressed interest in interacting with members across a range of styles (Figure 5). There was a significant difference in interest level among the four interaction styles (X2=14, p<0.003). Pairwise comparisons show significantly less interests in “one shot” style, than other styles, including “buddy” style, “support group” style, and “group campaign” style. When asked about additional ways they wished to interact with online peers, participants reported email (33%), phone (25%), around specific topics (17%), through social media (17%), such as Pinterest or Blogger, and in disease-specific groups (8%).
Figure 5.
Mean interest level in interaction styles
Discussion and Conclusion
Substantial opportunities exist to leverage the wealth of unstructured PGHD available in emerging technologies that patients regularly use, yet few efforts examine the validity and use of health profiles extracted from PGHD from the patient’s perspective. Although findings point to the perceived value of health interest profiles, we will evaluate their actual value in connecting peers in our future work. Findings from our user evaluation of health interest profiles extracted from online community posts demonstrates value in augmenting detailed health profiles through text extraction applied to unstructured PGHD in multiple ways.
First, health interest profiles not only align closely with members’ stated health interests, but expand upon those interests with little user effort. Automated extraction can populate profiles with content that community members accepted—when given the opportunity to add or remove health interests, few changes were made. Further, extracted content appears to overlap little with the content that members manually enter. This finding suggests that automated text extraction captures new and different kinds of interests and experiences than community members generally recall. In addition, extracted content appears to not only encourage members to refine their profile through manual removal of unsuitable content, but also act as a prompt for users to enter additional content they might not otherwise consider. Extraction is necessarily limited to the text users choose to post, which makes manual profile entry an important option for users. Thus machines can assist, but not necessarily replace, the user when processing PGHD.
Second, our findings illustrate positive attitudes of community members toward the use and expansion of health interest profiles with additional personal characteristics to connect with peers for support (e.g., common health interests). Community members expressed comfort in disclosing a number of personal characteristics to community members. They also expressed interest in viewing those characteristics of others. These findings provide support for expanding our automated extraction of detailed user profiles with additional characteristics of interest that can be used to facilitate peer matching and interaction. Important matching characteristics (i.e., someone who is trustworthy, has cancer experience, and has a similar diagnosis) and preferred interaction styles provide insight for future work in which we will use those profiles to match and help connect users for peer support. Although individuals with similar personal characteristics are likely to be attracted to each other (i.e., “birds of a feather flock together”), there may be merit in exploring “difference matching” (e.g., “opposites attract”).
Although our findings provide new insight into the value of machine-assisted processing of PGHD, our text extraction was most effective when sufficient content, but not inappropriate content, was extracted. Finding this sweet spot poses a challenge. Whereas users can edit out irrelevant terms extracted by an overzealous machine, the inability of existing text extraction tools to capture important content is problematic. Topics such as provider care and genetics are clearly important to patients, but represent gaps in MetaMap and UMLS. Despite improvements in mapping UMLS to consumer-oriented terms,36–37 effectively processing PGHD requires enhancements.
Emerging trends in PGHD present significant promise for shaping health care, self-management, population health, and policy. Our findings offer insights that speak to the value of processing PGHD from the patient’s perspective. In particular, we illustrate one promising approach to leverage PGHD in the context of online communities. Substantial opportunities exist for capturing a wealth of unstructured PGHD available in emerging technologies that patients regularly use. As patient engagement in health grows and our desire to capture PGHD intensifies, it is critical that we prioritize development of technologies that can effectively process this unique and valuable resource.
Acknowledgments
We thank participants as well as Corey Shaffer, Alana Brody, and Meg Monday for community data and recruitment. This research was supported by National Science Foundation Smart Health & Wellbeing Award #1117187.
References
- 1.Wu AW. Advances in the use of patient reported outcome measures in electronic health records. Nov, 2013. Retrieved Mar 3 2014 from: www.pcori.org/assets/2013/11/PCORI-PRO-Workshop-EHR-Landscape-Review-111913.pdf.
- 2.Cella D, Riley W, Stone A, Rothrock N, Reeve B, Yount S, et al. The Patient-Reported Outcomes Measurement Information System (PROMIS) developed and tested its first wave of adult self-reported health outcome item banks: 2005–2008. J Clin Epidemiol. 2010 Nov;63(11):1179–94. doi: 10.1016/j.jclinepi.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backonja U, Kim K, Casper GR, Patton T, Ramly E, Brennan PF. Observations of daily living: putting the “personal” in personal health records. 2012 Jun;2012:6. eCollection. [PMC free article] [PubMed] [Google Scholar]
- 4.Choe EK, Lee NB, Lee B, Pratt W, Kietnz JA. Understanding quantified-selfers’ practices in collecting and exploring personal data. Proc. SIGCHI Conference on Human Factors in Computing Systems (CHI ’14); pp. 1143–1152. [Google Scholar]
- 5.Sharf BF, Vanderford ML. Illness narratives and the social construction of health. In: Thompson TL, Dorsey A, Parrott R, Mille K, editors. Handbook of health communication. Francis & Taylor e-library; 2008. pp. 9–34. [Google Scholar]
- 6.Swan M. Health 2050: the realization of personalized medicine through crowdsourcing, the Quantified Self, and the participatory biocitizen. J Pers Med. 2012;2(3):93–118. doi: 10.3390/jpm2030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wicks P, Vaughan TE, Massagli MP, Heywood J. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat Biotechnol. 2011 May;29(5):411–414. doi: 10.1038/nbt.1837. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura C, Bromberg M, Bhargava S, Wicks P, Zeng-Treitler Q. Mining online social network data for biomedical research: a comparison of clinicians’ and patients’ perceptions about amyotrophic lateral sclerosis treatments. JMIR. 2012;14(3):e90. doi: 10.2196/jmir.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartzler A, McDonald D, Park A, Huh J, Pratt W. Mentor matching in peer health communities. Proc AMIA; 2012; p. 1764. [Google Scholar]
- 10.Eysenbach G, Powell J, Englesakis M, Rizo C, Stern A. Health related virtual communities and electronic support groups: systematic review of the effects of online peer to peer interactions. BMJ. 2004;328:1166. doi: 10.1136/bmj.328.7449.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro M, Johnston D, Wald J, Mon D. Patient-generated health data: White paper prepared for the Office of the National Coordinator for Health IT by RTI International. Apr, 2012. Retreived Mar 3 2014 from: www.rti.org/pubs/patientgeneratedhealthdata.pdf.
- 12.Deering MJ. ONC Issue brief: Patient-generated health data and health IT. Office of the National Coordinator for Health Information Technology. Retrieved Mar 3 2014 from: www.healthit.gov/sites/default/files/pghd_brief_final122013.pdf.
- 13.Hoey LM, Ieropoli SC, White VM, Jefford M. Systematic review of peer-support programs for people with cancer. Patient Educ Couns. 2008;70(3):315–37. doi: 10.1016/j.pec.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 14.Van Uden-Kraan CF, Drossaert CH, Taal E, Seydel ER, van de Laar MA. Participation in online patient support groups endorses patients’ empowerment. Patient Educ Couns. 2009;74(1):61–69. doi: 10.1016/j.pec.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 15.Berry DL, Blumenstein BA, Halpenny B, Wolpin S, Fann JR, Austin-Seymour M, et al. Enhancing patient-provider communication with the electronic self-report assessment for cancer: A randomized trial. J Clin Oncol. 2011;29(8):1029–35. doi: 10.1200/JCO.2010.30.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry DL, Hong F, Halpenny B, Patridge AH, Fann JR, et al. Electronic self-report assessment for cancer and self-care support: Results of a multicenter randomized trial. J Clin Oncol. 2014;32(3):199–205. doi: 10.1200/JCO.2013.48.6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourgeois FT, Porter SC, Valim C, Jackson T, Cook EF, Mandl KD, et al. The value of patient self-report for disease surveillance. J Am Med Informatics Assoc. 2007;14(6):765–771. doi: 10.1197/jamia.M2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ralston JD, Coleman K, Reid Robert J, Handley MR, Larson EB. Patient experience should be part of meaningful-use criteria. Health Affairs. 2010;29(4):607–613. doi: 10.1377/hlthaff.2010.0113. [DOI] [PubMed] [Google Scholar]
- 19.Frost J, Okun S, Vaughan T, Heywood J, Wicks P. Patient-reported outcomes as a source of evidence in off-label prescribing: Analysis of data from PatientsLikeMe. J Med Internet Res. 2011;13(1):e6. doi: 10.2196/jmir.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bove R, Secor E, Healy BC, Musallam A, Vaughan T, Glanz BI, et al. Evaluation of an online platform for multiple sclerosis research: Patient description, validation of severity scale, and exploration of BMI effects on disease course. PloS one. 2013;8(3):e59707. doi: 10.1371/journal.pone.0059707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park A, Hartzler A, Huh J, McDonald D, Pratt W. Extracting everyday health interests from online communities. Proc. AMIA; 2012; p. 1889. [Google Scholar]
- 22.Fox S, Jones S. The social life of health information. Pew Internet & American Life (2009) Retreived Mar 3 2014 from: http://www.pewinternet.org/Reports/2009/8-The-Social-Life-of-Health-Information.aspx.
- 23.Hartzler A, Pratt W. Managing the personal side of health: How patient expertise differs from the expertise of clinicians. J Med Internet Res. 2011;13(3):e62. doi: 10.2196/jmir.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarasohn-Kahn J. The wisdom of patients: Health care meets online social media. California HealthCare Foundation; 2008. Retireveid Mar 3 2014 from: http://www.chcf.org/documents/chronicdisease/HealthCareSocialMedia.pdf. [Google Scholar]
- 25.Rimer BK, Lyons EJ, Ribisl KM, Bowling JM, Golin CE, Forlenza MJ, Meier A. How new subscribers use cancer-related online mailing lists. J Med Internet Res. 2005;7(3):e32. doi: 10.2196/jmir.7.3.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Civan-Hartzler A, McDonald D, Powell C, Skeels M, Mukai M, Pratt W. Bringing the field into focus: User-centered design of a patient expertise locator. Proc. Conference on Hum Fac in Comput Systems (CHI’10); 2010; pp. 1675–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.boyd dm, Ellison NB. Social networking sites: Definition, history, and scholarship. J Computer-Mediated Communication. 2007;13:210–230. [Google Scholar]
- 28.Stecher K, Counts S. Thin slices of online profile attributes. Proc. ICWSM’08; 2008. [Google Scholar]
- 29.boyd dm, Heer J. Profiles as conversation: Networked identity performance on Friendster. Proc. HICSS-39; 2006; p. 59c. [Google Scholar]
- 30.Nuschke P, Holmes T, Qadah Y. My health, my life: a web-based health monitoring application. Extended Abstracts on Human Factors in Computing Systems(CHI ’06); 2006; pp. 1861–1866. [Google Scholar]
- 31.Frost J, Massagli M. Social uses of personal health information within PatientsLikeMe, an online patient community: What can happen when patients have access to one another’s data. J Med Internet Res. 2008;10(3):e15. doi: 10.2196/jmir.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aronson AR, Lang F-M. An overview of MetaMap: Historical perspective and recent advances. JAMIA. 2010;17:229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphreys B, Lindberg D, Schoolman H. The Unified Medical Language System: an informatics research collaboration. J Am Med Informatics Assoc. 1998;5(1):1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Perl Y, Geller J, Cimino JJ. Analysis of a study of the users, uses, and future agenda of the UMLS. J Am Med Informatics Assoc. 2007;14:221–31. doi: 10.1197/jamia.M2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCray AT, Nelson SJ. The representation of meaning in the UMLS. Methods Inf Med. 1995;34:193–201. [PubMed] [Google Scholar]
- 36.Zeng QT, Tse T. Exploring and Developing consumer health vocabularies. J Am Med Informatics Assoc. 2006;13:24–30. doi: 10.1197/jamia.M1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doing-Harris KM, Zeng-Treitler Q. Computer-assisted update of a consumer health vocabulary through mining of social network data. J Med Internet Res. 2011 May 17;13(2):e37. doi: 10.2196/jmir.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. JBI. 2003;36(6):414–432. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]