Abstract
Infobuttons have proven to be an increasingly important resource in providing a standardized approach to integrating useful educational materials at the point of care in electronic health records (EHRs). They provide a simple, uniform pathway for both patients and providers to receive pertinent education materials in a quick fashion from within EHRs and Personalized Health Records (PHRs). In recent years, the international standards organization Health Level Seven has balloted and approved a standards-based pathway for requesting and receiving data for infobuttons, simplifying some of the barriers for their adoption in electronic medical records and amongst content providers. Local content, developed by the hosting organization themselves, still needs to be indexed and annotated with appropriate metadata and terminologies in order to be fully accessible via the infobutton. In this manuscript we present an approach for automating the annotation of internally-developed patient education sheets with standardized terminologies and compare and contrast the approach with manual approaches used previously. We anticipate that a combination of system-generated and human reviewed annotations will provide the most comprehensive and effective indexing strategy, thereby allowing best access to internally-created content via the infobutton.
Introduction
Information needs and infobuttons
Information gaps at the point-of-care have been well documented in the medical literature, and they pose a real risk in ensuring that current best practice is reflected the care that patients receive1,2. Several approaches to addressing these information needs have been presented in the informatics literature3,4. One of the more notable means for addressing this issue is the infobutton, a context-aware linking resources that points users to relevant clinical reference materials at the point of care, typically accessed in clinical workflows while users are engaged with routine tasks in electronic health records5,6. The profile of the infobutton has been elevated in recent years through the development and refinement of international standards and implementation guides in Health Level Seven (HL7) as well as its inclusion in the Meaningful Use criteria, as part of the United States government incentive program for broadening the uptake and usage of key features within electronic health records7.
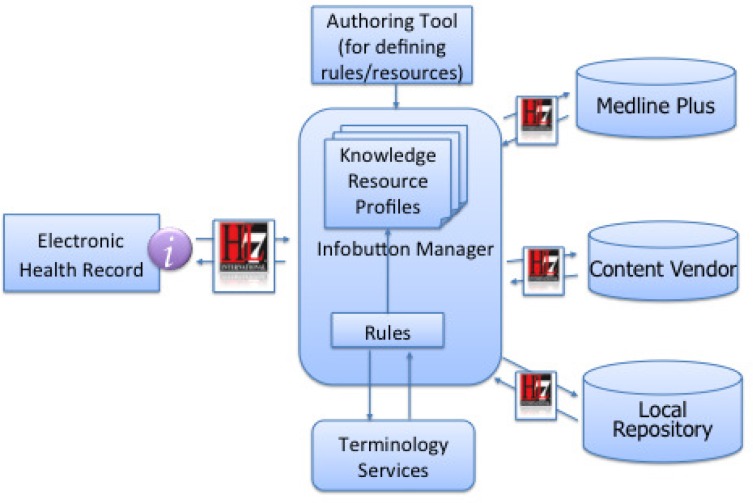
Figure 1 illustrates the flow of information within a typical infobutton exchange, as prescribed by the HL7 standard. In this type of exchange, a request is manifest from the electronic health record (EHR), passing several key parameters including 1) the primary (and secondary if applicable) concept of interest, the age and sex of the patient, the role of the intended recipient, the care setting, and even the user’s current task in the EHR. As information moves to the infobutton manager, key tasks including logging, selection rule logic, translation of internal codes to reference terminologies (like SNOMED, LOINC, ICD-9, etc), conversion to appropriate URLs to request further information take place. As these requests propagate outward to content repositories, including externally licensed and internally developed content, each repository is tasked with identifying relevant content and responding with metadata that can allow the users at the point of care to access that relevant content. Most content providers do so by building upon a combination of intelligent text parsing and indexing materials with relevant concepts from standardized terminologies. The approach and challenges faced by content providers in dealing with these terminology issues and aligning their content and services with these systems have been well-documented by Strasberg et al8.
Figure 1-.
Flow showing data exchange and processing through infobutton manager. Request is passed from EHR to infobutton manager, where matching, code translation, and request output send queries to content providers. Content providers receive requests, process them using standardized parameters, and return best results by means of search algorithms and indices using standardized terminologies.
Although the infobutton standard allows for flexibility by enabling requests to pass free text in the place of coded concepts, anecdotal evidence has consistently shown that infobutton performance improves greatly when content providers index their materials appropriately using relevant terminologies, instead of just relying fully upon free text queries. This poses some unique challenges for content providers in that they must index content using appropriate terminologies and granularity, as well as deal with the important issue of ‘rank’ among results that come back from these queries. It also poses a unique challenge for institutions who have infobuttons, but also want to implement access to local content repositories from the infobutton. These organizations must address the infrastructure necessary for centralizing, searching, and providing standards-based responses against their own content in order to enable this type of access from the infobutton. Open-source efforts like the Infobutton Responder within the OpenInfobutton open-source project are reflective of this need9. Local content owners need the ability to centralize their content, assign appropriate metadata, and include keywords indices from relevant reference terminologies in order to make their local content visible from these frameworks.
Background
Infobutton use at Intermountain Healthcare
Intermountain Healthcare is a not-for-profit integrated healthcare delivery system based in Salt Lake City, Utah. It provides healthcare for the entire state of Utah and parts of southeastern Idaho. Intermountain maintains 22 inpatient hospitals (including a children’s hospital, an obstetrical facility and a dedicated orthopedic hospital), more than 185 outpatient clinics, and 18 community clinics serving uninsured and low-income patients. Intermountain provides primary and specialty care for approximately half of the residents of the state of Utah. Its clinical information systems are internally built and maintained, dating back several decades. They include inpatient and outpatient EHRs, a decision support framework, an infobutton manager platform, and a patient portal. They have been widely-regarded as ‘state-of-the-art’ and are well recognized in the literature for supporting best care practice with clinical decision support interventions10. Intermountain’s infobutton manager (a key component of this effort) is used regularly and its development and uptake have been detailed previously11–13.
Infobuttons have been in use at Intermountain Healthcare for over 14 years. They have been integrated in two separate clinical systems, including usage from 4 major modules within these systems including medication ordering, lab result review, problem list, and microbiology. Usage has steadily increased over the years, with over 1,700 unique monthly users, accounting for over 18,000 infobutton sessions per month. They have also recently been made available for patient use from our patient portal in the same modules. At present, we are transitioning our production infobutton manager environment from a legacy, internally-built system over to a local implementation of the OpenInfobutton initiative.
At Intermountain Healthcare, we have maintained an enterprise clinical knowledge repository designed for centralizing, versioning, and distributing internally-developed clinical content for the past 15 years14. In the past, a very limited subset of these materials have been made available through our infobutton manager, but the process involved to do so was fairly arduous. Terminology engineers, knowledge engineers, and content experts had to work in concert with one another to assign specific terminology concepts to the metadata for these documents in our knowledge repository, as well as in corresponding domains in our healthcare data dictionary. The tasks involved were such that a content expert would be ill-equipped to index and maintain content without substantial technical help. Furthermore, the content authors who create and maintain many of these content resources may or may not be well-versed in the relevant terminology standards for indexing that content, and as such, may not be best suited for that type of task. As a result, only small portions of Intermountain’s internal content library have been accessible via the infobutton.
Current needs and strategic directions
As part of a broader effort at Intermountain to make internally-developed content more accessible to users through channels like the infobutton, we have recently engaged with a group of content authors who maintain a library of patient education materials. The collection includes over 1,600 locally-authored patient education handouts, worksheets, forms, and we are exploring means for automating the annotation process described above. In current processes, content authors can and do assign free-text keywords to their content to facilitate local search, but have never had need to index the content using standardized terminologies. We envision an environment in which automated content analysis and suggested codes for annotation accompany the normal content publication and versioning processes typical in knowledge management. As such, we hope to draw upon the unique strengths of both computer-based and human annotators in indexing our local content libraries with relevant, appropriate metadata.
Methods
Terminology selection
We prefer to use an annotation approach that is as generalizable as possible, and be capable of producing ICD-9 and SNOMED-CT codes. Our previous efforts at annotating and exposing content through our infobutton manager have been based on our local terminology, the Healthcare Data Dictionary. This terminology, however, is not intended to be an independent reference standard. Our contracts with external providers have shown that these are the two terminologies that have the most support among content providers who index their content accordingly. Since our content of choice was most applicable for requests from the problems domain, we deemed that these were appropriate choices for the effort.
Tools and Services
We analyzed several annotation engines for possible inclusion in our effort, including Metamap and the NCBO BioPortal Annotator. Although both have their strengths, we opted to use the BioPortal Annotator, which is based on the Mgrep concept recognizer15. It has been shown to have high precision in a study that used it for recognizing disease terms in descriptions of clinical trials (87%) and has been characterized as a lightweight concept recognizer that can provide fast, good-enough concept recognition in clinical texts16–17. It also already contains lexicons for both SNOMED and ICD-9, an important feature in the overall effort we were pursuing.
We selected a convenience sample of 59 documents (from a subcollection of the library) to test our approach. In our content repository, all documents consist of two artifacts, a standardized XML header containing uniform metadata about each document, as well as the document body itself. For the annotation effort itself, we decided to focus strictly on the body of the document. Although the metadata in the document header could be useful for indexing, we felt that annotating the corpus of the knowledge itself would be a more generalizable approach, with fewer dependencies on the local approach to metadata management.
All of the 59 documents were stored in PDF format. In order to extract and preserve the content of these documents for analysis and annotation, we used the open-source Apache Tika software, a toolkit that detects and extracts metadata and structured text content from various documents using parser libraries18. After processing these documents through Tika, the resultant text streams files were stored to memory and streamed into a local instance of the NCBO Annotator.
The annotation engine produces XML files for each document analyzed, containing a rank-ordered list of identified concepts, links back to the NCBO website for more detailed views about each concept, character mappings for each concept identified and match types associated with each reference. Figure 2 illustrates the flow of information from the documents, through TIKA, the NCBO Annotator and the resultant output. After processing each file and creating an XML file with the annotations data, we extracted the relevant concept terminologies, identifiers, and text representations and consolidated them into a text file for analysis. All 59 documents were able to successfully be processed and annotated. The NCBO Annotator found one or more matching concepts for all of the documents in either SNOMED-CT or ICD-9.
Figure 2-.

Illustration depicting flow of information from PDF files through Tika software, the resultant text stream feeding through the NCBO annotator, and the outputted XML annotations files
Separately, we extracted all of the human-derived keywords for each document from our clinical knowledge repository. As mentioned earlier, each document has a corresponding header document which contains metadata about the document, including a keywords section that accommodates both free text and coded keywords. Although most of the keywords were free text, a subset of these were ICD-9 codes. For purposes of comparison, we normalized all the concepts to free text and organized them for analysis.
Results
Keyword Overloading by authors in manual keyword entry
One of the early results of our analysis is that our authors have found ways of ‘overloading’ or misusing the existing metadata fields in the header. They have developed an internal identification pattern and have used the keyword field as if they served as an ‘alternate ID’ field. Presumably, they have done so in ways that allow them to search and retrieve content using these identifiers. For example, a document about depression would contain expected keywords like “clinical depression”, “mental health”, “major depression” but also internal codes such as “CPM019”. This did prove problematic in the analysis of the data, so we removed these data points from consideration.
Dealing with ‘noise’ concepts in the annotation space
One of the interesting results of processing the document set through the NCBO toolset was that raw processing of the documents against a concept space like ICD-9 or SNOMED resulted in large concept sets. Collectively, annotating the 59 documents against ICD-9 resulted in over 1138 ICD-9 codes (roughly 19 per document) and over 35,000 SNOMED-CT codes (338 per document). Some of the ICD-9 codes were deemed to be irrelevant in the context of concepts relevant to the problem list (the proposed point of integration via infobutton for this content). Concepts such as ‘process’, ‘hospital facility’ and ‘time’ were clearly concepts found in the document, but were deemed as less relevant in the context of the specific task of indexing content for linking from the problem list. This number was cut somewhat by further processing the data to remove category or ‘container’ concepts in ICD-9 (e.g. T052, or ‘Activity’).
One of the reason for which there were a surprisingly large number of concepts in the SNOMED-CT annotation space is that the annotator included retired concepts in the output. We were able to consolidate this list by programmatically reducing the search space by collapsing identical ‘concept term’ entries. We also leveraged an existing mapping table to our local terminology space to identify active SNOMED terms. This reduced the overall SNOMED-CT concept space by more than 50%.
Given that the practicalities of annotating content have shown that assigning that many identifiers to the content is not useful in making search results precise, we employed other methods to further narrow the concept space. We preprocessed the documents by first annotating the content using a MedLine Plus ontology available in the NCBO annotator, and then subsequently annotated that concept space using the ICD-9 and SNOMED-CT lexicons. This was particularly useful in that it focused the output concept space to be relevant to concepts in the MedLine Plus ontology, one that is more tightly focused on diseases and conditions. This further reduced the concept space of the SNOMED-CT codes by almost 80%.
Collectively, after accounting for retired concepts in SNOMED-CT, removing some of the less-relevant categorical ICD-9 codes and pre-processing the annotation space against the MedLine Plus ontology, we derived a unique concept set of 117 ICD-9 concepts and 741 SNOMED-CT concepts, some of which that were identified in multiple documents in the set we tested.
Similarity between human-identified keywords and annotated concepts
The analysis of similarity between the two concept spaces (human-identified keywords and machine-derived codes) made for a somewhat difficult process. Automatic transformation of free-text entries to coded elements resulted in poor translation and very scant results overall. Translating codes back to text representations at least allowed for each identified concept to have a representation, but made the comparison between concept spaces somewhat difficult, due to the inherent nature of free text. The differences in the overall number of concepts identified per document between the human and computer raters, combined with the non-categorical data made kappa analysis not a good option. In the end, we opted to convert coded entries to free text and present some metrics of similarity based off some computed and some human-analyzed metrics.
For purposes of illustration, we present our analysis of a random sample document that we reviewed manually, aligning and matching computer-derived and human-derived concepts to compare and contrast the concepts identified by each respective sets. This document focused on the management of low back pain, including assessment, treatment options and goals. Table 1 contains keywords identified by the content authors, ICD-9 and SNOMED-CT. Although we present data specific to this particular document, the observations hold true across multiple documents in the set.
Table 1-.
Human derived keywords, and computer-derived ICD-9 and SNOMED-CT codes for a patient education sheet dealing with low back pain.
| Human Keywords | ICD9CM | ICD9 text | SNOMEDCT | SNOMEDCT text |
|---|---|---|---|---|
| lumbar | 338–338.99 | PAIN | 279039007 | LOW BACK PAIN |
| radiculopathy | 338.2 | CHRONIC PAIN | 62482003 | LOW |
| mechanical back pain | 780–789.99 | SYMPTOMS | 161891005 | BACK PAIN |
| start back | 733.0 | OSTEOPOROSIS | 22253000 | PAIN |
| cauda equina | 782.3 | EDEMA | 82423001 | CHRONIC PAIN |
| stenosis | 756.12 | SPONDYLOLISTHESIS | 90734009 | CHRONIC |
| Disc | 800–829.99 | FRACTURES | 257733005 | ACTIVITY |
| sacroiliac | 344.61 | CAUDA EQUINA SYNDROME | 363679005 | IMAGING |
| Facet | 7396004 | DIAGNOSTIC IMAGING | ||
| CPM009 and CPM009a | 261004008 | DIAGNOSTIC | ||
| CPM009d | ||||
| CPM009b and CPM009c | ||||
| Low back pain | ||||
| Low back pain, chronic | ||||
| Low back pain, acute | ||||
| DJD - Degenerative joint disease | ||||
| Degenerative L-S Spine Disease | ||||
| Degeneration of intervertebral disc | ||||
| Spinal osteoarthritis | ||||
| Degenerative Arthritis-Low Back | ||||
| Degeneration of lumbar intervertebral disc | ||||
| Degenerative spondylolisthesis | ||||
| Herniated intervertebral disc L3, L4 | ||||
| Herniated intervertebral disc L4,L5 | ||||
| Herniated intervertebral disc L5,S1 | ||||
| Spondylolisthesis, grade 1 | ||||
| Spondylolisthesis, grade 2 | ||||
| Spondylolisthesis, grade 3 | ||||
| Herniated Disc | ||||
| Sacroiliitis | ||||
Human coded keywords
Free-text captured keywords often have special characters. It is often hard for machine to analyze and match to terms. The data not normalized and standardization of capitalization, and the approach to using nouns vs. adjectives is often haphazard. As noted earlier, most of the documents had keyword entries that serve as internal identifiers, such as CMP009d. Human assigned identifiers include no rank of relevance and appear in apparently random order.
Another interesting observation is the inclusion of concepts derived from knowledge outside the space of the concept domain. The content authors referenced a “start back protocol” while annotation engine missed the term completely. The referenced concept is the Keele STarT back tool that was developed by the Primary Care Centre at Keele University, with funding from Arthritis Research UK. It is well validated for low back pain, but is also being used widely for other spinal pains. Other documents contained similar examples, including references to instruments like the PHQ-9 for mental health.
Coded concepts
The ICD9 code list provided a much smaller concept space. In an extreme example, the ICD-9 annotation provided 20 concepts while the SNOMED-CT annotation gave 2087. The ICD-9 annotations often don’t have as much detail as the SNOMED-CT codes, in part due to the granularity of the terminologies. The ICD-9 codes sometimes included concepts from unexpected domains, (including activities like ‘yoga’ that were found) but the tools allowed us the flexibility to include or restrict those as we saw fit. The pre-annotation against the MedLine Plus ontology helped significantly with the large numbers of concepts we derived when using SNOMED-CT. We opted not to index the content directly against the MedLine Plus ontology because these codes aren’t typically used as reference terminologies in infobutton implementations.
In all three sets of concepts, the overarching concepts of ‘pain’ and ‘chronic pain’ are present in all three, but the SNOMED-CT matching offered more specificity by listing ‘low back pain’ as its top concept. The ICD-9 annotation found ‘cauda equina syndrome’ which was not found in the SNOMED-CT set, but was present in the human keyword space. Overall the major, expected concepts were present in most annotation sets.
Discussion
Feasibility and performance
One of the key points we established early in the effort is that we were not interested in implementing a full-fledged natural language processing environment. In order to index the content of these documents, we wanted a system that was 1) capable of identifying key concepts, 2) able to detect and present overall concept relevance in ranked order, and 3) able to connect these concepts with identifiers in relevant reference terminologies. We believe that we have succeeded in these objectives. The system was able to perform these annotations quickly, giving us some hope that this type of analysis might be feasible at runtime, when a user is publishing a new document or updated version of existing content. If this proves to be the case, we may be able to leverage this system while the users are updating other required portions of metadata, giving them a chance at the end of that session to review, and accept or reject the proposed coded concepts that may be deemed relevant in the document by the NCBO Annotator. An approach like this would still allow human review of the concepts, but would push much of the burden of identifying and embedding relevant terminology codes in the metadata.
We have not yet conducted an analysis of determining whether the machine-identified concepts are acceptable for use from our clinical partners. Our limited matching analyses presented above suggest that while there is significant overlap in the human and computer-derived concept spaces, there are key differences as well. We plan to present our findings to our clinical education partners (the owners of the content set that is desired to be ‘infobutton friendly’. Assuming that our clinical domain experts agree with the approach, we anticipate that we should be able to scale the process up quickly to index the full set of 1600+ patient education sheets. For each document, we would add the coded concepts from SNOMED and/or ICD-9 to the header metadata document in the knowledge repository. With this information in place, we could configure infobuttons to point to these resources in relatively quick fashion, provided a brief update to the enabled profiles in our infobutton manager configuration center.
Limitations
We have not accounted for negation (e.g. through a NegEx type of implementation) in the early phases of our project19. While this is not likely to play as important of a role in concept identification for educational materials as it does in understanding the meaning behind clinical notes (where negative findings and ‘rule-outs’ are common) we anticipate that implementing its inclusion would likely enhance the performance of the system. We intend to explore this in subsequent phases of our implementation
We have not been able to calculate and share Kappa statistics, since the computer and the users annotated content in different ways (coded elements vs. free text). We anticipate that we could further analyze similarity using ontology-based similarity tools that match and assign probabilistic similarity metrics to concepts under comparison. We expect that as we do so we will continue to find some overlap, but also areas where both the human and computer codes differ from each other, due to their different approaches in annotating content. The differences between tacit and explicit knowledge will likely be exhibited in the differences that analysis would show.
Even with coded concepts in place for all the documents in the collection, our approach doesn’t yet provide a satisfactory way of addressing the important issue of presenting ‘rank’ and overall relevance back to infobutton users. It is highly likely that indexing all 1600 documents in this collection will result in some significant overlap in terms of multiple documents that are indexed with common concepts like ‘diabetes’ and ‘obesity’. While the NCBO toolset does output a rank-ordered list of concepts in rendering its output, our current approach for storing metadata for coded concepts does not allow for capturing ‘relative relevance scores’ or the like. We anticipate that in providing a service that is capable of presenting multiple results back to an infobutton, ranked by relevance, we will need to further capture this detail either in the metadata, a separate index file, or through other search engine types of techniques to apply weights to the various results.
Another limitation to our approach is that at present, we are not indexing the content with relevant terminologies in other domains like medications and lab orders. Often, the document owners would insert text-based keywords like amoxicillin, doxycycline and others, indicative of treatment patterns for the condition being discussed. We can look to perform similar indexing using these types of terminologies in future efforts.
Our approach to annotation also does not account for materials which are written in Spanish. This will be less of a problem going forward in our local knowledge repository, since English and Spanish versions of the same document are semantically linked in our database. As such, annotating these concepts in the English narrative could be used to encode concepts in the corresponding Spanish documents as well. In other document repositories where these types of linkages don’t exist, the alternate language documents may not be able to be processed and annotated automatically.
Conclusion
We have demonstrated the feasibility of using existing annotators in the biomedical informatics research domain to identify key concepts of patient education materials, using ICD-9 and SNOMED-CT as reference standards. Our system was able to extract and analyze concepts for all documents we analyzed in the set of 59 patient education sheets sampled. In a parsimonious effort to do so, we were able to identify more than 117 ICD-9 codes and 741 SNOMED-CT codes to index the starter set of content analyzed. When projected back to the complete set of more than 1600 documents, we anticipate that we will be able to present this content from hundreds of relevant entries in the problem list, expanding the reach of this content without the need of extensive, time-intensive human-based annotation. We expect that combining the strengths of computer-based annotation and human review will allow for the best approach for combining strengths and saving time in annotating knowledge content for better access from tools like the infobutton.
References
- 1.Covell DG, Uman GC, Manning PR. Information needs in office practice: are they being met? Ann Intern Med. 1985;103(4):596–9. doi: 10.7326/0003-4819-103-4-596. [DOI] [PubMed] [Google Scholar]
- 2.Ely JW, Osheroff JA, Chambliss ML, Ebell MH, Rosenbaum ME. Answering physicians’ clinical questions: obstacles and potential solutions. J Am Med Inform Assoc. 2005;12(2):217–24. doi: 10.1197/jamia.M1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauser SE, Demner-Fushman D, Jacobs JL, Humphrey SM, Ford G, Thoma GR. Using wireless handheld computers to seek information at the point of care: an evaluation by clinicians. J Am Med Inform Assoc. 2007 Nov-Dec;14(6):807–15. doi: 10.1197/jamia.M2424. Epub 2007 Aug 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldbach H, Chang AY, Kyer A, Ketshogileng D, Taylor L, et al. Evaluation of generic medical information accessed via mobile phones at the point of care in resource-limited settings. J Am Med Inform Assoc. 2014 Jan-Feb;21(1):37–42. doi: 10.1136/amiajnl-2012-001276. Epub 2013 Mar 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cimino JJ, Elhanan G, Zeng Q. Supporting infobuttons with terminological knowledge. Proc AMIA Annu Fall Symp. 1997:528–32. [PMC free article] [PubMed] [Google Scholar]
- 6.Maviglia SM, Yoon CS, Bates DW, Kuperman G. KnowledgeLink: Impact of context-sensitive information retrieval on clinicians' information needs. J Am Med Inf Assoc. 2006;13:67–73. doi: 10.1197/jamia.M1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Del Fiol G, Huser V, Strasberg HR, Maviglia SM, Curtis C, Cimino JJ. Implementations of the HL7 Context-Aware Knowledge Retrieval (“Infobutton”) Standard: challenges, strengths, limitations, and uptake. J Biomed Inform. 2012 Aug;45(4):726–35. doi: 10.1016/j.jbi.201112.006. Epub 2012 Jan 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strasberg HR, Del Fiol G, Cimino JJ. Terminology challenges implementing the HL7 context-aware knowledge retrieval (‘Infobutton’) standard. J Am Med Inform Assoc. 2013 Mar-Apr;20(2):218–23. doi: 10.1136/amiajnl-2012-001251. Epub 2012 Oct 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Infobutton Responder - OpenInfobutton Project. [Accessed on March 1, 2014]. Available at: http://www.openinfobutton.org.
- 10.Clayton PD, Narus SP, Huff SM, et al. Building a comprehensive clinical information system from components. The approach at Intermountain Health Care. Methods Inf Med. 2003;42:1–7. [PubMed] [Google Scholar]
- 11.Reichert JC, Glasgow M, Narus SP, Clayton PD. Using LOINC to link an EMR to the pertinent paragraph in a structured reference knowledge base. Proc AMIA Annu Fall Symp. 2002:652–6. [PMC free article] [PubMed] [Google Scholar]
- 12.Del Fiol G, Rocha RA, Clayton PD. Infobuttons at Intermountain Healthcare: utilization and infrastructure. Proc AMIA Annu Fall Symp. 2006:180–4. [PMC free article] [PubMed] [Google Scholar]
- 13.Del Fiol G, Haug PJ, Cimino JJ, Narus SP, Norlin C, Mitchell JA. Effectiveness of topic-specific infobuttons: a randomized controlled trial. J Am Med Inform Assoc. 2008 Nov-Dec;15(6):752–759. doi: 10.1197/jamia.M2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulse NC, Galland J, Borsato EP. Evolution in Clinical Knowledge Management Strategy at Intermountain Healthcare. AMIA Annu Symp Proc. 2012;2012:390–399. Published online 2012 November 3. [PMC free article] [PubMed] [Google Scholar]
- 15.NCBO Bioportal Annotator. [Accessed on March 1, 2014]. Available at: http://bioportal.bioontology.org/annotator.
- 16.Shah NH, Bhatia N, Jonquet C, Rubin D, Chiang AP, Musen MA. Comparison of concept recognizers for building the Open Biomedical Annotator. BMC Bioinformatics. 2009;10(Suppl 9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LePendu P, Iyer SV, Fairon C, Shah NH. Annotation Analysis for Testing Drug Safety Signals using Unstructured Clinical Notes. Journal of Biomedical Semantics. 2012;3(Suppl 1):S5. doi: 10.1186/2041-1480-3-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apache Tika Project. [Accessed on March 1, 2014]. Available at: https://tika.apache.org/
- 19.Chapman WW, Bridewell W, Hanbury P, Cooper GF, Buchanan BG. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001 Oct;34(5):301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]