Abstract
Online health communities play an increasingly prevalent role for patients and are the source of a growing body of research. A lexicon that represents the sublanguage of an online community is an important resource to enable analysis and tool development over this data source. This paper investigates a method to generate a lexicon representative of the language of members in a given community with respect to specific semantic types. We experiment with a breast cancer community and detect terms that belong to three semantic types: medications, symptoms and side effects, and emotions. We assess the ability of our automatically generated lexicons to detect new terms, and show that a data-driven approach captures the sublanguage of members in these communities, all the while increasing coverage of general-purpose terminologies. The code and the generated lexicons are made available to the research community.
Introduction
As online health communities like forums, blogs, and mailing lists become increasingly prevalent, patients are turning to these resources for information exchange and interaction with peers [1]. Patients with breast cancer, in particular, rely on cancer-specific online health communities for both informational and emotional support [2–6]. While this type of social networking has become central to the daily lives and decision-making processes of many patients, there are still many research questions open. For many research activities, capturing domain knowledge about topics discussed in a community and organizing terms and concepts discussed into lexicon and terminologies is needed for knowledge discovery and information extraction [5,7]. Designing automated tools to build these lexicons is a challenging task, however, because the language used in online health communities differs drastically from the genres traditionally considered in the field of information processing and from the sublanguages already investigated in the biomedical domain [8,9]. Health community vocabulary is characterized by abbreviations and community-specific jargon [10], and posts are authored in a style-free and unedited manner, with often informal and ungrammatical language. In addition, the content of the posts is both emotionally charged and dense with factual pieces of information, indicating that specific semantic types of information, like emotions, are more prevalent than in traditional biomedical texts.
In the biomedical domain, there are several clinical terminologies available that provide candidate keywords for lexicons, such as names of diseases, procedures, and drugs [11,12]. There exist health-consumer oriented terminologies, but their focus is on health consumers rather than patients in online communities, which have a different level of health literacy and convey at once a larger and more granular vocabulary for health terms than general health consumers [13]. More recently, researchers experimented with crowdsourcing to identify medical terms in patient-authored texts, but show that further processing and supervised learning is still needed to achieve acceptable results [14]. Finally, like for methods trained in other genres, existing terminologies, whether clinical or health consumer, do not cover the many misspellings and abbreviations typical of a given online health community and require manual updating to capture new terms introduced into the sublanguage [15,16].
Automated creation of lexicons has a long history in natural language processing. Unsupervised named entity recognition, and the use of seed terms in particular as a starting point for lexicon building, is a practical and promising method because it does not require a corpus, manually annotated with examples of terms [17]. Instead, seed terms are leveraged by looking for candidate terms with high context similarity to the seed terms. In the biomedical domain, such an approach has been used to identify disease names in medical mailing lists [15], recognize clinical and biological terms [18,19]. The approach is rooted in the Distributional Hypothesis, which states that words with similar contexts tend to have similar meanings [20]. In the biomedical domain, distributional semantics has been used for a wide range of tasks, such as matching MEDLINE abstracts to terms in an ontology [21], automatic generation of synonyms for gene and protein names [22], evaluation of language incoherence in patients with schizophrenia [23], and identifying semantically similar concepts in clinical texts [24,25], to name just a few [26–28].
In this paper, we describe an unsupervised method to generate lexicons representing the sublanguage of an online health community focusing on specific semantic types. Starting from a seed set of terms, all in the same semantic category (like medication names), it computes a typical context in which terms of that category occur. The context representative of a semantic category is then leveraged to identify new terms, which can augment the lexicon. To assess the value of our method and the generated lexicons, we ask the following research questions: (i) Can the method identify new terms to augment a seed set, and if so how accurately? (ii) How well does the method perform on generating lexicons for different semantic categories? And (iii) how stable is the method with respect to the quality of the underlying seed set?
Methods
The procedure for detecting terms in a forum that are representative of a specific semantic category is outlined in Figure 1. A seed set is gathered (either from an existing lexicon or from a small manually created one) representative of a given semantic category. Seed terms and their context, as defined from their occurrences in the online forum, are aggregated into a representative context vector, which reflect the typical context for terms in the category. As such, the representative vector acts as an implementation of the distributional hypothesis, where a word is defined by the context in which it is conveyed. To identify new terms for the semantic category, candidate terms from the forum are selected and an individual context vector is defined for each. Determining whether a candidate term belongs to a semantic category is achieved by computing the similarity between its individual context vector and the semantic category’s representative vector. If a candidate term is used with words and patterns similar to the ones of the semantic category, it is likely the candidate term belongs. In our methods, we focus on three semantic categories of interest: (i) medications, (ii) signs and symptoms, and (iii) emotions and mental states.
Figure 1.

Overall pipeline to identify in an online health forum the terms representative of a specific semantic category.
Dataset
Following ethical guidelines in processing of online patient data, we focus on a popular, publicly available breast cancer forum with a large number of participants and obtained IRB approval. Posts from the publicly available discussion board breastcancer.org were collected [29,30]. At the time of collection, there were more than 60,000 registered members posting in 60 sub-forums. Our dataset consists of the most popular sub-forums. The extracted corpus for our analysis contains 26,153 threads corresponding to 253,231 posts. Overall, the corpus has 25.8M words for a vocabulary of 145K unique words. When considering only words that appear at least twice in the corpus, the vocabulary consisted of 75K words.
Lexicon Building
Choosing Seeds and Candidates
For each semantic category, we use an existing lexicon or a manually curated list of terms to gather a set of seed terms that are known to belong to the target category (e.g., medications). Using the forum corpus, we also extract a large number of candidate terms that may or may not be members of the target semantic category. We customize our framework to our three semantic categories: medications, signs and symptoms, and emotions. The creation of these sets is described in Seed and Candidate Sets section below.
Constructing Context Vectors
Once the sets of seed and candidate terms have been selected, we employ a vector-space distributional similarity method to create context vectors for each term. The context vectors are derived from the vocabulary V found in the forum posts with the constraint that a word appears at least twice in the corpus. Each element in a term’s vector contains a count of the number of times a word in V appeared in a certain context, such as directly preceding our term of interest. Because we use 5 contextual feature types, as described in Table 1, each context vector consists of 5|V| elements. We chose a set of local, highly specific contextual features to capture similarity in meaning and usage. For instance, in the example given in Figure 2, we can capture some of the data contained in exact patterns such as “been on X”, as well as more general contextual features, such as the presence of the word started somewhere within 3 words of our target. This information helps the method find candidate terms that exhibit similar behavior to our seed terms, and are therefore likely to be in the same semantic category. These context vectors form the underlying representation in our method.
Table 1.
Feature types used in the vector space model.
| preceding word | A cell for each word in the vocabulary, indicating the number of times it appeared directly before the target term |
| word at -2 | A cell for each word in the vocabulary, indicating the number of times it appeared 2 words before the target term |
| following word | A cell for each word in the vocabulary, indicating the number of times it appeared directly following the target term |
| word at +2 | A cell for each word in the vocabulary, indicating the number of times it appeared 2 words after the target term |
| word within 3 | A cell for each word in the vocabulary, indicating the number of times it appeared within 3 words before or after the target term |
Figure 2.
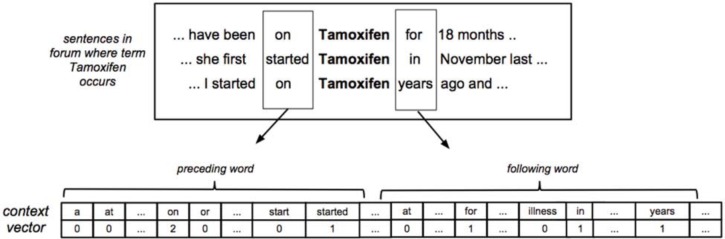
Part of the context vector for the seed term Tamoxifen. Each term context vector has a separate set of counts for preceding word, following word, word at -2, word at +2, and word within 3.
Creating a Representative Vector
In order to create a unified representation of a semantic category, the context vectors of the seed terms are merged into a representative vector for the category. Using vector addition, the individual context vectors are added. The vector is normalized by the number of seeds, producing a vector containing the average value for each of the seed vectors. A smoothing step is then performed, in which any values of the representative vector that are specific to only one seed are set to zero. This is intended to remove any contextual information that is unique to a single seed term and does not represent the semantic category as a whole. For example, assuming Arimidex is a seed term for medications, and it appears in the sentence “I have been on Arimidex (an aromatose inhibitor)”, we will want to make use of the feature “preceding word on”, since it is an indicator of a medication term, and will likely be shared by other seeds. However, the word aromatose is specific to the seed Arimidex, and we will discard the associated data unless it is shared by at least one other seed.
To further reduce noise and ensure a high-quality representative vector, a pre-filtering step is employed. The initial representative vector as created above is compared with each of the original seed term vectors using a cosine similarity metric [31]. If the similarity is below a certain threshold (.6 in our experiments), the seed term is considered an outlier, and is removed. The representative vector is then re-created as described above using the filtered group of seed terms.
Calculating Similarity
A candidate term t is more likely to belong to a semantic category if its context vector is similar to the representative vector r for the category. Similarity is computed as the cosine metric between the two vectors. If the vector t is composed of (t1, t2, …, tn) and r is composed of (r1, r2, …, rn) then, their cosine is defined as
The values of cosine range from zero, indicating no similarity, similarity to 1, indicating maximal similarity. Thus, our procedure scores each candidate term according to the similarity of its vector to the representative vector for the semantic category. The candidates can then be ranked in descending order of their similarity scores.
Seed and Candidate Sets
Seed sets are collected separately for each of the three semantic categories as described below.
Medications
To create a set of seed terms denoting names of medications, we use the comprehensive list of medications provided by RxNorm [32]. The list is then ordered by frequency of occurrence in the corpus, and terms appearing with low frequency in our corpus are removed (less than 50 in our experiments), resulting in a seed set of 137 medication terms.
The set of candidate terms for the medication category is defined initially as all out-of-vocabulary words in a standard English dictionary (dictionary from the Aspell program was used in our experiments), following the assumption that medication names are proper names, and thus not part of the standard English vocabulary. We only considered out of vocabulary terms from our corpus, which were frequent enough (50 times at least). This resulted in a set of 1,131 words as potential candidates for medication names.
Signs and Symptoms
We experiment with two medical lexicons for the construction of a set of seed terms denoting signs & symptoms. The first is the Unified Medical Language System (UMLS), where we use a list of all terms assigned to the ‘sign or symptom’ semantic type [11]. The second resource is SIDER, a list of terms denoting side effects extracted from FDA drug labels [33]. For each of these lists, we filter out all terms that are more than two words long. We then search for occurrences of the remaining terms in our data and extract all single word terms occurring more than 50 times, and all two-word terms occurring more than 20 times. This procedure provides the four seed sets described in Table 2. Despite the fact that both UMLS and SIDER seed lists share the most frequent term, there is relatively low overlap between them amongst these high-frequency terms (17 single-word terms, and 21 two-word terms).
Table 2.
Number and average frequency of the terms in the four seed sets employed for detecting Signs & Symptoms, before and after (in parenthesis) the filtering procedures, along with the most frequent term in each seed set. The rightmost column specifies the coverage (cumulative frequency of all the terms inside the set) of each unfiltered seed set.
| Seed Set | Size | Avg. Frequency | Most Frequent | Coverage |
|---|---|---|---|---|
| UMLS single word | 84 (45) | 1,205 (1,577) | pain | 103,695 |
| UMLS two words | 136 (63) | 134 (228) | hot flashes | 37,702 |
| SIDER single word | 88 (51) | 918 (1,418) | pain | 80,780 |
| SIDER two words | 92 (38) | 166 (335) | hot flashes | 31,926 |
In the case of signs and symptoms, we cannot restrict candidates to out-of-vocabulary terms, as we did for medications, since signs and symptoms are often conveyed using standard-English words and are often multi-words. Instead, we consider any single-word or two-word term as a potential candidate, provided it appears frequently in our data (more than 50 times for single words, and more than 20 times for two-word terms), and consists of well-formed words (does not include numbers or other non-alphabetic characters).
In addition, for two-word terms, we perform another filtering step to reduce the number of candidates and improve quality. This filter is designed to remove multi-word terms that are very common in the data as a result of the frequency of the component words, rather than the term as a whole. For instance, the two-word term “and I” appears frequently in our data, but has little meaning as a unit, and its frequency is due to it being composed from two very common words. To filter such cases, we compare the probability of the term as a whole to the expected probability of the component words appearing in adjacent positions by chance, according to their individual probabilities, as shown in Equation 1. The ratio r between these probabilities is compared to a manually specified threshold t (in our experiments, t = 20), and terms with ratios below the threshold are removed from the candidate list. After the selection and filtering procedures, we were left with a candidate list of 10,844 single-word candidates, and 37,015 two-word candidates.
Emotions
While there exist terminologies for emotions [34], we experimented with a very small seed set for emotions. Part of our motivation is to test the robustness of our method to discovering new terms when a limited terminology or none is available. Given the most frequent words in the corpus of posts, we manually selected 10 adjectives as a seed set, which conveyed an emotional state randomly: scared, grateful, sorry, fatigued, guilty, comfortable, nervous, confused, afraid, and happy. Following the filtering step described above to compute the representative vector, there were six emotion seed terms left: scared (frequency of 5,512 occurrences in the corpus), grateful (frequency 1,445), sorry (frequency 20,768), confused (frequency 1,807), afraid (frequency 3465), and happy (frequency 11,338). For the sake of reproducibility, we replicated the experiments with different seed sets chosen randomly and obtained very similar results to the ones given this instance of seed set, and thus only report on these results.
Experimental Setup
The output of our method for a given semantic category is a ranked list of terms, which can augment a terminology of known lexical variants for the category (ranking is based on the terms’ similarity scores to the given semantic category). We asked domain experts (two clinicians and one health psychologist) to review the lists for each of the three categories and tag each ranked term as a true positive (indeed a term that belongs to the semantic category) or a false positive (a term that does not belong to the semantic category). We report on the Precision at K[31], a standard evaluation metric for retrieval tasks in which the overall gold standard is unknown in advance – with different values of K for the top-K returned results, from K=10 to 50. We also report the cumulative coverage of the true-positive terms retrieved at the different K – that is, considering only terms that are not seeds. The coverage is a sanity check that the effort spent on discovering these terms pays off in terms of content that would have been ignored otherwise. For medications, the experts also encountered a number of terms that fell in a gray area. For instance, terms which were general names of treatments, or categories of medications, such as anthracyclines, a class of antibiotics. There were also terms indicating various drug cocktail treatments, as well as names of dietary supplements alternative treatments. Thus, for medication, we report two types of Precisions at K: a strict evaluation, which represents whether the ranked terms were medication names indeed, and one with a less strict definition of medication, which includes medication classes and drug cocktails.
Results
The code and the generated lexicons are available to the research community at people.dbmi.columbia.edu/noemie.
Augmenting an Existing Lexicon
Medications
In Table 3 we list the top ten terms according to the similarity with the representative vector for the medication category, along with their similarity score and frequency in the corpus. For the most part, the system correctly identifies terms indicating medications. There are misspellings (e.g., tamoxifin, benedryl, femera) and abbreviations (e.g., tamox) of medication names. The terms bisphosphonates and hormonals indicate classes of medications.
Table 3.
List of top 10 retrieved medication terms not included in seed set, along with their similarity score and their frequency.
| Retrieved term | Sim. score | Freq. | Retrieved term | Sim. score | Freq. |
|---|---|---|---|---|---|
| Tamox | 0.888 | 6,107 | bisphosphonates | 0.821 | 549 |
| Hormonals | 0.888 | 1,012 | carbs | 0.821 | 326 |
| Tamoxifin | 0.880 | 666 | mammos | 0.817 | 704 |
| Benedryl | 0.831 | 402 | femera | 0.815 | 452 |
| Fatique | 0.827 | 108 | lymphedema | 0.815 | 2,656 |
We can see four classification errors: fatique, carbs, mammos, and lymphedema. The first is a rare misspelling of fatigue in the dataset, with thus little power to be categorized correctly. The term carbs is used in a similar fashion to many medications, since it is an ingested compound and forum users often discuss its effect on their health, much like they discuss medications. In general, we observed that various types of dietary supplements were common in our results for this reason.
Figure 3 shows the precision of the classification as we go down the list of retrieved terms (and following our experimental setup where only words outside of RxNorm were assessed for validity). Coverage ranged from 9,188 for K=10 to 11,191 occurrences for K=50 for medications and treatments, and ranged from 7,627 (K=10) to 8,859 occurrences (K=50) for medication names alone.
Figure 3.
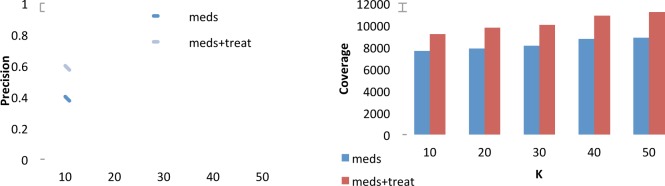
Precision (left) and number of instances covered (right) for the top K={10,20,30,40,50} retrieved terms not in the seed set for medication and treatments (meds+treat) and medication names only (meds).
Signs and Symptoms
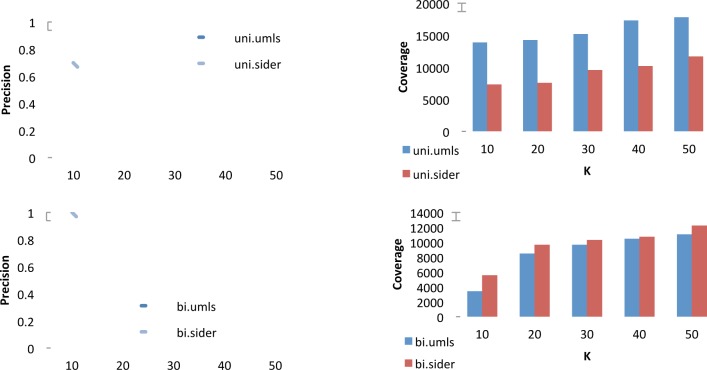
Table 4 shows the top 10 single-word and two-word terms retrieved as Signs and Symptoms retrieved when using SIDER as seed set. Figure 4 shows precision and coverage at K for the Signs and Symptoms category using either UMLS or SIDER as seed set.
Table 4.
Top 10 single- and two-word terms retrieved as Signs & Symptoms using SIDER as a seed set.
| Retrieved term | Sim. score | Freq. | Retrieved term | Sim. score | Freq. |
|---|---|---|---|---|---|
| itching | 0.954 | 807 | joint pain | 0.985 | 2,213 |
| caffeine | 0.950 | 342 | mouth sores | 0.966 | 604 |
| chemo | 0.950 | 76,737 | body aches | 0.959 | 221 |
| depression | 0.950 | 2,575 | acid reflux | 0.958 | 205 |
| discomfort | 0.945 | 1,520 | nose bleeds | 0.954 | 131 |
| bleeding | 0.942 | 1,376 | hair loss | 0.952 | 1,549 |
| bruising | 0.942 | 336 | bone aches | 0.949 | 119 |
| soreness | 0.935 | 476 | stomach problems | 0.948 | 101 |
| exhaustion | 0.935 | 248 | extreme fatigue | 0.947 | 110 |
| surgery | 0.934 | 35,831 | mood swings | 0.945 | 309 |
Figure 4.
Precision (left) and number of instances covered (right) for the top K={10,20,30,40,50} retrieved single-word signs & symptoms (top) and two-word signs & symptoms (bottom), reported for UMLS and Sider as seed set.
As mentioned in the Methods section, we made use of two resources to develop two separate seed sets for this semantic category. In the figure, we see that the different characteristics of the seed set (see Table 2), result in differences in performance for our system. The UMLS seed set has better coverage than Sider on single-word terms, for a similar number of words. This means that the single-word terms in the UMLS are more suited to our domain, and this results in higher coverage and precision for the output of our system. For two-word terms the situation is reversed. The SIDER seed set has similar coverage, but is significantly smaller than the UMLS one (see Table 2). This means that the seed terms are more suited to our domain. For two-word terms, we get better coverage and precision when using SIDER as a seed.
There is another important difference worth noting between single-word terms and two-word ones. In the case of single word terms, the coverage of both the lexicons we employ is quite high. This means it is difficult to find new terms not mentioned in the lexicon, and these are found with lower confidence. This is also the reason for relatively low precision for single-word terms in this semantic category (the precision is measured only for the new terms). For two-word terms, on the other hand, initial coverage of the seed sets is quite low. There are many terms in the data that are strong members of this semantic category, but are not mentioned in the lexicons. This means the system can discover high quality new terms, with higher coverage and better precision.
Emotions
Table 5 shows the list of top-10 retrieved emotion terms from the small seed set of six emotion terms. All terms are high-frequency terms in the corpus, except for greatful. Interestingly, the misspelled greatful, despite its low frequency had a high similarity to emotions probably because of its correct spelling grateful was one of the seed term. The precision is much higher with emotions than with the other two semantic categories medications and signs and symptoms, starting at 100% at K=10 and decreasing to 78% at K=50. For this category, we evaluated up to K=100, with a precision of 64%. Moreover, the coverage of the true-positive emotion terms ranged from 20,076 for K=10 to 51,281 for K=50. This indicates two findings: (i) terms relating emotional states are highly frequent in our corpus, confirming that much emotional support is exchanged amongst the forum members; and (ii) our method is particularly good at discovering new terms when provided with a very small seed set (in this case a set of 6 chosen terms).
Table 5.
List of top 10 retrieved emotion terms not included in seed set, along with their similarity score and their frequency.
| Retrieved term | Sim. score | Freq. | Retrieved term | Sim. score | Freq. |
|---|---|---|---|---|---|
| glad | 0.878 | 12,414 | thankful | 0.741 | 1,273 |
| relieved | 0.847 | 922 | desperate | 0.721 | 252 |
| excited | 0.780 | 1,035 | delighted | 0.719 | 152 |
| thrilled | 0.769 | 779 | greatful | 0.716 | 80 |
| sad | 0.745 | 2,994 | saddened | 0.698 | 175 |
Sensitivity to Choice of Seed Set
A common concern when using statistical methods that rely on seed terms is the sensitivity of the method to the choice of seeds. To investigate this issue in our framework, we compared the results of using a variety of different seeds, and examined the effect on the terms retrieved by the system.
First, we compared the output of the system when using the seed set based on UMLS terms to the output when using seeds from SIDER. Despite low overlap between the two seed sets, the output of the system was similar for both. When comparing the top hundred most highly scored terms, we found an overlap of 91% in the output for two-word terms, and 89% for single word terms. This indicates that the semantic category we are looking for – terms indicating signs and symptoms – is a well-defined one, with specific usage patterns in the data. A practical implication is that any seed set containing good representatives of the semantic category can be used to successfully retrieve other terms in a fairly robust fashion.
We also experimented to discover if single-word terms could be used as seeds to retrieve multi-word terms in the same semantic category. We used the SIDER single-word seed set to rank the two-word candidates. In this case, however, we found much lower correspondence with the output of the two-word seeds (60% when compared to the UMLS two-word seed group, and 57% compared to the Sider seed). These findings indicate that single-word terms describing side effects are used in a different manner than multi-word expressions, in terms of immediate context, and that it is important to use a seed set of the same type as the candidates that are being ranked (single-word seeds for single word candidates, and multi-word terms as seeds for multi-word candidates).
Finally, on the basis of the success of a small, manually selected seed set for the emotion category, we experimented with using a similar strategy for the medication and signs and symptoms categories. We randomly shuffled the posts in our data and manually selected the first ten terms we saw that belonged to each category – i.e., without any reliance on any dictionary. We re-ran our method by filtering these small seed sets and constructing context vectors, and thus the resulting seed sets were at most ten words randomly chosen for each category. Table 6 shows the random seed sets in each category. The starred terms were filtered out automatically at the pre-filtering step when creating the representative vector for a given category.
Table 6.
Random seed set for Medications, Signs and Symptoms single words, and Signs and Symptoms two words. The terms with asterix were filtered out automatically during the step for construction of the representative vector.
| Medications | Signs & Symptoms Single word | Signs & Symptoms Two words |
|---|---|---|
| tamoxifin | Pain | allergic reactions |
| herceptin | Leakage | mood swings |
| taxol | Cyst | * distended abdomen |
| carboplatin | Nausea | mouth ulcers |
| taxotere | neuropathy | hot flashes |
| tylenol | Baldness | high fever |
| xeloda | Blisters | scar tissue |
| zofran | Fatigue | * temple pain |
| percoset | headaches | abdominal pain |
| avastin | exhaustion | back pain |
For medication names, using a small set of random seeds was very successful, achieving 66% precision on the top 50 ranked results (74% if names of treatments are included), as compared to 44% and 62% when using RxNorm as basis for the seed. This demonstrates that if the target class is well defined, our method can learn accurate information from only a small number of examples, and a large, manually compiled lexicon is not necessary. For the category of signs and symptoms, the small randomly selected seed sets were also very effective. For single-word terms, the small seed set achieved 44% precision on the top 50 ranked results, significantly higher than that achieved by using UMLS and SIDER as seed sets, where the accuracy was 38% and 34%, respectively. For two-word terms, the randomly selected seed set achieved similar precision to using UMLS (62% on the top 50), but was not as effective as using SIDER (88%).
Discussion
Principal Findings
The primary objective of this study was to evaluate the use of lexical semantics in creating lexicons for use in content analysis of online health communities. Existing lexicons, like RxNorm, UMLS, and SIDER are fairly static resources, with potentially low coverage of the particular sublanguage of online health communities, whose informality often includes unique jargon, misspellings and abbreviations created by community members. Our method aims to fill in these gaps, by generating lexicons to represent the language of members in a given community with respect to different semantic categories.
Our study suggests that using context vectors trained on a small seed set is a viable, robust method to expand existing medical lexicons across a range of potential semantic categories. The method was robust across semantic categories as long as seeds were good representatives of those categories. Furthermore, we showed that the seed set can be very small (e.g., six terms like in our experiments with detecting emotion terms) and still generate viable lexicons with good coverage. Finally, our experiments with UMLS and SIDER suggest that seed set selection should take into account surface characteristics like number of words in phrase. Finally, our study’s experimental setup assessed the validity of only terms that were not already covered by existing lexicons. Thus, in the case of a semantic category and a lexicon with good coverage, our method has less opportunity to identify new terms (e.g., RxNorms and medications), but when the existing lexicons are scarce, our method identifies new terms with high accuracy (e.g., emotions).
Implications for Quantitative Research on Online Health Communities
As online health communities become a standard data source for mining information about patients, the underlying lexicons used to retrieve or assess prevalence of different terms must be representative of the way community members communicate. The lexicons we generated contain variations of known terms, which would be difficult to discover otherwise, as well as terms, which are not covered by existing lexicons. By making our code and generated lexicons available, we hope to contribute a valuable resource to the research community.
Limitations
Although the current work can be viewed as an important first step for augmenting lexicons to reflect online health community sublanguage, the results do not have high enough precision to be used without a manual annotator in the loop. Our hope is that the generated lexicons are still useful to researchers, since it is much easier to cross off terms in a generated list that should not in the lexicon, than it is to browse through thousands of posts manually to identify terms representative of the way members communicate (in our experiments, manual review was short and easy, on average 20 minutes per 100 terms). In our future work, we plan to experiment with other unsupervised methods and improve the accuracy of our generated lexicons. Second, our experiments focused on three specific semantic categories. While we chose them, because they are important types of content to know about for the content analysis of a breast cancer community and results indicate that a small seed set can generate valid lexicons for all three categories, we have not generalized our work to other types of semantic categories. Finally, the experiments presented in this paper focus on a single community as underlying corpus. In the future, we would want to compare lexicons learned from different communities specific to the same disease. We have conducted preliminary experiments indicating that, for instance, the lexicon learned in one breast cancer community is a useful resource for another, but further work is needed to generalize this finding. We also plan to test our method on other health communities specific to diseases different from breast cancer, to test the generalizability and robustness of our method.
Conclusion
This paper describes a method to generate a lexicon to represent the language of patient users in a given online health community with respect to specific semantic types. We experiment with a breast cancer forum and detect terms that belong to three semantic types: medications, symptoms and side effects, and emotions. Experimental results show that our method captures the sublanguage of members in these communities with more coverage than existing, general-purpose terminologies do. Furthermore, even with a very small number of seed terms, the method can generate reliable lexicons. This work contributes a building block to quantitative research on online health communities.
Figure 5.
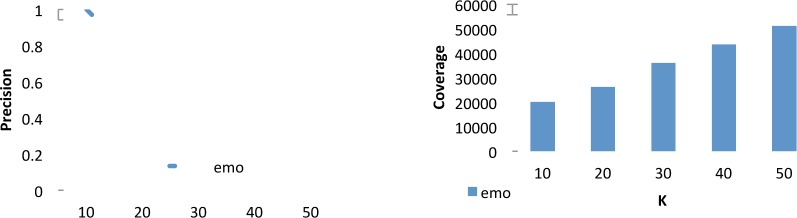
Precision (left) and number of instances covered (right) at K={10,20,30,40,50} for the retrieved emotion terms not in the seed set.
Acknowledgments
This work is supported by an NSF (National Science Foundation) award #1027886 (NE).
References
- 1.Fox S, Duggan M. Health online 2013. Pew Research Center’s Internet & American Life Project 2013. http://pewinternet.org/Reports/2013/Health-online.aspx.
- 2.Rozmovits L, Ziebland S. What do patients with prostate or breast cancer want from an Internet site? A qualitative study of information needs. Patient Educ Couns. 2004;53:57. doi: 10.1016/S0738-3991(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 3.Civan A, Pratt W. Threading together patient expertise. Proc AMIA Annual Symposium. 2007:140–4. [PMC free article] [PubMed] [Google Scholar]
- 4.Meier A, Lyons EJ, Frydman G, et al. How cancer survivors provide support on cancer-related Internet mailing lists. J Med Internet Res. 2007;9 doi: 10.2196/jmir.9.2.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overberg R, Otten W, De Man A, et al. How breast cancer patients want to search for and retrieve information from stories of other patients on the internet: an online randomized controlled experiment. J Med Internet Res. 2010;12 doi: 10.2196/jmir.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Kraut R, Levine J. To stay or leave? The relationship of emotional and informational support to commitment in online health support groups; Proc ACM Conference on Computer-Supported Cooperative Work (CSCW); 2011. [Google Scholar]
- 7.Portier K, Greer GE, Rokach L, et al. Understanding topics and sentiment in an online cancer survivor community. J Natl Cancer Inst Monogr. 2013;47:195–8. doi: 10.1093/jncimonographs/lgt025. [DOI] [PubMed] [Google Scholar]
- 8.Harris ZS. A theory of language and information: a mathematical approach. Clarendon Press; Oxford: 1991. [Google Scholar]
- 9.Friedman C, Kra P, Rzhetsky A. Two biomedical sublanguages: a description based on the theories of Zellig Harris. J Biomed Inform. 2002;35:222–35. doi: 10.1016/s1532-0464(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 10.Nguyen D, Rosé CP. Language use as a reflection of socialization in online communities. Proc Workshop on Language in Social Media (LSM) 2011:76–85. [Google Scholar]
- 11.Unified Medical Language System (UMLS) http://www.nlm.nih.gov/research/umls/
- 12.NCI Metathesaurus. http://ncim.nci.nih.gov/ncimbrowser/
- 13.Zeng QT, Tse T, Divita G, et al. Term identification methods for consumer health vocabulary development. J Med Internet Res. 2007;9:e4. doi: 10.2196/jmir.9.1.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maclean D, Heer J. Identifying medical terms in patient-authored text: a crowdsourcing-based approach. J Am Med Inform Assoc. 2013;20:1120–7. doi: 10.1136/amiajnl-2012-001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yangarber R, Lin W, Grishman R. Unsupervised learning of generalized names. Proc International Conference on Computational Linguistics (COLING) 2002:1–7. [Google Scholar]
- 16.Slaughter L, Ruland C, Rotergard AK. Mapping cancer patients’ symptoms to UMLS concepts. Proc AMIA Annual Symposium. 2005:699–703. [PMC free article] [PubMed] [Google Scholar]
- 17.Alfonseca E, Manandhar S. Extending a lexical ontology by a combination of distributional semantics signatures. Knowledge Engineering and Knowledge Management: Ontologies and the Semantic Web. 2002:281–93. [Google Scholar]
- 18.Zhang S, Elhadad N. Unsupervised biomedical named entity recognition: experiments with clinical and biological texts. J Biomed Inform. 2013;46:1088–98. doi: 10.1016/j.jbi.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonnalagadda S, Cohen T, Wu S, et al. Using empirically constructed lexical resources for named entity recognition. Biomed Inform Insights. 2013;6:17–27. doi: 10.4137/BII.S11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris ZS. Mathematical structures of language. 1968. [Google Scholar]
- 21.Vanteru BC, Shaik JS, Yeasin M. Semantically linking and browsing PubMed abstracts with gene ontology. BMC Genomics. 2008;9:S10. doi: 10.1186/1471-2164-9-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen A, Hersh W, Dubay C, et al. Using co-occurrence network structure to extract synonymous gene and protein names from MEDLINE abstracts. BMC Bioinformatics. 2005;6:103. doi: 10.1186/1471-2105-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elvevaag B, Foltz PW, Weinberger DR, et al. Quantifying incoherence in speech: An automated methodology and novel application to schizophrenia. Schizophr Res. 2007;93:304–16. doi: 10.1016/j.schres.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pivovarov R, Elhadad N. A hybrid knowledge-based and data-driven approach to identifying semantically similar concepts. J Biomed Inform. 2012;45:471–81. doi: 10.1016/j.jbi.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garla VN, Brandt C. Semantic similarity in the biomedical domain: an evaluation across knowledge sources. BMC Bioinformatics. 2012;13:261. doi: 10.1186/1471-2105-13-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen T, Widdows D. Empirical distributional semantics: Methods and biomedical applications. J Biomed Inform. 2009;42:390. doi: 10.1016/j.jbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turney PD, Pantel P, others From frequency to meaning: Vector space models of semantics. J Artif Intell Res. 2010;37:141–88. [Google Scholar]
- 28.Erk K. Vector Space Models of Word Meaning and Phrase Meaning: A Survey. Lang Linguist Compass. 2012;6:635–53. [Google Scholar]
- 29.Jha M, Elhadad N. Cancer stage prediction based on patient online discourse. Proc BioNLP ACL Workshop. 2010:64–71. [Google Scholar]
- 30.Driscoll P, Lipsky Gorman S, Elhadad N. Learning Attribution Labels for Disorder Mentions in Online Health Forums. Proc SIGIR Workshop on Health Search and Discovery. 2013:3–6. [Google Scholar]
- 31.Manning CD, Raghavan P, Schutze H. Introduction to Information Retrieval. Cambridge University Press; 2008. [Google Scholar]
- 32.Nelson SJ, Zeng K, Kilbourne J, et al. Normalized names for clinical drugs: RxNorm at 6 years. J Am Med Inform Assoc. 2011;18:441–8. doi: 10.1136/amiajnl-2011-000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn M, Campillos M, Letunic I, et al. A side effect resource to capture phenotypic effects of drugs. Mol Syst Biol. 2010;6:343. doi: 10.1038/msb.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pennebaker JW, Francis ME, Booth RJ. Linguistic inquiry and word count: LIWC 2001. 2001. [Google Scholar]