Abstract
Condition-specific registries are essential resources for supporting epidemiological, quality improvement, and clinical trial studies. The identification of potentially eligible patients for a given registry often involves a manual process or use of ad hoc software tools. With the increased availability of electronic health data, such as within Electronic Health Record (EHR) systems, there is potential to develop healthcare standards based approaches for interacting with these data. Arden Syntax, which has traditionally been used to represent medical knowledge for clinical decision support, is one such standard that may be adapted for the purpose of registry eligibility determination. In this feasibility study, Arden Syntax was explored for its ability to represent eligibility criteria for a registry of very low birth weight neonates. The promising performance (100% recall; 97% precision) of the Arden Syntax approach at a single institution suggests that a standards-based methodology could be used to robustly identify registry-eligible patients from EHRs.
Introduction
The increased adoption of Electronic Health Record (EHR) systems for the management of patient data has largely been touted for the ability to improve health care outcomes1–4 as well as to support research endeavors5–7. An essential first step in the enablement of EHR data to support research is the identification of patient cohorts that match a specified set of criteria8,9. Myriad approaches that leverage healthcare standards have been described for identifying such types of patient cohorts within EHR systems to serve as subjects of prospective clinical trials8,10. By contrast, there has been limited description of such types of vendor-agnostic approaches that may also be used to populate condition-specific registries from EHR systems.
Condition-specific registries provide a population-level view of retrospective data that may originate from clinical encounters11,12. Approaches that may be used for identification of eligible patients for clinical trials could potentially be used for identifying patients that fit a specified set of criteria for inclusion in a registry. Previous work has demonstrated the potential to develop automated approaches for identifying patients eligible for clinical trials based on data that are available in an EHR system13. The success of such approaches may also support the development of systems that can further automate the process of populating registries, taking advantage of data that are available within contemporary EHR systems.
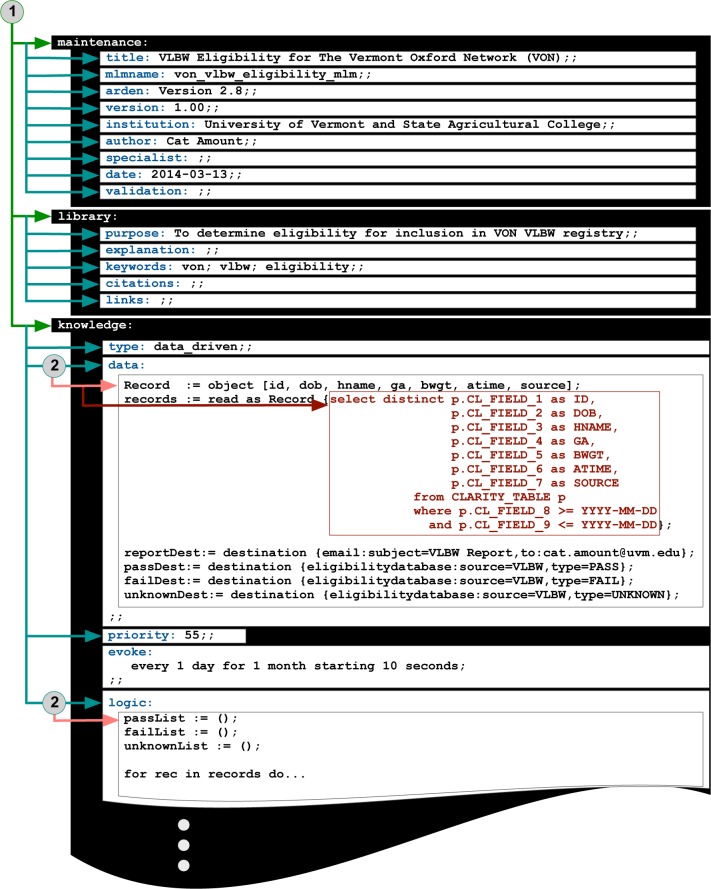
Arden Syntax, which dates back to 1989, is a Health Level 7 (HL7) maintained standard for representing clinical knowledge such that it may be used to support clinical decision-making14. As an HL7 standard, it is regularly updated and supported by a formal working group that oversees the advancement of the standard in accordance with HL7 processes15,16. By maintaining algorithms in self-contained Medical Logic Modules (MLMs), Arden Syntax provides a means to share decision-making rules independent from technical implementation across institutions or environments17,18. MLMs are organized into three major categories (maintenance, library, and knowledge) that store information into “slots.”18 The maintenance and library categories have slots for metadata associated with the management (maintenance category) and categorization (library category) of a given MLM. The knowledge category contains slots that are used for representing the actual clinical knowledge. For example, the data and logic slots within the knowledge category define the variables that will be used within the MLM (data slot) and the procedural logic that is required for representing the clinical knowledge for the MLM (logic slot).
For an MLM written in Arden Syntax to function, it requires that the source data conform to a usable format. This has historically required custom interfaces for accessing EHR-based data17,19–21. By contrast, the secondary use of EHR-based data for research purposes commonly involves the extraction of data into research data repositories. Perhaps the most successful system demonstrating the value of EHR extracted data for secondary use is the Informatics for Integrating Biology and the Bedside (i2b2) platform, which provides a standard and portable interface for browsing health data that may have originated from an EHR22. The use of external frameworks, such as i2b2, therefore helps position research-motivated clinical enterprises to utilize health data for secondary uses5. There remain other contexts, however, where the incorporation of such external systems into the health data ecosystem of a clinical enterprise is not feasible or perhaps even permissible. To address this challenge, many EHR vendors provide a “reporting database” that enables one to query data from the EHR using Structured Query Language (SQL). Similar to how previous studies have demonstrated the potential to leverage Arden Syntax for identifying clinical trial patient cohorts from EHR data23, there may be an opportunity to use similar techniques to identify registry-eligible patient populations.
The Vermont Oxford Network (VON) is a non-profit collaboration that gathers and enables the study of neonatology data from over 900 Neonatal Intensive Care Units (NICUs) that span the globe24,25. The data are gathered for neonates from VON members that meet specified eligibility criteria into de-identified registries that have been used to support a range of activities, including quality improvement projects, clinical trials, and outcomes research25. VON members each develop a process for identifying eligible neonates according to specified VON criteria and provide data systematically using common formats or interfaces that are maintained by VON. The source systems increasingly include healthcare enterprises that utilize an EHR for primary clinical data gathering and analysis. Due to the potential range of EHR options, including both vendor and homegrown systems, there is motivation to develop a systematic and uniform process to identify eligible neonates whose data may be contributed into the VON registries.
This study explored the potential to leverage Arden Syntax for identifying eligible patients from the EHR at an academic health center EHR. Here, the process for adapting Arden Syntax to be used for identifying cohorts of eligible records from contemporary EHR systems with reporting interfaces is described. The performance of the approach is quantified based on an evaluation relative to a reference standard that included previously identified records for a VON registry. The promising results provide the motivation for developing a comprehensive data abstraction tool that can help automate the process of populating condition-specific registries.
Materials and Methods
The overall goal of this study was to explore the potential of using Arden Syntax for representing eligibility criteria to identify patients whose data might be included in a condition-specific registry. The system and approach were developed and evaluated in accordance with a human subjects protocol that was reviewed and approved by the Committee of Human Research in the Medical Sciences at the University of Vermont. The proposed approach was implemented using Java, making use of the ANother Tool for Language Recognition (ANTLR26,27) parser generator to enable the processing of Arden Syntax. The evaluation of the system was carried out using data from Fletcher Allen Health Care (FAHC), the clinical affiliate of the College of Medicine at the University of Vermont.
VON VLBW Eligibility Criteria
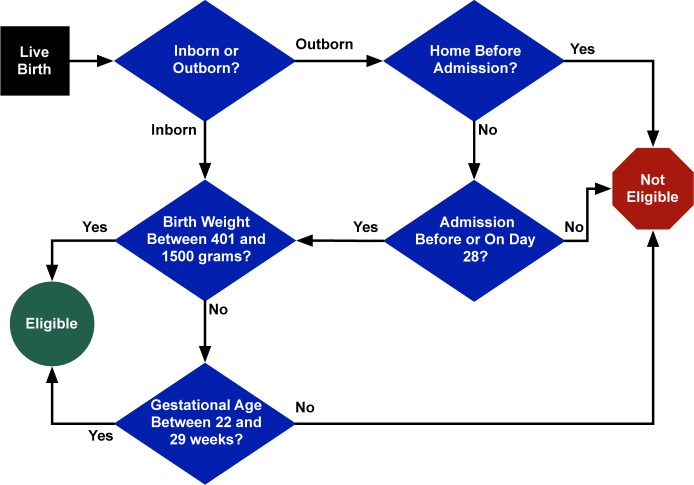
This study focused on the VON Very Low Birth Weight (VLBW) registry, which gathers information on approximately 85% of all VLBW infants born in the US each year. The VON VLBW criteria for eligibility are graphically depicted in Figure 1. Briefly, the registry is focused on gathering data for live-born infants, where status of life is defined as a neonate who breathes or has any evidence for living (e.g., heart beating, umbilical cord pulsation, or definite voluntary muscle movement). Neonates are then categorized into two groups: (1) inborn (where the birth occurs within a particular hospital) and (2) outborn (where the birth occurs outside this hospital but is admitted within 28 days of birth without first having gone home). Additional criteria for inclusion into the VON VLBW registry are defined by birth weight (between 401 and 1500 grams) or gestational age (between 22 and 29 weeks).
Figure 1:
VON VLBW Eligibility Criteria.
Identifying Eligibility Data Fields in the HER
The framework for the approach developed in this study utilized a reporting database that contains data extracted from the EHR. First, all the data elements required for determining eligibility within the MLM were manually identified from within the reporting database (i.e., date of birth, hospital name, gestational age, birth weight, admission date/time, and admission source). The chosen data fields were determined based on a manual review of the available data fields in the EHR and a validation of the chosen data fields relative to the FAHC obstetric and neonatology workflows that have been in place for more than a decade. A SQL statement was then created that retrieved all the data elements in a single query from the FAHC EHR reporting database (equivalent to generating a ‘report’ that included the required data elements).
Representing VON VLBW Eligibility Criteria in Arden Syntax
Within the VLBW MLM data slot, a patient record object was created for storing the eligibility data elements needed as well as links to other data needed for retrieving additional information about the patient (i.e., patient identifier). The object was then populated using the aforementioned SQL statement. The eligibility logic as represented by the flow diagram shown in Figure 1 was then encoded into the logic slot of the VLBW MLM using Arden Syntax. Conversions between local system units to those used in the eligibility criteria were also done within the logic slot (e.g., conversion of the birth weight from imperial ounces to metric grams). To ensure processing of all records retrieved from the EHR, a default categorization of “unknown” was added for patients for whom inborn/outborn status could not be determined (the first step in the eligibility determination as shown in Figure 1 after birth); these patients were automatically deemed not eligible.
Developing a System to Process the VLBW MLM
After eligibility criteria were encoded into the MLM, a Java-based system was developed to interact with the FAHC EHR reporting database using the jTDS28 Java database connectivity application programming interface. Eligibility status for records subjected to the VLBW MLM was then stored into an eligibility database (either “pass”[eligible], “fail”[not eligible], or “unknown”[requires manual review]). For records deemed eligible, an additional Java routine was used to gather electronically available data from the FAHC EHR reporting database using another set of SQL queries. The resulting records were then transformed into a format that could be transmitted to the VON VLBW registry. The overall approach is shown in Figure 2.
Figure 2:
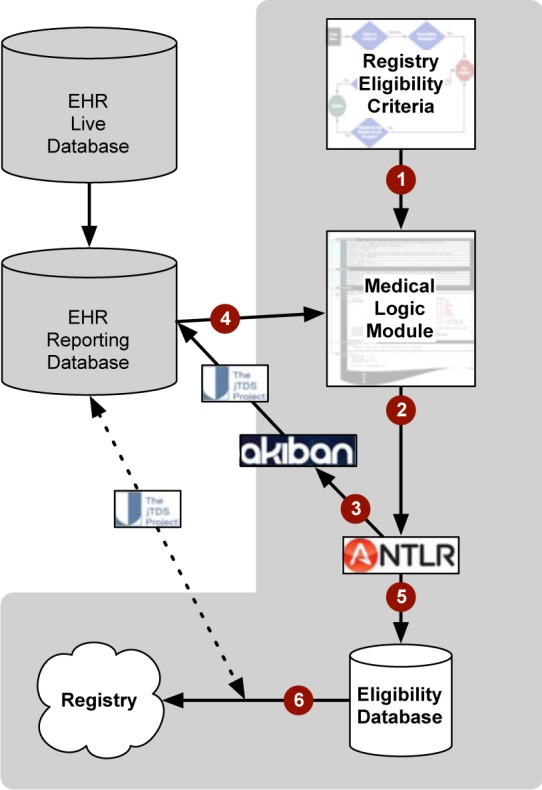
Overview of Approach. (1) Eligibility criteria are manually encoded into an Medical Logic Module (MLM); (2) an ANTLR parser is used to interpret the Arden Syntax; (3) SQL statements are parsed by the Akiban SQL parser and used to send queries to an SQL-92 compliant database using the jTDS Application Program Interface (API), such as an EHR reporting database that uses a relational database model (which is populated from the live EHR database that uses a hierarchical database model); (4) the returned results of the SQL query are then used by the MLM to determine eligibility; (5) eligible records are recorded in a Eligibility Database; (6) the records in the Eligibility Database are enhanced with additional data needed for the registry by additional SQL queries using the jTDS API.
The actual processing of the VLBW MLM was done in a two-phased process, as depicted in Figure 3. First, an ANTLR parser generator was used to extract the MLM categories and associated slots. ANTLR uses a context-free grammar (expressed in Extended Backus-Naur Form) to recognize language, generate syntax trees, and process those trees to create machine-interpretable actions26. An additional ANTLR parser was then used to process the syntax embedded within specific slots (e.g., the data and logic slots). For this study, version 2.8 of Arden Syntax was used as the basis for the parsing grammar. Additionally, the Akiban SQL parser29 was used to interpret embedded SQL-92 compliant SQL statements. The MLM parsing resulted in a parse tree for which a Java routine was developed to interpret the retrieved data from the source database and apply the encoded eligibility rules.
Figure 3:
MLM Parsing. A system was developed that (1) used an ANTLR parser generator to identify the MLM categories (maintenance, library, and knowledge) and their contained slots; and (2) used an additional ANTLR parser generator to process the slots and leveraged the Akiban SQL Parser to process SQL statements embedded within the data slot. For this feasibility study, a SQL statement was used to retrieve data from the FAHC EHR reporting database that were used for eligibility determination (ID = Patient identifier; DOB = patient date of birth; HNAME = hospital name; GA = gestational age; BWGT = birth weight; ATIME = admission time; and SOURCE = admission source) according to the criteria depicted in Figure 1.
System Evaluation
A reference standard was generated from VON VLBW registry records associated with eligible records from the time that the Epic EHR at FAHC was fully deployed (2010) to a full complete calendar year prior to the time of this study (2012). The records contained within the VON VLBW registry for this time period were based on manual chart review and guided by a process workflow that has been in place for over a decade. The VLBW MLM was then configured to determine the eligibility of infants born between January 1, 2010 and December 31, 2012. True Positives (TP) were defined as patients who were deemed eligible by the VLBW MLM and in the reference standard; False Positives (FP) were defined as patients who were deemed eligible by the VLBW MLM but not in the reference standard; and False Negatives (FN) were defined as those patients who were in the reference standard but not deemed eligible by the VLBW MLM. The metrics of precision (TP/TP+FP) and recall (TP/TP+FN) were then used to assess the overall performance of the approach.
Results
All of the necessary fields for determining neonate eligibility for inclusion in the VON VLBW registry were identified within the relational data schema used for the FAHC EHR reporting database. The SQL statement that was used to identify the data for all neonates was then embedded within the Arden Syntax within the data slot of the knowledge category of an MLM. The VON VLBW eligibility criteria were then manually encoded into the MLM (within the logic slot of the knowledge category). Finally, the instructions for writing the status of a given processed record were defined within the action slot of the knowledge category.
A Java-based system was developed to interpret MLMs written in Arden Syntax (version 2.8) as well as embedded SQL-92 compliant SQL statements, utilizing a combination of the ANTLR parsing generator and Akiban SQL parser. The developed VLBW MLM was then used to determine the eligibility of patients born between January 1, 2010 and December 31, 2012. In total, the system processed 12,025 neonates who were either born at or transferred to FAHC for that period, of whom the VLBW MLM deemed 192 to be eligible for inclusion into the VON VLBW registry. The evaluation was done relative to the reference standard, which contained 187 neonates that were manually identified and entered in the VON VLBW registry. The total processing time used for the system to determine the eligibility for all newborn records spanning the three-year period was less than five minutes. Of the eligible patients, 187 were in agreement with the reference standard (TP), five were not found in the reference standard (FP), and no patients in the reference standard were missing from the predicted eligible patient list (FN). The overall precision and recall of the system was thus determined to be 97.4% and 100.0%, respectively.
Discussion
The primary motivation for use of Arden Syntax has historically been described in the context of clinical decision support systems14. Previous studies have explored other potential applications for Arden Syntax, such as for clinical trial eligibility23 and facilitating clinical quality studies30; however, the majority of systems developed to date for processing Arden Syntax have been done within the context of clinical decision support31–34. This study thus reflects a first application of Arden Syntax for the identification of patient cohorts for inclusion in population registries. A recent survey suggests complex clinical trial eligibility representation may require a combination of multiple available formal standards8. In this study, the potential to leverage Arden Syntax for identifying registry-eligible patients was explored, where the eligibility rules could be expressed through procedural statements. The promising results suggest that the proposed approach can be used in place of time-consuming and manual processes to identify registry-eligible patients using eligibility criteria that are compatible with machine-encoded logic (e.g., as can be represented in Arden Syntax).
The use of an algorithm-based approach may provide a means to uniformly apply eligibility criteria for reliable data gathering as required for population-level registries that support public health initiatives. Such uniform application of eligibility algorithms may also be used to identify potential challenges in the use of applications. For example, in the evaluation performed for this study, there were five patients identified by the VLBW MLM but not reported as eligible to the VON VLBW registry (classified in the evaluation as “False Positives”). On closer examination of the data associated with these patients, it was determined that the strict definition of the VON VLBW criteria should have included these individuals. These patients may have been excluded from the registry due to difficult situations (e.g., how are stillborns or planned terminations registered?). While each institution may establish a culture around certain criteria, it can be difficult to ensure consistency when gathering data across many sites. An MLM-driven registry eligibility approach may provide a foundation for high-quality and consistent data acquisition. An interesting future study might be to use the developed VLBW MLM across VON data providers and quantify any possible effect of inclusion/exclusion of certain patient types based on hospital culture. Such a study may help to refine the inclusion/exclusion criteria used in the algorithm to better reflect accepted and consistent practices for a given patient population.
A major potential challenge that has been noted in general deployment of MLMs is the difficulty in accommodating the heterogenic nature of EHR implementations across institutions. The ability to use MLMs is often challenged by the difficulty in completely representing all the local variables needed for the execution. As Arden Syntax specifies that local data variables are to be referenced between a set of curly braces (“{}”), which has led to an referred to as the “curly-brace problem”35,36. Amidst some description of systems that are able to directly interact with transactional database systems37, the majority of previous implementations of MLM driven decision support systems leveraged local relational databases that contained information from EHR systems36. Contemporary solutions have also included the translation of Arden Syntax MLMs into intermediate formalisms (e.g., Drools38). In the approach presented here, EHR data was mapped through the use of SQL-92 compliant statements to acquire data from an EHR reporting database. This SQL statement, as well as the system-specific databases for tracking pass/fail/unknown records, was the major “curly brace” aspect of the developed approach. The increasing availability of SQL-compliant reporting databases with contemporary EHR systems (in many cases to facilitate the ability to meet Meaningful Use requirements) provides a common interface that allows for the development of an MLM that is built around the processing of a set of records that could be generated as part of an EHR report. Still, the approach described here requires database experience with the local EHR reporting database and the ability to generate a SQL statement that contains mappings to the required elements for determining eligibility. Additionally, the same challenges that one might have within a reporting context (e.g., identifying which fields are the relevant ones to retrieve) remain the same in the context of the approach developed here. It is also important to underscore that because the context for which Arden Syntax was used here is for the population of a registry that is based on information that is commonly captured within a clinical chart, it was conceivable that many of the requisite data elements would be available within a reporting interface that is updated nightly. This is in contrast to more typical scenarios where MLMs are used for more real-time clinical decision support, and would thus need to interface directly with the live EHR system. Nonetheless, the promising results demonstrate the application of Arden Syntax for retrospective patient eligibility determination and the potential utility for other contexts, such as retrospective determination of patient cohorts that may be eligible for clinical trials or who may need to be systematically alerted about a potential adverse event.
It must be acknowledged that other formalisms may be able to represent the logic for cohort identification using procedural logic. However, a significant reason for exploring the potential utility of Arden Syntax over other possible (generally more business analytic centric solutions, such as Pentaho) formalisms is because it is a maintained HL7 healthcare standard. As an established and maintained healthcare standard, Arden Syntax is likely to be more readily accepted for integration into healthcare environments. Furthermore, it is likely that one could embody the logic for cohort identification using a set of boutique SQL statements that encode the eligibility criteria, but the portability to another institution would likely be challenged by the need to modify a possibly lengthy SQL script to address variations in schema design. This could be especially difficult when considering that different reporting databases may have very different data structures and implementations of SQL dialects that may not necessarily be able to handle the full logic required for eligibility determination. The variation of SQL implementations and customization could then challenge the ability to centrally manage the process for consistent eligibility determination, since each institution will be required to maintain local customizations of the criteria to meet their data structure and SQL environment. In the context of international registries, such as the VON VLBW registry, these technical considerations are important in light of the range of technical capabilities across data contributors. Nonetheless, the minimal technical requirement that the solution presented here does require is the ability to generate a SQL statement that gathers the fields required to determine eligibility. Future studies that directly benchmark standards based (e.g., Arden Syntax) approaches against alternative approaches for patient cohort identification are needed.
The intent of the system described here was two fold: (1) identify eligible patients for inclusion in a registry; and (2) gather electronically available data from the EHR to transmit for eligible patients to the registry. In this study, the primary focus was on exploring the feasibility of developing a system that was able to identify eligible patients. The evaluation was thus focused on the ability to use the VLBW MLM to reliably identify potentially eligible patients for the VON VLBW registry. While not described here, the system is also able to gather additional data beyond the information needed for determining eligibility using SQL statements and export the full set of gathered data into a number of consumable formats (e.g., as eXtensible Markup Language or a Comma-Separated Value file). However, it should be noted that a remaining major challenge is the identification of the needed data elements from within an EHR system that should be targeted for inclusion into a registry. This requires study of the workflow at a given institution for data gathering from the respective EHR and developing appropriate data transformation techniques. One must also consider that many elements within a registry may not be available as discrete elements (e.g., they may be in natural language form) and thus advanced processing (e.g., using natural language processing [NLP]) may be required before data can be appropriately formatted into the requisite format for the target registry. Currently, if a data element required for eligibility is either inaccessible or not interpretable through simple processing (e.g., regular expressions), a record will be labeled as “unknown” and require manual review to override the status. As such, a user interface is being developed that enables manual review and override of the MLM based eligibility determination that also fits the workflow currently used to populate the VON VLBW registry. A major intent of developing the system described in this study in Java was thus to enable the potential leveraging of Java-based resources that are available for such processing (e.g., NLP could be integrated into the system using the Unstructured Information Management Architecture framework39).
The positive results of the performed evaluation for the VON VLBW registry using the VLBW MLM support the intention to deploy the developed system into production. It is anticipated that the system can be run in production (and set to run on a defined periodic basis through the evoke slot of the knowledge category) parallel to the current method for populating the registry. Additional evaluations will be done to compare differentials in patient classifications, and it is anticipated that if similar performance benchmarks as presented here are achieved for a defined period that the system will be used as the primary mode for identification of eligible patients for inclusion in the VON VLBW registry. While it is conceivable that the approach described here could lead to a fully automated system, it is expected that the system in practice will remain semi-automated; that is, the eligibility status of patients will be presented to users through a curation interface that allows for manual override of status before submission to the registry. Further evaluations are also being planned at other institutions that have a similar SQL-compliant reporting environment as leveraged in this study. As part of the process of deploying the process at other institutions, it is expected that a version of the system described here will be made available for other applications. In the meantime, the corresponding author (INS) may be contacted for the most recent version of the system.
Conclusion
Standards traditionally used for clinical decision support may be utilized to support the population of condition-specific registries. In this study, Arden Syntax was explored as a possible health standard for representing eligibility criteria for a neonatology registry. Based on a system developed to interact with the reporting framework associated with a reporting database that is based on data from an EHR, it was found that a Medical Logic Module written in Arden Syntax was able to perform well relative to a reference standard representing three years of registry-eligible patients. The promising results suggest that Arden Syntax may used to represent eligibility criteria and thus provide a mechanism to leverage electronically available health data (e.g., as encapsulated within an EHR) for supporting the population of patient cohort databases such as condition-specific registries.
Acknowledgments
This work was funded by a contract from the Vermont Oxford Network. Beth Anderson and Jeffrey D. Horbar are employees of the Vermont Oxford Network. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Vermont Oxford Network.
References
- 1.Herrin J, da Graca B, Aponte P, et al. Impact of an EHR-Based Diabetes Management Form on Quality and Outcomes of Diabetes Care in Primary Care Practices. American journal of medical quality : the official journal of the American College of Medical Quality. 2014 Jan 7; doi: 10.1177/1062860613516991. [DOI] [PubMed] [Google Scholar]
- 2.Hunt JS, Siemienczuk J, Gillanders W, et al. The impact of a physician-directed health information technology system on diabetes outcomes in primary care: a pre- and post-implementation study. Informatics in primary care. 2009;17(3):165–74. doi: 10.14236/jhi.v17i3.731. [DOI] [PubMed] [Google Scholar]
- 3.Shekelle PG, Morton SC, Keeler EB. Costs and benefits of health information technology. Evid Rep Technol Assess (Full Rep) 2006 Apr;(132):1–71. doi: 10.23970/ahrqepcerta132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudhry B, Wang J, Wu S, et al. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Annals of internal medicine. 2006 Jun 16;144(10):742–52. doi: 10.7326/0003-4819-144-10-200605160-00125. [DOI] [PubMed] [Google Scholar]
- 5.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nature reviews Genetics. 2011 Jul;12(6):417–28. doi: 10.1038/nrg2999. [DOI] [PubMed] [Google Scholar]
- 6.Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. Journal of the American Medical Informatics Association : JAMIA. 2013 Dec;20(e2):e206–11. doi: 10.1136/amiajnl-2013-002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clinical pharmacology and therapeutics. 2011 Apr;89(3):379–86. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng C, Tu SW, Sim I, Richesson R. Formal representation of eligibility criteria: a literature review. J Biomed Inform. 2010 Jun;43(3):451–67. doi: 10.1016/j.jbi.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abhyankar S, Demner-Fushman D, Callaghan FM, McDonald CJ. Combining structured and unstructured data to identify a cohort of ICU patients who received dialysis. J Am Med Inform Assoc. 2014 Jan 2; doi: 10.1136/amiajnl-2013-001915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuggia M, Besana P, Glasspool D. Comparing semi-automatic systems for recruitment of patients to clinical trials. International journal of medical informatics. 2011 Jun;80(6):371–88. doi: 10.1016/j.ijmedinf.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Collen MF. Computer Medical Databases: The First Six Decades (1950–2010) New York: Springer-Verlag; 2012. [Google Scholar]
- 12.Flagg EW, Datta SD, Saraiya M, et al. Population-based surveillance for cervical cancer precursors in three central cancer registries, United States 2009. Cancer causes & control : CCC. 2014 Feb 28; doi: 10.1007/s10552-014-0362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Embi PJ, Jain A, Clark J, Harris CM. Development of an electronic health record-based Clinical Trial Alert system to enhance recruitment at the point of care; AMIA Annu Symp Proc; 2005. pp. 231–5. [PMC free article] [PubMed] [Google Scholar]
- 14.Samwald M, Fehre K, de Bruin J, Adlassnig KP. The Arden Syntax standard for clinical decision support: experiences and directions. Journal of biomedical informatics. 2012 Aug;45(4):711–8. doi: 10.1016/j.jbi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 15. https://http://www.hl7.org/implement/standards/
- 16.Kawamoto K, Honey A, Rubin K. The HL7-OMG Healthcare Services Specification Project: motivation, methodology, and deliverables for enabling a semantically interoperable service-oriented architecture for healthcare. J Am Med Inform Assoc. 2009 Nov-Dec;16(6):874–81. doi: 10.1197/jamia.M3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poikonen J. Arden Syntax: the emerging standard language for representing medical knowledge in computer systems. American journal of health-system pharmacy : AJHP : official journal of the American Society of Health-System Pharmacists. 1997 Mar 01;54(3):281–4. doi: 10.1093/ajhp/54.3.281. [DOI] [PubMed] [Google Scholar]
- 18.Hripcsak G. Arden Syntax for Medical Logic Modules. MD computing : computers in medical practice. 1991 Mar-Apr;8(2):76–78. [PubMed] [Google Scholar]
- 19.Hripcsak G, Johnson SB, Clayton PD. Desperately seeking data: knowledge base-database links; Proceedings/the Annual Symposium on Computer Application [sic] in Medical Care Symposium on Computer Applications in Medical Care; 1993. pp. 639–43. [PMC free article] [PubMed] [Google Scholar]
- 20.Ohno-Machado L, Parra E, Henry SB, Tu SW, Musen MA. AIDS2: a decision-support tool for decreasing physicians’ uncertainty regarding patient eligibility for HIV treatment protocols; Proc Annu Symp Comput Appl Med Care; 1993. pp. 429–33. [PMC free article] [PubMed] [Google Scholar]
- 21.Pryor TA. The use of medical logic modules at LDS hospital. Comput Biol Med. 1994 Sep;24(5):391–5. doi: 10.1016/0010-4825(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 22.Murphy SN, Mendis ME, Berkowitz DA, Kohane I, Chueh HC. Integration of clinical and genetic data in the i2b2 architecture; AMIA Annu Symp Proc; 2006. p. 1040. [PMC free article] [PubMed] [Google Scholar]
- 23.Ohno-Machado L, Wang SJ, Mar P, Boxwala AA. Decision support for clinical trial eligibility determination in breast cancer; Proceedings/AMIA Annual Symposium AMIA Symposium; 1999. pp. 340–4. [PMC free article] [PubMed] [Google Scholar]
- 24. http://www.vtoxford.org/
- 25.Horbar JD, Soll RF, Edwards WH. The Vermont Oxford Network: a community of practice. Clinics in perinatology. 2010 Mar;37(1):29–47. doi: 10.1016/j.clp.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 26.Parr T. The Definitive ANTLR 4 Reference. Dallas: Pragmatic Programmers; 2013. [Google Scholar]
- 27. http://www.antlr.org/
- 28. http://jtds.sourceforge.net/
- 29. http://www.akiban.com/
- 30.Jenders RA. Suitability of the Arden Syntax for representation of quality indicators; AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium; 2008. p. 991. [PubMed] [Google Scholar]
- 31.Gietzelt M, Goltz U, Grunwald D, et al. ARDEN2BYTECODE: a one-pass Arden Syntax compiler for service-oriented decision support systems based on the OSGi platform. Computer methods and programs in biomedicine. 2012 Jun;106(2):114–25. doi: 10.1016/j.cmpb.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 32.Karadimas HC, Chailloleau C, Hemery F, Simonnet J, Lepage E. Arden/J: an architecture for MLM execution on the Java platform. Journal of the American Medical Informatics Association : JAMIA. 2002 Jul;9(4):359–68. doi: 10.1197/jamia.M0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn RA, Reider RS. A C++ framework for developing Medical Logic Modules and an Arden Syntax compiler. Computers in biology and medicine. 1994 doi: 10.1016/0010-4825(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 34.Jenders RA, Shah A. Challenges in using the Arden Syntax for computer-based nosocomial infection surveillance; Proceedings/AMIA Annual Symposium AMIA Symposium; 2001. pp. 289–93. [PMC free article] [PubMed] [Google Scholar]
- 35.Jenders RA, Corman R, Dasgupta B. Making the standard more standard: a data and query model for knowledge representation in the Arden syntax; AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium; 2003. pp. 323–30. [PMC free article] [PubMed] [Google Scholar]
- 36.Jenders RA, Dasgupta B. Challenges in implementing a knowledge editor for the Arden Syntax: knowledge base maintenance and standardization of database linkages; Proceedings/AMIA Annual Symposium AMIA Symposium; 2002. pp. 355–9. [PMC free article] [PubMed] [Google Scholar]
- 37.Liang YC, Chang P. The development of variable MLM editor and TSQL translator based on Arden Syntax in Taiwan; AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium; 2003. p. 908. [PMC free article] [PubMed] [Google Scholar]
- 38.Jung CY, Sward KA, Haug PJ. Executing medical logic modules expressed in ArdenML using Drools. Journal of the American Medical Informatics Association : JAMIA. 2012 Jul;19(4):533–6. doi: 10.1136/amiajnl-2011-000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. http://uima.apache.org/