Abstract
Natural language processing has been successfully leveraged to extract patient information from unstructured clinical text. However the majority of the existing work targets at obtaining a specific category of clinical information through individual efforts. In the midst of the Health 2.0 wave, online health forums increasingly host abundant and diverse health-related information regarding the demographics and medical information of patients who are either actively participating in or passively reported at these forums. The potential categories of such information span a wide spectrum, whose extraction requires a systematic and comprehensive approach beyond the traditional isolated efforts that specialize in harvesting information of single categories. In this paper, we develop a new integrated biomedical NLP pipeline that automatically extracts a comprehensive set of patient demographics and medical information from online health forums. The pipeline can be adopted to construct structured personal health profiles from unstructured user-contributed content on eHealth social media sites. This paper describes key aspects of the pipeline as well as reports experimental results that show the system’s satisfactory performance in accomplishing a series of NLP tasks of extracting patient information from online health forums.
Introduction
Natural language processing (NLP) has been widely leveraged in the biomedical domain in recent years. It has been shown effective in extracting biomedical information from free-structured text data [1,2,3]. Many applications have been developed to enhance the performance of clinical information retrieval. The extracted information is sometimes restructured or encoded in a special machine-friendly format for automatic computer processing. Most biomedical NLP applications focus on specific areas and thus extract information from text data in single clinical application areas such as radiology reports [4], discharge summaries [5], medication instructions [6,7] and nursing narratives [8]. Typical input to these text extraction tasks is formal medical reports that contain precise clinical information about patients. However these reports do not comprehensively reveal the overall health conditions of a patient because of their narrow focus, definitive purpose, and limited scope of information collection aimed at by a single medical report in a specific area. To better understand patient conditions with more depth and breadth, we need to extract health-related information of patients more richly and diversely. A good class of information sources useful for this purpose is the increasingly emerging and popular online health forums, which contain posts in large numbers written by a variety of patients, caregivers, and supporters. Many of these posts voluntarily offer, through narrative text, a board range of information regarding the personal background and medical conditions of patients, including patients’ demographic and health-related information, such as patients’ living habits, working circumstances, and family history. Such an enriched body of information is typically not included in formal clinical reports. Having access to the personal medical information through the non-traditional communication channel can be a valuable additional resource for clinical and epidemiological research. By extracting the comprehensive scope of patients’ health-related information from the relevant posts on online health forums, this study exploits a new opportunity to access patient information that differs from traditional information extraction approaches relying on formal and specialized clinical reports.
It is noted that many existing studies have attempted to extract information from online forums. For example, Ritter and Mausam propose a method to extract open domain event information from Twitter [9] through topic modeling. Jung propose a name entity recognition method for microtexts in social networks such as Twitter [10]. In [11] Sondhi and Gupta use support vector machine and conditional random field to classify sentences in health forums into two medical categories, including physical examination/symptoms and medications. However they neither extract detailed attributes from those sentences nor attempt to structure or restructure the extracted information. In this paper we propose an algorithmic pipeline that automatically extracts a comprehensive set of patient demographic and health-related information about their posts on online health forums. The goal is to construct structured patient health profiles from unstructured user-contributed content on eHealth social media sites. The derived structured patient health profiles are encoded in the XML format, containing structured and categorized patient information that is easy for computer to read, process, retrieve, and analyze. Such output format can also be easily transformed into different representation models and formats for automated consumption by various third-party systems, e.g. Entity-Relationship (ER) and UML models.
Methods
System Framework
The proposed system comprises a sequence of modules, each responsible for a type of NLP function, which collectively composes our overall application pipeline. The initial input to the pipeline is a corpus of webpages downloaded from a collection of online health forums. The system output is a set of structured patient health profiles. Each module transforms its input data according to its functional role and then subsequently passes the intermediate output to its following module until the final form of the target structured patient profile is generated. The first module is a preprocessor, which extracts the text content of all posts written by or about a user and then merges the result into one user profile. This module also detects and segments the post text into individual sentences for further processing. The second module is a classifier. It assigns each sentence a set of class labels that specify the particular category of patient information delivered by the sentence. The next module is a parser, which utilizes multiple NLP tools to extract health-related attributes from each category of sentences. The last module is an encoder. It generates the final form of the profile of patient information in the XML format. Figure 1 illustrates the overall framework of the proposed system pipeline.
Figure 1:
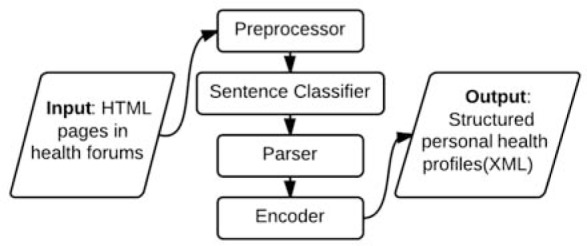
The overview framework of our system pipeline.
Preprocessing
The raw data downloaded from the Internet is a corpus of HTML webpages collected from a range of online health forums. These raw HTML files cannot be directly fed into the text mining modules of our pipeline since they carry lots of text irrelevant to our text mining purpose such as HTML tags and Javascripts codes. To address this problem, we develop an HTML parser to filter out such irrelevant content from the raw webpage downloads. The remaining text is retained for the subsequent analysis. The parser has different parsing rules according to different HTML patterns exhibited by various health forums. To identify and gather all posts written by or about a specific patient in the cyberspace, we perform link analysis on each online forum. All analysis results derived for the same user are then merged into a single file for the person. When identifying a user, we also use the HTML parser to detect the user’s name according to different tag patterns exhibited in HTML pages downloaded from different forums. For example, posts on the American Cancer Society forum express a user’s name through the following string pattern: “<div class=“author”>.*</div>”, where “.*” means a character string of arbitrary letters and of any length. If we encounter a string in the pattern of “<div class=“author”>mxperry</div>”, we can parse this string and extract the user name as “mxperry”. Each forum adopts its own source code pattern. We respectively explore each site’s particular encoding pattern for string parsing when extracting user names. For the tracking purpose, each user is assigned with a unique patient ID number. As mentioned earlier, all detected content corresponding to a common user is then consolidated into a single text file for the user, which we call the user’s online content file. For each resultant online content file corresponding to a specific user, we then apply a sentence splitter to segment the consolidated post content into its constituent sentences. Overall, the preprocessing module transforms the original unorganized collection of webpages downloaded from the Internet to a corpus of personal information files. Each online content file of a user contains the user’s patient ID, name or nickname, followed by a list of sentences written by or about the user on online health forums.
Extracting and Classifying Sentences Containing Patient Demographic and Health-Related Information
Our classification procedure regards each sentence in a user’s online content file as the basic processing element. By classifying every such sentence, the method offers a finer granularity for information access and processing. For example, different researchers may be interested in different aspects of a patient’s information for knowledge discovery and pattern mining. Our method recognizes 11 basic classes of sentences for those appearing on online health forums. Some classes also have sub-classes. The 11 classes are selected based on the medical concepts from the Medical Entity Dictionary (MED) [12], Unified Medical Language System (UMLS) [13], and some extension recommended by the domain experts we consulted with for the project. These resources collectively cover nearly all categories of health-related information supplied by online health forums that are encountered in this study. Table 1 lists all supported classes and sub-classes in this study.
Table 1:
The list of sentence classes and subclasses on eHealth forums supported in this work.
| Class | Description | Subclass | Example |
|---|---|---|---|
| DMO | Demographic information | AGE (age) | 11 years old |
| GEN (gender) | male, female | ||
| ETH (ethnic) | Asian, Hispanic | ||
| BVR | Patient’s behavior | WOK (work) | I was a teacher at 2009 |
| LIF (living habit or behavior) | I have been smoking for 8 years | ||
| STU (study) | I studied for my master degree | ||
| CLI (clinical response) | My back pain relieved after the therapy | ||
| FMA | Family information | My father died of cancer | |
| SPM | Symptom | chest pain, cough | |
| ETR | Exam/Test and Results | TES (test) | chest x-ray |
| RES (result) | Blood pressure is normal | ||
| DIG | Diagnosis | I was diagnosed with stage IV lung cancer | |
| PRO | Medical Procedure/Treatment | surgery, chemotherapy | |
| MED | Medication | I took Lisinopril one time a day | |
| PHY | Physical State | tired, sleepy | |
| PSY | Psychological State | sad, stressed | |
| NOC | Not belonging to any class |
Every sentence in the training set is manually labeled with one or multiple applicable class labels and their corresponding subclass labels, if the latter type of finer-level sentence classification is available. Each sentence may have multiple labels since these sentence classes are not necessarily mutually exclusive, given that a sentence may convey one or more information classes simultaneously. We explore a number of text classification methods such as Naive Bayesian, Bayesian Net, Adaboost, K-Nearest Neighbor, Support Vector Machine (SVM), Sequential Minimal Optimization (SMO), Logistic Regression, Neutral Network, and Decision Tree. Multiple types of text features such as TF-IDF, unigram, bigram, trigram, and words sequence are extracted for input to each candidate classification method. Our implementation chooses the Naive Bayesian classifier as its multi-class sentence classifier in the end because of its robustness and efficiency for short text classification according to empirical evidence. Note that the sequence of sentences may bear an important impact on sentence labeling outcome. For example, a sentence that states a patient’s medical test results (RES) is very likely to follow a sentence that states the patient’s medical test (TES). According to this observation, we utilize the Hidden Markov Model (HMM) to boost our basic Naive Bayesian classifier. Our implementation utilizes the HMM boosting procedure proposed in [14]. The states in the HMM model correspond to the 11 classes of sentences recognized in our method. Experimental results show that our Naive Bayesian classifier boosted by HMM outperforms other candidate classifiers explored. Using the multi-label classifier mentioned in the above, the method can automatically assign one or multiple class labels for each sentence in the patient’s online content file. Those sentences labeled with “NOC,” which stands for “not belonging to any class,” are filtered out.
Parsing Sentences using NLP Tools to Extract Detailed Patient Information
After we extract sentences that contain a patient’s demographic and health-related information using the Naive Bayes classifier, the method can further extract detailed attributes of patient information from each sentence. For sentences of different classes, different natural language processing toolkits are adopted for the parsing task. The patient information extraction result is formatted following the inline XML schema.
Extracting Patient Demographic Information
We use and extend DIVER, a tool developed by Hsieh and Doan [15] that identifies and standardizes demographic variables, to extract patient demographic information from sentences belonging to the class of Demographic information (DMO). In particular, we extract three types of attributes: age, ethnic, and gender. An example is as follows:
Input: <DMO:AGE>I am 35 years old now.</DMO:AGE>
Output: <DMO:AGE>I am <Age>35 years old</Age>now. </DMO:AGE>
Extracting Heath-Related Patient Information
We use MedLEE [1], a medical natural language processing system, to parse sentences that belong to the classes of SPM, ETR, DIG, and PRO. MedLEE has been proved to yield satisfactory performance in processing free-structured clinical text, such as X-ray reports, discharge summaries, sign-out notes, and nursing narratives [2,8]. MedLEE covers a broad range of concepts and semantic rules in the medical domain such as medication, procedure, body measurement, and laboratory test. The output of MedLEE is represented using a frame-based format. Each frame contains the frame type, frame value, and several modifier slots, which are also represented following a frame-based format. We show an example below that demonstrates how MedLEE structures a sentence that belongs to the symptom class.
Input: severe pain in back
Output: [problem, pain, [degree, severe, [bodyloc, back]]
We can easily transform the frame-based output of MedLEE into its corresponding XML format by extracting each value of the frame as an attribute with the type of the frame used as the tag name. An example is shown below on the transformation process.
Input: <SYP>I suffered from a severe pain in my back.</SYP>
Output: <SYP>I suffered from a <Degree>severe</Degree> <Problem>pain</Problem> in my <Bodyloc>back</Bodyloc>.</SYP>
As medical information is often related to time, organization, and location, we also need to extract those attributes. For such purpose, we adopt the Stanford Core NLP toolkit [16]. It contains a named entity recognition tool that can tag seven types of entities, including Time, Location, Organization, Person, Money, Percent, and Date. Leveraging this tool, we can extract and tag the time, date, organization, and location attributes from a sentence. Below is a parsing example on a sentence that discusses a medical procedure:
Input: <PRO>I received chemotherapy at CTCA in Oklahoma in 2009.</PRO>
Output: <PRO>I received <Procedure>chemotherapy</Procedure> at <Organization>CTCA</Organization> in <Location>Oklahoma </Location> in <Date>2009</Date>.</PRO>
Extracting Medication Information
For sentences that discuss medication information, we use the MedEX, a medication NLP tool introduced in [6], to extract concrete attributes involved, such as drugName, strength, route, frequency, form, dose amount, intake time, duration, and dispense amount. MedEx can extract more detailed attributes than MedLEE in the above area. Below is an example:
Input: <MED>I was discharged on Lopressor 50 mg, take 1 Tablet by mouth two times a day.</MED>
Output: <MED>I was discharged on <DrugName>Lopressor</DrugName> <Strength>50 mg</Strength>, take <Dose>1 Tablet</Dose> <Route>by mouth</Route> <Frequency>two times a day<Frequency>. </MED>
Extracting Patient Behavior
For sentences that describe patients’ behaviors, physical and psychological states, as well as family information, the involved information often does not fall into any single domain. To extract those attributes, we need a generalpurpose parsing tool. In this subsection, we are concerned with the method for extracting patient behaviors. Our system implementation utilizes the part-of-speech parser provided by the Stanford Core NLP toolkit, which can generate a parse tree for each input sentence. This parser has a good reputation in processing text in the biomedical domain [17]. An example of the generated parse tree is as follows:
Input: <BHV:LIF>I smoked very often in the last 10 years</BHV:LIF>.
Parse Tree: (ROOT (S (NP (PRP I)) (VP (VBD smoked) (ADVP (RB very) (RB often)) (PP (IN in) (NP (DT the) (JJ last) (CD 10) (NNS years))))))
We first identify the subject of the sentence by finding the noun phrase (NP) that appears immediately before the verb phrase (VP), where both phrases are in a simple declarative clause (S), which must be the child node of the sentence root. In this example, we identify “I” as the subject following the above heuristic. For sentences that describe patient behaviors, our approach assumes that the verb phrase in a predicate usually contains the direct description of patient behavior. Such verb phrase usually locates at the right sibling of the sentence subject. After identifying the verb phrase, we then filter out the descriptive words in it, including the adverb phrase (ADVP) and preposition phrase (PP), so that only verbs would remain. We then tag each remaining verb as a patient behavior attribute. We also use the Stanford named entity parser mentioned in the previous section to label the time, date, organization, and location appearing in an input sentence. An example output is shown below for the same input displayed in the above:
Output: <BHV:LIF>I <Behavior>smoked</Behavior> very often in the <Date>last 10 years</Date> </BHV:LIF>.
Extracting Patient Physical and Psychological States
To extract patient physical and psychological states, we use the same parse tree method mentioned in the above. The difference is that this time the main focus is on extracting adjective phrases in an input sentence instead of verb phrases because adjective phrases are generally more descriptive of patients’ physical and psychological states. In particular, by identifying the adjective words (JJ) in adjective phrases (ADJP), we can extract attributes regarding patient states. An example is as follows:
Input: <PYS>I feel scared and stressed in the first two weeks.</PYS>
Parse Tree: (ROOT (S (NP (PRP I)) (VP (VBP feel) (ADJP (JJ scared) (CC and) (JJ stressed)) (PP (IN in) (NP (DT the) (JJ first) (CD two) (NNS weeks))))))
Output: <PSY>I feel <Psy-state>scared</Psy-state> and <Psy-state>stressed</Psy-state> in the first two weeks.</PSY>
Extracting Patient Family Information
For those sentences that contain family information (FMA) such as family medical history, we extract any mentioning of family member(s) in the sentences. We first use the parse tree to find the subject of an input sentence. We then search the appearance of any subject word according to a vocabulary list regarding different roles of family members according to English Grammar Online [18]. The list contains 45 words. If the subject word appears in the list, we then label it using the tag “Member.” An example is as follows:
Input: <FMA>My father died of lung cancer at the age of 83. </FMA>
Output: <FMA>My <Member>father </Member> died of lung cancer at the age of 83. </FMA>
Results
Data
We downloaded a total of 11274 webpages from a number of US health forums and hospital association websites, such as American Cancer Society’s Cancer Survivors Network, eHealth Forum, PatientsLikeMe, etc. After the pre-processing stage, we constructed 3196 patients’ personal online content files using the pipeline discussed earlier. After segmenting these content files into individual sentences, we obtained 9730 unique sentences as our full experimental data set. We then invited 4 researchers in the domain of medical informatics to manually assign sentence class labels for all these sentences with the help of MED and UMLS. Table 2 shows the 10 online health forums where we collect our data and their size of patient population respectively.
Table 2:
Ten online forums and their respective URLs and user populations.
| Forum Name | URL | Population |
|---|---|---|
| American Cancer Society | http://www.cancer.org | 401 |
| eHealth Forum | http://ehealthforum.com | 319 |
| PatientsLikeMe | http://www.patientslikeme.com | 222 |
| HealingWell.com | http://www.healingwell.com | 335 |
| Seattle Cancer Care Alliance | http://www.seattlecca.org | 380 |
| Gibis Cancer Center | http://www.gibbscancercenter.com | 303 |
| Team Draft | http://www.teamdraft.org | 346 |
| American Lung Association | http://www.lung.org | 298 |
| HuffPost Impact | http://www.huffingtonpost.com/impact | 323 |
| APIAHF | http://www.apiahf.org | 296 |
Sentence Classification
To explore the effectiveness of the new pipeline, we tested the overall performance of different classifiers mentioned in Section 3. We use Weka [19], a widely used machine learning software, to run our experiments. From the whole dataset we randomly chose 7000 sentences as the training data and the rest 2730 sentences as the testing data. We adopted the cross-validation schema to benchmark the learning performance. After we train our sentence classifier through the training dataset, we measured its performance on the testing dataset. We performed ten-fold cross validation to measure the precision, recall, and F-rate of the method.
Table 3 shows that in terms of the sentence classification performance, the Naive Bayesian classifier boosted by HMM outperforms all other classifiers. Although the rest of the classifiers may be more effective for tackling certain text classification tasks, for the particular sentence classification task concerned in this study, we adopted Naive Bayes as the optimal learning device for our implementation. Table 4 shows that the sentence classification module implemented in our pipeline yields generally satisfactory performance in terms of its precision, recall, and F-rate, especially in the medically related areas. In such areas, text tends to be written using more medical terms and formal language than those in the non-medical areas. This particular language characteristic may affect the choice of the optimal text classifier. Figure 2 shows the percentages of sentences in each class. According to the statistics, each sentence class has enough samples for training the corresponding classifier.
Table 3:
Performance comparision regarding the 9 sentence classifiers: Naive Bayes boosted by HMM (NB-HMM), Naive Bayes (NB), Bayes Network (BN), Adaboost (AD), K-Nearest Neighbor (KNN), Support Vector Machine (SVM), Neutral Network (NN), Sequential Minimal Optimization (SMO), Logistic, and Decision Tree (DT).
| Classifier | Precision (%) | Recall (%) | F-rate (%) | Classifier | Precision (%) | Recall (%) | F-rate (%) |
|---|---|---|---|---|---|---|---|
| NB-HMM | 93.2 | 90.9 | 91.7 | SVM | 82.2 | 70.3 | 75.8 |
| NB | 91.3 | 89.6 | 90.4 | NN | 76.3 | 74.5 | 75.4 |
| BN | 90.8 | 87.6 | 89.1 | SMO | 77.9 | 70.8 | 73.8 |
| AD | 86.6 | 83.2 | 80.1 | DT | 87.5 | 81.1 | 84.2 |
| KNN | 82.7 | 68.9 | 75.2 | ||||
Table 4:
Performance of different sentence classifiers.
| Sentence Class | Precision (%) | Recall (%) | F-rate (%) | Sentence Class | Precision (%) | Recall (%) | F-rate (%) |
|---|---|---|---|---|---|---|---|
| DMO | 90.6 | 89.6 | 90.1 | SPM | 85.7 | 84.3 | 85.0 |
| DMO:AGE | 89.6 | 87.2 | 88.3 | ETR | 91.1 | 91.5 | 91.3 |
| DMO:GEN | 93.3 | 90.6 | 91.9 | ETR:TES | 91.1 | 89.8 | 90.4 |
| DMO:ETH | 87.0 | 85.6 | 86.3 | ETR:RES | 91.3 | 91.1 | 91.1 |
| BHV | 81.7 | 78.0 | 79.8 | DIG | 93.2 | 90.2 | 91.6 |
| BHV:WOK | 78.8 | 75.7 | 77.2 | PRO | 82.9 | 79.3 | 81.1 |
| BHV:LIF | 78.1 | 74.3 | 76.2 | MED | 95.5 | 91.1 | 93.2 |
| BHV:STU | 82.5 | 80.0 | 81.2 | PHY | 80.3 | 78.8 | 79.5 |
| BHV:CLI | 85.3 | 81.2 | 83.1 | PSY | 89.1 | 85.4 | 87.2 |
| FMA | 88.6 | 85.8 | 87.2 | NOC | 80.2 | 86.5 | 83.2 |
Figure 2:
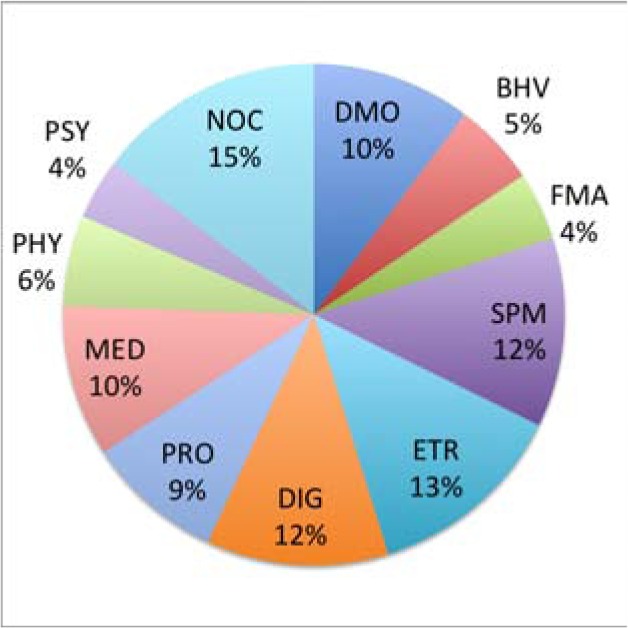
Percentage of sentences in each class.
Extracting Detailed Patient Information
For the 9730 sentences analyzed in our experiments, those labeled with “NOC” are first excluded. We then utilize a set of NLP tools integrated in our prototype system implementation to extract attributes containing detailed patient information for different classes of sentences respectively. For the evaluation and benchmarking purpose, the information extraction accuracy is manually verified by four domain experts according to MED, UMLS, and some extensions made by them based on the sentence classes not related to MED and UMLS directly. Since the demographic information usually requires more precise extraction, we further report the information extraction performance for each demographic attribute. To further demonstrate the effectiveness of the information extraction module, we compare its performance with that of another peer concept extraction tool—BioTagger-GM [20]. The peer tool uses machine learning techniques to train semantic taggers for extracting medical concepts from clinical documents. We apply both our method and the peer method to extract ten classes of information concerned in this study, i.e. all sentence classes except for the “NOC” class. The results yielded by both methods are then compared. Figure 3 shows that our proposed information extraction pipeline, which collaboratively leverages multiple NLP tools, consistently outperforms the peer method.
Figure 3:
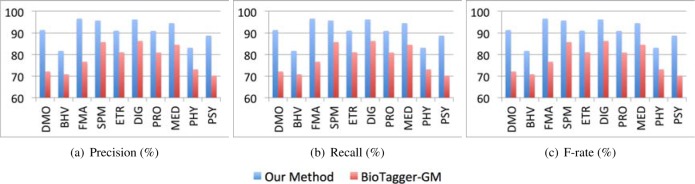
Comparison of information extraction performance between our proposed method and the peer method.
Table 5 shows that the information extraction module of our implemented pipeline performs satisfactorily for all sentence classes. Some categories received relatively low F-rates such as life behavior. A potential reason was due to the more complex language and ambiguous vocabularies often used for conveying information of these categories.
Table 5:
Performance of information extraction in different sentence classes.
| Class | Precision(%) | Recall(%) | F-rate(%) | Class | Precision(%) | Recall(%) | F-rate(%) |
|---|---|---|---|---|---|---|---|
| DMO | 91.3 | 86.3 | 88.7 | SPM | 95.7 | 87.3 | 91.3 |
| DMO:AGE | 87.2 | 83.3 | 85.2 | ETR | 91.0 | 88.5 | 89.7 |
| DMO:GEN | 97.6 | 96.1 | 96.8 | ETR:TES | 89.1 | 86.8 | 87.9 |
| DMO:ETH | 90.6 | 87.6 | 89.0 | ETR:RES | 91.9 | 88.2 | 90.0 |
| BHV | 81.7 | 78.0 | 79.8 | DIG | 96.2 | 91.3 | 93.6 |
| BHV:WOK | 88.8 | 79.7 | 84.0 | PRO | 90.9 | 85.4 | 88.0 |
| BHV:LIF | 78.0 | 76.3 | 77.1 | MED | 94.5 | 92.1 | 93.1 |
| BHV:STU | 81.5 | 78.1 | 79.8 | PHY | 83.1 | 79.9 | 81.4 |
| BHV:CLI | 89.3 | 86.2 | 87.7 | PSY | 88.7 | 84.2 | 86.3 |
| FMA | 96.6 | 95.8 | 96.2 |
Coverage of Patient Information on Different Health Forums
We examine the distributions of different categories of patient information on different online health forums and websites. The amount and range of demographic and health-related patient information available on a forum indicate the forum’s richness of information on certain patient conditions. We measure the information coverage of a forum using the ratio of the users on the forum whose extracted health profiles contain the desired class of information. Table 6 shows the respective sizes of patient populations on top ten most popularly encountered health forums in our study. We can see that although the information coverage varies among different domains and health forums, our system can extract a relatively comprehensive set of patient information since the information coverage ratios for all classes on each of the top ten forums consistently exceed 60%.
Table 6:
Coverage of different classes of information on top ten most popular online health forums in this study.
| Forum | Class | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DMO | BHV | FMA | SPM | ETR | DIG | PRO | MED | PHY | PSY | |
| Coverage(%) | ||||||||||
| American Cancer Society | 77.6 | 70.3 | 65.5 | 86.6 | 83.2 | 87.9 | 74.3 | 79.6 | 68.1 | 70.0 |
| eHealth Forum | 83.6 | 70.8 | 61.4 | 80.4 | 71.9 | 86.3 | 86.4 | 84.4 | 73.9 | 63.2 |
| PatientsLikeMe | 80.3 | 87.4 | 62.9 | 86.0 | 79.0 | 78.6 | 86.5 | 71.6 | 72.6 | 73.4 |
| HealingWell.com | 77.8 | 66.6 | 86.0 | 71.2 | 77.9 | 80.5 | 78.3 | 81.3 | 72.5 | 65.8 |
| Seattle Cancer Care Alliance | 78.6 | 70.3 | 59.6 | 73.3 | 72.1 | 77.4 | 73.9 | 79.7 | 66.7 | 69.0 |
| Gibis Cancer Center | 78.4 | 61.0 | 84.7 | 75.3 | 78.4 | 80.9 | 88.8 | 78.3 | 69.6 | 66.0 |
| Team Draft | 84.0 | 63.3 | 60.7 | 83.9 | 83.4 | 73.5 | 72.5 | 89.9 | 63.4 | 70.6 |
| American Lung Association | 81.2 | 87.6 | 83.3 | 73.8 | 77.3 | 79.2 | 89.6 | 73.1 | 67.1 | 72.8 |
| HuffPost Impact | 78.5 | 69.4 | 62.4 | 81.7 | 74.5 | 77.6 | 81.6 | 75.0 | 75.8 | 82.3 |
| APIAHF | 70.0 | 78.2 | 56.7 | 88.1 | 80.3 | 76.7 | 77.5 | 70.3 | 69.9 | 65.8 |
Distribution of Patient Demographic Information on Different Forums
By extracting patient’s demographic information we can further analyze the demographical composition of different patient groups, which can help medical researchers more insightfully analyze the common characteristics and behavior features of the group of the patients. For this need, the implemented prototype system also aims to extract comprehensive patient demographic information from online health forums. Figures 4 and 5 respectively present the age, gender, and ethnic distribution of users on the top ten most popular health forums concerned in this study. Figure 4 shows the age curves of patients extracted from the ten forums respectively. Their peaks fall into the range of 30 to 55 years old. We assume that the sparse presence of old patients might be partially caused by the inability of those people in accessing and participating on online health forums. We also find that the average age of patients on cancer forums is greater than that on general health forums, which indicates cancer may be more likely to affect older people than young people. In general, the patient demographic statistics extracted can provide valuable clues for analyzing online patient content in an age-adjusted manner.
Figure 4:
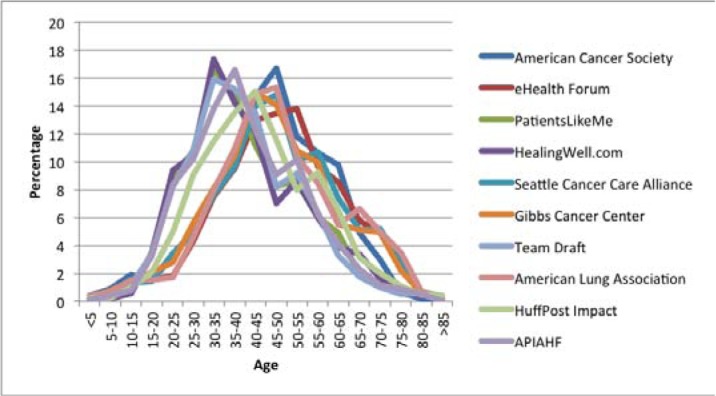
Patient age distributions on each of the ten most popular forums analyzed in this study.
Figure 5:
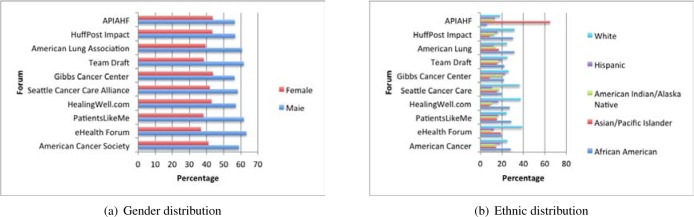
Patient gender and ethnic distributions on each of the ten most popular forums analyzed in this study.
Discussion
The proposed biomedical NLP pipeline and its prototype system implementation extract patients demographic and health-related information in a wide range of categories from online health forums. For each category of patient information present on health forums, a corresponding set of NLP procedures are applied to extract values of detailed category attributes. The set of supported information categories is also adaptive, which can be adjusted and expanded over the time to extract more comprehensive patient information. One potential limitation of this work is that a patient may register on mutiple health forums and possibly using different user names or nicknames, none of which may be their real names. How to detect and merge different profiles of the same patient into one consolidatd record is a meaningful algorithmic problem deserving immediate future studies.
Conclusion
We proposed and developed a software pipeline for extracting and structuring patient’s personal and health related information from online health forums. Our experimental results demonstrated the usability and effectiveness of the prototype implementation. The automatically acquired health profiles of online patients can provide valuable informational aid for cyber-mining based eHealth research.
Acknowledgments
This study was performed under the Protocol, F 186-14, approved by the Institutional Review Board (HHS FWA #00003246) as well as the Protocal, ORNL(12)-130, approved by the Oak Ridge Site-Wide IRB (HHS FWA #00005031). The study was funded in part by the National Cancer Institute (Grant #: 1R01CA170508).
References
- [1].Friedman Carol. Towards a comprehensive medical language processing system: methods and issues; Proceedings of the AMIA annual fall symposium; American Medical Informatics Association; 1997. p. 595. [PMC free article] [PubMed] [Google Scholar]
- [2].Friedman Carol. A broad-coverage natural language processing system; Proceedings of the AMIA Symposium; American Medical Informatics Association; 2000. p. 270. [PMC free article] [PubMed] [Google Scholar]
- [3].Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. Journal of the American Medical Informatics Association. 1994;1(2):142–160. doi: 10.1136/jamia.1994.95236145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Friedman C, Alderson Philip O, Austin John HM, Cimino James J, Johnson Stephen B. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association. 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sigfried GM, Elhadad N, Zhu XX, Cimino JJ, Hripcsak G. Extracting structured medication event information from discharge summaries. 2008. [PMC free article] [PubMed] [Google Scholar]
- [6].Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. Medex: a medication information extraction system for clinical narratives. Journal of the American Medical Informatics Association. 2010;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deléger L, Grouin C, Zweigenbaum P. Extracting medical information from narrative patient records: the case of medication-related information. Journal of the American Medical Informatics Association. 2010;17(5):555–558. doi: 10.1136/jamia.2010.003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hyun S, Johnson SB, Bakken S. Exploring the ability of natural language processing to extract data from nursing narratives. Computers Informatics Nursing. 2009;27(4):215–223. doi: 10.1097/NCN.0b013e3181a91b58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ritter A, Etzioni O, Clark S, et al. Open domain event extraction from twitter; Proceedings of the 18th ACM SIGKDD international conference on Knowledge discovery and data mining; ACM; 2012. pp. 1104–1112. [Google Scholar]
- [10].Jung Jason J. Online named entity recognition method for microtexts in social networking services: A case study of twitter. Expert Systems with Applications. 2012;39(9):8066–8070. [Google Scholar]
- [11].Sondhi P, Gupta M, Zhai CX, Hockenmaier J. Shallow information extraction from medical forum data; Proceedings of the 23rd International Conference on Computational Linguistics: Posters; Association for Computational Linguistics; 2010. pp. 1158–1166. [Google Scholar]
- [12].JJ Cimino, PD Clayton, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology. Journal of the American Medical Informatics Association. 1994;1(1):35–50. doi: 10.1136/jamia.1994.95236135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bodenreider Olivier. The unified medical language system (umls): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl 1):D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Xu R, Supekar K, Huang Y, Das A, Garber A. Combining text classification and hidden markov modeling techniques for structuring randomized clinical trial abstracts; AMIA Annual Symposium Proceedings; American Medical Informatics Association; 2006. p. 824. [PMC free article] [PubMed] [Google Scholar]
- [15].Hsieh A, Doan S, Conway M, Lin KW, Kim H. Demographics identification: Variable extraction resource (diver); Healthcare Informatics, Imaging and Systems Biology (HISB), 2012 IEEE Second International Conference on; IEEE; 2012. pp. 40–49. [Google Scholar]
- [16].NLP Stanford. Toolkits.
- [17].Huang Y, Lowe HJ, Klein D, Cucina RJ. Improved identification of noun phrases in clinical radiology reports using a high-performance statistical natural language parser augmented with the umls specialist lexicon. Journal of the American Medical Informatics Association. 2005;12(3):275–285. doi: 10.1197/jamia.M1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].English Grammar Online. http://www.ego4u.com/. [Online; accessed 1-March-2014]
- [19].Witten IH, Frank E, Trigg LE, Hall MA, Holmes G, Cunningham SJ. Weka: Practical machine learning tools and techniques with java implementations. 1999. [Google Scholar]
- [20].Torii M, Wagholikar K, Liu HF. Using machine learning for concept extraction on clinical documents from multiple data sources. Journal of the American Medical Informatics Association. 2011:amiajnl–2011. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]


