Abstract
In this paper, we present the results of a method using undirected paths to determine the degree of semantic similarity between two concepts in a dense taxonomy with multiple inheritance. The overall objective of this work was to explore methods that take advantage of dense multi-hierarchical taxonomies that are more graph-like than tree-like by incorporating the proximity of concepts with respect to each other within the entire is-a hierarchy. Our hypothesis is that the proximity of the concepts regardless of how they are connected is an indicator to the degree of their similarity. We evaluate our method using the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT), and four reference standards that have been manually tagged by human annotators. The overall results of our experiments show, in SNOMED CT, the location of the concepts with respect to each other does indicate the degree to which they are similar.
1 Introduction
The automated discovery of groups of semantically similar concepts and terms is critical to improving the retrieval1 and clustering2 of biomedical and clinical documents, and the development of biomedical terminologies and ontologies3. Additionally, semantic similarity measures could be used indirectly in applications such as finding articles with similar content in PubMed4, and clustering symptoms and disorders found in the text of clinical reports for post-marketing medication safety surveillance56. Similarity measures quantify the degree to which two concepts are similar based on their taxonomical proximity through the type-of (or is-a) relationships that exist between them. This is often referred to as a hyponym relationship where one term’s ancestral pedigree is included within that of another term. The path passing through a common descendant would link the two concepts. The undirected path (u-path) measure is a method to obtain the degree of semantic similarity between two concepts in lexical resources with significant multiple inheritance and are more graph-like than tree-like. This measure quantifies this degree based on the reciprocal of the shortest path between two concepts regardless of the direction of graph traversal.
Other similarity measures use the shortest path in their calculation but require that the concepts be connected through their least common subsumer (LCS) such as the conceptual distance measure proposed by Rada, et. al1 and subsequently implemented by Caviedes and Cimino7, the measure proposed by Leacock and Chodorow8, and the path measure, which we refer to as lcs-path, as implemented by Pedersen, et. al9. The lcs-path measure and the measure proposed by Leacock and Chodorow8 were initially developed with WordNet10 as their lexical resource. Concepts in WordNet are primarily organized in a acyclic hierarchy, free from multiple inheritance, therefore the shortest path between any two concepts would contain the LCS. This is not the case though for other taxonomies such as the Systematized Nomenclature of Medicine Clinical Terms11 (SNOMED CT), which is a dense multi-hierarchical taxonomy of clinical terms.
The u-path measures relaxes the requirement that the path between the two concepts must go through the LCS. Figure 1 shows the individual shortest path lengths used by the lcs-path and u-path measures to calculate the degree of similarity between Neuropathy and Paralysis in SNOMED CT. Neuropathy and Paralysis are both disorders that may involve the peripheral nervous system. In this example, the length of the shortest path is four but increases to ten when requiring the path to be connected through the concepts’ LCS.
Figure 1:
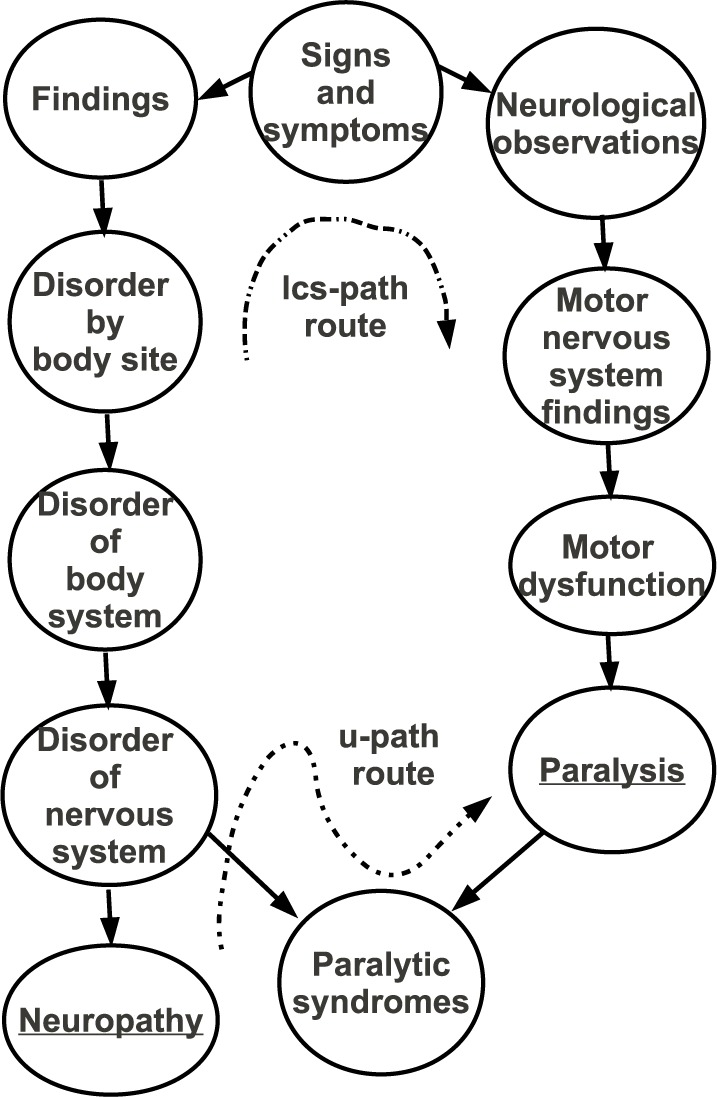
Example of the lcs-path and u-path measures
The overall objective of this work is to begin exploring methods that take advantage of dense multi-hierarchical taxonomies. Our hypothesis is that the proximity of the concepts, regardless of how they are connected, is an indicator of the degree of their similarity. Towards this end, we evaluate u-path using SNOMED CT and four reference standards that have been manually tagged by human annotators. The results show, in SNOMED CT, the location of the concepts with respect to each other regardless of their LCS can be used to indicate the degree to which they are similar.
2 Semantic Similarity Measures
Existing semantic similarity measures can be categorized into two groups: path-based and information content (IC)-based. Path-based measures rely solely on the shortest path information, whereas IC-based measures incorporate the probability of the concept occurring in a corpus of text.
Path-based Measures
Rada, et al.1 introduces the conceptual distance measure (c-dist), which is calculated as the length of the shortest path between two concepts that connects the concepts through their least common subsumer (LCS). The LCS is the most specific ancestor shared by two concepts. The length is calculated by counting the number of nodes between the two concepts. The lcs-path measure is a modification of this and is calculated as the reciprocal of the length of the shortest path as shown for concepts c1 and c2 in Equation 1.
| (1) |
Wu and Palmer12 extend this measure by incorporating the depth of the LCS. In a multi- hierarchical taxonomy, we define the depth to be the minimum path between the concept and the root. In this measure, the similarity is twice the depth of the two concepts LCS divided by the sum of the depths of the individual concepts as defined in Equation 2.
| (2) |
Leacock and Chodorow8 extend the path measure by incorporating the depth of the taxonomy. Here, the similarity is the negative log of the shortest path between two concepts divided by twice the total depth of the taxonomy (D) as defined in Equation 3.
| (3) |
Nguyen and Al-Mubaid13 incorporate both the depth and LCS in their measure. In this measure, the similarity is the log of two plus the product of the shortest distance between the two concepts minus one and the depth of the taxonomy (D) minus the depth of the concepts LCS (d). Its range depends on the depth of the taxonomy.
| (4) |
Batet, et al.14 introduce a measure that takes in account the common concepts shared (referred to as shared superconcepts) between the two concepts (ci and cj) and their LCS (lcs(ci, cj). A concept’s (ci) set of superconcepts (T (ci)) consist of all of the concepts found in all of the shortest paths between ci and the LCS. In this measure the log ratio of the shared superconcepts as defined in Equation 5 where T (ci) = {cj ∈ C|cj is a superconcept of ci}.
| (5) |
Information Content-based Measures
Information content (IC) measures the specificity of a concept in a hierarchy. The fundamental assumption with the IC measures is that the more frequent a concept is, the less specific it is. Therefore, a concept with a high IC value is more specific to a topic than one with a low IC value. IC is formally defined as the negative log of the probability of a concept (c*) as shown in Equation 6.
| (6) |
The probability of a concept is determined by summing the probability of the concept (P (c)) occurring in some text plus the probability its descendants (P (d)) occurring in some text as shown in Equation 7
| (7) |
The initial probability of a concept (P (c)) and its descendants (P (d)) is obtained by dividing the number of times a concept is seen in the corpus (freq(d)) by the total number of concepts (N) as seen in Equation 8.
| (8) |
Resnik15 modified IC to be used as a similarity measure. He defined the IC of two concepts (c1 and c2) to be the IC of their least common subsumer (LCS) as seen in Equation 9. The LCS is the most specific concept two concepts share as an ancestor; if two concepts have more than one LCS, we use the most specific one.
| (9) |
Jiang and Conrath16 and Lin17 extended Resnik’s IC-based measure by incorporating the IC of the individual concepts. Lin defined the similarity between two concepts by taking the quotient between twice the IC of the concepts’ LCS and the sum of the IC of the two concepts as seen in Equation 10.
| (10) |
Jiang and Conrath defined the distance between two concepts to be the sum of the IC of the two concepts minus twice the IC of the concepts’ LCS. We modify this measure to return a similarity score by taking the reciprocal of the distance as seen in Equation 11.
| (11) |
3 Method
As discussed in Section 2, lcs-path is the reciprocal of the length of the shortest path between two concepts in a hierarchy in which the shortest path is calculated by first finding the LCS of the two concepts and then aggregating the distance of all paths that connect the concepts through the LCS. In the u-path measure, the paths are not required to contain the LCS and may meander through the hierarchy. We view u-path as a measure of similarity because it is based strictly on path information found in is-a relations, but we have relaxed the requirement that the path between the two concepts must go through the LCS.
The u-path measure is related to the measure proposed by Hirst and St. Onge18 (hso) that, like u-path, quantifies the strength of similarity between two concepts based on their closeness within a hierarchy. The measure assumes that two concepts are semantically close in a taxonomy if they are connected by a path that is neither too long nor too meandering and the relations between them are either is-a or has-a relations. The u-path measure has a similar assumption but does not put restrictions on the length or meandering of the path, only the type of relation used.
In a subsequent studies using WordNet, Budanitsky and Hirst19 report that hso obtains similar results to the path-based measure proposed by Leacock and Chodorow8, which is similar to the results reported by Patwardhan, et. al20. We believe this is because hso, like u-path, relies on the concepts being densely connected. Unfortunately, the disadvantage of using hso on a densely connected graph is that it requires keeping track of the length and direction changes of all of the paths between the two concepts. This becomes difficult and in some cases infeasible when using dense multi-hierarchical structures, like SNOMED CT. The u-path measure presented here, does not have this limitation.
4 SNOMED CT
The Systematized Nomenclature of Medicine–Clinical Terms11 (SNOMED CT) is a comprehensive clinical terminology created for the electronic representation of clinical health information and is one of the terminology sources in the Unified Medical Language System (UMLS) Metathesaurus. The 2010AB version of the Metathesaurus contains over 1.7 million biomedical and clinical concepts from over 100 different terminology sources that have been semi-automatically integrated into a single resource. The terminology sources in the Metathesaurus can be treated independently or in combination with other sources. Currently, SNOMED CT is the largest hierarchical terminological source in the Metathesaurus.
The Metathesaurus contains a variety of different links between concepts specifying their relationship. The two hierarchical relations, used in this study, are parent/child (PAR/CHD) and broader/narrower (RB/RN) relations. A PAR/CHD is a hierarchical relation between two concepts that has been explicitly defined by the source. An RB/RN relation is a hierarchical relation that does not explicitly come from a source but is created by the UMLS editors during the integration process. In the case of SNOMED CT, the PAR/CHD relations are strictly is-a relations and the RB/RN relations contain part-of and was-a relations. We use the PAR/CHD relations for our experiments.
In version 2010AB, SNOMED CT contains 280,695 concepts with a PAR/CHD relation; 198,241 leaf nodes and 82,454 non-leaf nodes. The depth of the taxonomy is 34, the average depth of a non-leaf node is 9.2 and the average depth of a leaf node is 11.8. The average branching factor of a non-leaf node is 5.1 and on average each node (concept) has 51 distinct paths to the root.
5 Reference Standards
We use four reference standards a to evaluate the u-path measure: the UMNSRS tagged for similarity, the UMNSRS tagged for relatedness, the MayoSRS tagged for relatedness and the MiniMayoSRS tagged for relatedness. We include reference standards tagged for relatedness in our evaluation in order to conduct a comparison with previous work and due to the scarcity of datasets tagged strictly for similarity. Relatedness measures quantify the relationship between two concepts that are not necessarily in a strict is-a or hyponym relationship; it is domain-dependent and grounded in human perception which takes into account that two concepts may be related in other ways. For example up is the opposite of down, an elbow is part-of an arm, and a scalpel cuts tissue. In this section, we describe the reference standards and then briefly discuss some of their differences.
MayoSRS: MayoSRS, developed by Pakhomov, et al.21, consists of 101 clinical term pairs whose relatedness was determined by nine medical coders and three physicians from the Mayo Clinic. The relatedness of each term pair was assessed based on a four point scale: (4.0) practically synonymous, (3.0) related, (2.0) marginally related and (1.0) unrelated. We evaluate our method on the mean score of the physicians and medical coders as provided by Pakhomov, et al.21.
MiniMayoSRS: MiniMayoSRS is a subset of the MayoSRS and consists of 30 term pairs on which a higher inter-annotator agreement was achieved. The average correlation between physicians is 0.68. The average correlation between medical coders is 0.78. We evaluate our method on the mean of the physician scores and the mean of the coders’ scores in this subset in the same manner as reported by Pedersen, et al.22.
UMNSRS: UMNSRS, developed by Pakhomov, et al.23, consists of 725 clinical term pairs whose semantic similarity and relatedness was determined independently by four medical residents from the University of Minnesota Medical School. The similarity and relatedness of each term pair was annotated based on a continuous scale by having the resident touch a bar on a touch sensitive computer screen to indicate the degree of similarity or relatedness. The Intra-class Correlation Coefficient (ICC) for the reference standard tagged for similarity was 0.47, and 0.50 for relatedness. Therefore, as suggested by Pakhomov and colleagues, we use a subset of the ratings consisting of 401 pairs for the similarity set and 430 pairs for the relatedness set which each have an ICC of 0.73.
Comparison of Reference Standards
There are primarily two main differences between the UMNSRS, MayoSRS and MiniMayoSRS reference standards. The first difference is that the scores assigned to the UMNSRS term pairs by the human annotators are on a continuous scale, where the scores assigned to the MayoSRS and MiniMayoSRS are on a four point scale.
The second difference is the range of semantic groupings of the term pairs. A semantic group is a coarse-grained grouping of the semantic types in the UMLS developed by McCray, et al.24 to provide a coarse-grained distinction between UMLS concepts based on their semantic validity, parsimony, completeness, exclusivity, naturalness, and utility. Fifteen such semantic groups have been currently defined for the UMLS Metathesaurus concepts b.
Over half of the term pairs consist of Disorder-Disorder term pairs for each of the reference standards, although MayoSRS and MiniMayoSRS contain the largest percentage. The second largest percentage of term pairs in the UMNSRS reference standards are Disorder-Chemical & Drug term pairs which occur only a few times in MayoSRS and MiniMayoSRS. Examples of these type of term pairs such as Obesity (C0028754) and Orlistat (C0076275) which the annotators found more similar/related than the term pairs Glipizide (C0017642) and Haemophilia (C0684275). The third largest percentage of term pairs in the UMNSRS reference standards are Chemical & Drug term pairs which do not occur in MayoSRS or MiniMayoSRS. The MayoSRS and MiniMayoSRS reference standards contain the most diverse term pair groupings, eleven and eight respectively, where the UMNSRS reference standards only contain the three. Table 1 shows a breakdown of the semantic groups for each of the reference standards.
Table 1:
Semantic Groupings of Term Pairs in the Reference Standards
| Semantic Group | MayoSRS | MiniMayoSRS | UMNSRS | ||
|---|---|---|---|---|---|
|
| |||||
| Term 1 | Term 2 | relatedness | relatedness | similarity | relatedness |
|
| |||||
| Activities Behaviors | Phenomena | 1 | |||
| Anatomy | Anatomy | 1 | 1 | ||
| Chemical & Drug | Chemical & Drug | 77 | 82 | ||
| Chemical & Drug | Devices | 1 | |||
| Chemical & Drug | Procedures | 1 | 1 | ||
| Disorder | Anatomy | 4 | 2 | ||
| Disorder | Chemical & Drug | 10 | 1 | 113 | 126 |
| Disorder | Concepts & Ideas | 3 | 1 | ||
| Disorder | Disorder | 66 | 21 | 211 | 222 |
| Disorder | Devices | 1 | |||
| Disorder | Physiology | 5 | |||
| Disorder | Procedures | 7 | 1 | ||
| Physiology | Physiology | 1 | |||
|
| |||||
| Total | 101 | 30 | 401 | 430 | |
6 Experimental Framework
We conducted our experiments using the freely available open source software package UMLS::Similarity25 version 1.13 c. This package takes as input two terms or concepts and returns the similarity between any two concepts using the path information in any of the sources available in the UMLS, including SNOMED CT, for each of the measures discussed in Section 2.
For our experiments, the path information was obtained using the PAR/CHD relations between concepts in SNOMED CT from the 2010AB version of the UMLS. This work was initiated in 2010 and for the sake of continuity and comparability of results, the 2010AB version was used throughout.
We calculated the IC of a concept for the IC-based measures using frequency information obtained from the National Library of Medicine’s UMLSonMedline dataset. This dataset consists of concepts from the 2009AB UMLS and the number of times they occurred in a snapshot of Medline taken on 12/01/2009. The frequency counts were obtained by using the Essie Search Engine which queried Medline with normalized strings from the 2009AB MRCONSO table in the UMLS. The frequency of a CUI was obtained by aggregating the frequency counts of the terms associated with the CUI to provide a rough estimate of its frequency.
7 Results and Discussion
Table 2 shows the Spearman’s Rank Correlation coefficients between the human scores and the scores obtained by u-path, lcs-path, and the measures proposed by Wu and Palmer12 (wup), Nguyen and Al-Mubaid13 (nam), Resnik15 (res), Jiang and Conrath16 (jcn) and Lin17 (lin) for the four reference standards.
Table 2:
Spearman’s Rank Correlation Results
| Measure | Reference Standard | ||||
|---|---|---|---|---|---|
|
| |||||
| MiniMayoSRS | MayoSRS | UMNSRS | |||
|
| |||||
| coders | physicians | sim. | rel. | ||
|
| |||||
| u-path | 0.7142 | 0.5527 | 0.3187 | 0.4776 | 0.2725 |
| lcs-path | 0.5341 | 0.3628 | 0.2324 | 0.5182 | 0.2903 |
| wup | 0.5346 | 0.4139 | 0.2344 | 0.4912 | 0.2425 |
| nam | 0.4228 | 0.2985 | 0.1461 | 0.3252 | 0.1634 |
| res | 0.5150 | 0.3852 | 0.2549 | 0.4737 | 0.2550 |
| jcn | 0.5437 | 0.4298 | 0.3145 | 0.5132 | 0.3418 |
| lin | 0.5524 | 0.4315 | 0.2948 | 0.4981 | 0.2909 |
The results show that for MiniMayo and MayoSRS the u-path measure obtained a higher correlation with human judgments than the other measures, but this was not the case for the UMNSRS reference standards. For both the UMNSRS tagged for similarity and relatedness, lcs-path or jcn obtained the highest correlation scores.
The results also show that jcn and lin obtain a higher overall correlation on the UMNSRS reference standards tagged for similarity and relatedness than the u-path measure, but this is not the case for the res measure. The difference between res and the other IC-based measures is that jcn and lin incorporate the IC of the individual concepts in conjunction with the IC of the LCS, where res just uses the IC of the LCS. We believe the lower results of res may indicate that it is the IC of the individual concepts that are providing the additional relevant information rather than the IC of the LCS.
One main difference between the UMNSRS and the MayoSRS and MiniMayoSRS datasets are the semantic groupings of the term pairs. The MayoSRS and MiniMayoSRS contain only Disorders-Symptom pairs while the UMNSRS data contains a mixture of Disorder, Symptom and Drug pairs. Table 3 shows a breakdown of the correlation results based on the term pairs’ semantic groups in the UMNSRS tagged for relatedness; as well as the overall correlation score for reference. Table 4 shows the same results on the UMNSRS tagged for similarity; and Table 5 shows the results for the MiniMayoSRS and MayoSRS.
Table 3:
Spearman’s Rank Correlation of UMNSRS tagged for Relatedness
| Semantic Groups | u-path | lcs-path | jcn |
|---|---|---|---|
|
| |||
| Disorder-Disorder | 0.4028 | 0.4290 | 0.4383 |
| Disorder-Chemical&Drug | −0.1037 | −0.1188 | 0.1369 |
| Chemical&Drug-Chemical&Drug | 0.3925 | 0.3761 | 0.4356 |
|
| |||
| UMNSRS rel. | 0.2725 | 0.2903 | 0.3418 |
Table 4:
Spearman’s Rank Correlation of UMNSRS tagged for Similarity
| Semantic Groups | u-path | lcs-path | jcn |
|---|---|---|---|
|
| |||
| Disorder-Disorder | 0.4521 | 0.5055 | 0.4329 |
| Disorder-Chemical&Drug | 0.1449 | 0.1489 | 0.1783 |
| Chemical&Drug-Chemical&Drug | 0.5693 | 0.5977 | 0.6885 |
|
| |||
| UMNSRS sim. | 0.4776 | 0.5182 | 0.5132 |
Table 5:
Spearman’s Rank Correlation of MayoSRS and MiniMayoSRS
| Semantic Groups for MayoSRS | u-path | lcs-path | jcn |
|---|---|---|---|
| Disorder-Disorder | 0.3535 | 0.2649 | 0.2579 |
| MayoSRS | 0.3187 | 0.2324 | 0.3142 |
|
| |||
| Semantic Group for MiniMayoSRS | u-path | lcs-path | jcn |
| Disorder-Disorder (physicians) | 0.4223 | 0.2580 | 0.2733 |
| Disorder-Disorder (coders) | 0.5933 | 0.3958 | 0.3353 |
| MiniMayoSRS (physicians) | 0.5527 | 0.3628 | 0.4298 |
| MiniMayoSRS (coders) | 0.7142 | 0.5241 | 0.5437 |
For the UMNSRS tagged for relatedness, the results show that u-path obtains a higher correlation for Chemical & Drug term pairs than lcs-path but not for Disorder term pairs. The jcn measure obtains a higher correlation than either path-based measures for each of the semantic groups.
The results also show that for u-path, lcs-path and jcn the correlation results are low for Disorder-Chemical & Drug term pairs. We believe this is because in SNOMED CT there does not exist many is-a relationships between drugs and disorders which is the relationship all of the similarity measures are exploiting. We hypothesize that relatedness measures, rather than similarity measures, may be a better measure for these type of relations.
For the UMNSRS tagged for similarity, the results show that lcs-path and jcn obtain a higher correlation with humans than u-path over each of the different semantic groups except for Disorder pairs in which u-path obtains a higher correlation than jcn. The low correlation results for Disorder-Chemical & Drug term pairs confirms our previous analysis above that there is a limited number of is-a relations between drugs and disorders for the similarity measures to exploit.
Table 5 shows the correlation results for the semantic groups in the MiniMayoSRS and MayoSRS that have at least twenty term pairs. The results show that for Disorder term pairs, the u-path obtains a higher correlation with the human judgments than lcs-path and jcn for these two reference standards. We believe that this indicates that the related disorders in these reference standards are co-located in similar areas in the taxonomy and not necessarily in a direct path through the LCS.
To further analyze the difference between the u-path and lcs-path, Table 6 shows the number of term pairs in each of the reference standards, and the corresponding percentage whose shortest path did not go through the LCS. The percentage of non-LCS paths in the reference standards indicate that SNOMED CT is indeed a dense multi-hierarchical structure in which the shortest path between concepts does not always contain the LCS. The overall higher percentage of non-LCS paths in the MiniMayoSRS and MayoSRS may explain why u-path obtained a higher correlation with human judgments than lcs-path. Although, this is not seen when analyzing the semantic grouping results in the UMNSRS reference standards, where the Chemical & Drug pairs contain the lowest number of non-LCS paths and u-path obtains a higher correlation than lcs-path. However, we believe that given the relatively large percentage of concept pairs that have a path between them shorter than the LCS path indicates that u-path has certain possibilities and merits further exploration.
Table 6:
Non-LCS Shortest Paths
| reference standard | semantic groups | # term pairs | # non LCS paths | % of non LCS paths |
|---|---|---|---|---|
| MiniMayoSRS | all | 29 | 17 | 0.59 |
| MayoSRS | all | 101 | 57 | 0.50 |
| UMNSRS (relatedness) |
all Disorder pairs Disorder-Chemical&Drug Chemical&Drug pairs |
430 223 125 82 |
184 110 63 11 |
0.43 0.49 0.50 0.13 |
| UMNSRS (similarity) |
all Disorder pairs Disorder-Chemical&Drug Chemical&Drug pairs |
401 212 112 77 |
183 106 63 14 |
0.46 0.50 0.56 0.18 |
8 Conclusions and Future Work
In this paper, we present the results of a method that quantifies the degree of similarity between concepts in a dense taxonomy with multiple inheritance using undirected path information. We show that it obtains a higher correlation than other path-based measures on two reference standards. We also analyze the semantic groupings of the term pairs showing that different measures perform better on different groupings. In the future, we plan to explore more fully the impact of the semantic groups of term pairs on the similarity measures.
The overall objective of this work was to explore a method that takes advantage of dense multi-hierarchical taxonomies that are more graph-like than tree-like. Our hypothesis was that the proximity of the terms regardless of how they are connected is an indicator to the degree of their similarity. The overall results show, in SNOMED CT, the location of the concepts with respect to each other indicates the degree to which they are similar. In the future, we plan to explore more complex measures that take advantage of this information such as graph-based centrality metrics and the measure proposed by Batet et al.14 26.
The results showed that u-path obtained higher correlation results on the reference standards tagged for relatedness. The multiple inheritance within the taxonomy implies the potential existence of ancestral pedigrees that represent different, although related, dimensions of a concept. Therefore, a path passing through a common descendant would then link two concepts that belong to two dimensions allowing for the connection between the two concepts to fall outside the traditional strict is-a relations. In the future, we plan to explore this further comparing the results to relatedness measures such as those proposed by Lesk27; Patwardhan28; Dagan, et al.29; Workman, et al.30; and Pivovarov and Elhadad31.
We also plan to explore weighting the undirected path based on its turns within the path. Hirst and St. Onge18 limited the degree in which the path is allowed to meander based on the types of turns the path is taking. The underlying thought behind this is that the deeper the undirected path passes through a common descendant the less similar the two concepts would be, although the relatedness between the two concepts would still be maintained.
Acknowledgments
This work was supported in part by the National Institute of Health, National Library of Medicine (NLM) Grant #R01LM009623-01. We would like to thank Russel Loane, Jim Mork and Lan Aronson from NLM for providing the UMLSonMedline dataset.
Footnotes
References
- 1.Rada R, Mili H, Bicknell E, Blettner M. Development and application of a metric on semantic nets. IEEE Transactions on Systems, Man, and Cybernetics. 1989;19(1):17–30. [Google Scholar]
- 2.Lin Y, Li W, Chen K, Liu Y. A document clustering and ranking system for exploring MEDLINE citations. Journal of the American Medical Informatics Association. 2007;14(5):651–661. doi: 10.1197/jamia.M2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bodenreider O, Burgun A. Aligning knowledge sources in the UMLS: methods, quantitative results, and applications. Proc of the 11th World Congress on Medical Informatics; 2004. pp. 327–331. [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Jimmy, John Wilbur W. Pubmed related articles: a probabilistic topic-based model for content similarity. BMC bioinformatics. 2007;8(1):423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pearson RK, Hauben M, Goldsmith DI, Gould AL, Madigan D, OHara DJ, Reisinger SJ, Hochberg AM. Influence of the meddra hierarchy on pharmacovigilance data mining results. Intl journal of medical informatics. 2009;78(12):e97–e103. doi: 10.1016/j.ijmedinf.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Bill RW, Liu Y, McInnes BT, Melton GB, Pedersen T, Pakhomov S. Evaluating semantic relatedness and similarity measures with standardized meddra queries. AMIA Annual Symposium Proc, volume 2012, page 43 American Medical Informatics Association; 2012. [PMC free article] [PubMed] [Google Scholar]
- 7.Caviedes JE, Cimino JJ. Towards the development of a conceptual distance metric for the umls. Journal of Biomedical Informatics. 2004;37(2):77–85. doi: 10.1016/j.jbi.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Leacock C, Chodorow M. Combining local context and WordNet similarity for word sense identification. WordNet: An electronic lexical database. 1998;49(2):265–283. [Google Scholar]
- 9.Pedersen T, Patwardhan S, Michelizzi J. WordNet::Similarity – Measuring the Relatedness of Concepts. The Annual Meeting of the Human Language Techonology and North American Association of Computational Linguistics: Demonstration Papers, pages 38–41; Boston, Massachusetts, USA. May 2004. [Google Scholar]
- 10.Miller GA. WordNet: a lexical database for English. Communications of the ACM. 1995;38(11):41. [Google Scholar]
- 11.Spackman KA, Campbell KE, Côté RA. Snomed rt: a reference terminology for health care. Proc of the AMIA annual fall symposium, page 640; American Medical Informatics Association; 1997. [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z, Palmer M. Verbs semantics and lexical selection. Proc of the 32nd Meeting of Association of Computational Linguistics, pages 133–138; Las Cruces, NM. June 1994. [Google Scholar]
- 13.Nguyen HA, Al-Mubaid H. New ontology-based semantic similarity measure for the biomedical domain. Proc of the IEEE Intl Conference on Granular Computing, pages 623–628; Atlanta, GA: 2006. [Google Scholar]
- 14.Batet M, Sánchez D, Valls A. An ontology-based measure to compute semantic similarity in biomedicine. Journal of biomedical informatics. 2011;44(1):118–125. doi: 10.1016/j.jbi.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Resnik P. Using information content to evaluate semantic similarity in a taxonomy. Proc of the 14th Intl Joint Conference on Artificial Intelligence, pages 448–453; Montreal, Canada. August 1995. [Google Scholar]
- 16.Jiang J, Conrath D. Semantic similarity based on corpus statistics and lexical taxonomy. Proc on Intl Conference on Research in Computational Linguistics, pages 19–33; 1997. [Google Scholar]
- 17.Lin D. An information-theoretic definition of similarity. Intl Conf ML Proc, pages 296–304; 1998. [Google Scholar]
- 18.Hirst G, St-Onge D. Lexical chains as representations of context for the detection and correction of malapropisms. WordNet: An Electronic Lexical Database, pages 305–332. 1998 [Google Scholar]
- 19.Budanitsky A, Hirst G. Evaluating wordnet-based measures of lexical semantic relatedness. Computational Linguistics. 2006;32(1):13–47. [Google Scholar]
- 20.Patwardhan S, Banerjee S, Pedersen T. Using measures of semantic relatedness for word sense disambiguation. Computational Linguistics and Intelligent Text Processing, pages 241–257; 2010. [Google Scholar]
- 21.Pakhomov SVS, Pedersen T, McInnes B, Melton GB, Ruggieri A, Chute CG. Towards a Framework for Developing Semantic Relatedness Reference Standards. Journal of Biomedical Informatics. 2010 Oct; doi: 10.1016/j.jbi.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen T, Pakhomov SVS, Patwardhan S, Chute CG. Measures of semantic similarity and relatedness in the biomedical domain. Journal of Biomedical Informatics. 2007;40(3):288–299. doi: 10.1016/j.jbi.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Pakhomov S, McInnes BT, Adam T, Liu Y, Pedersen T, Melton GB. Semantic similarity and relatedness between clinical terms: An experimental study. Proc of the American Medical Informatics Association (AMIA) Symposium, pages 572–576; Washington, DC. November 2010; [PMC free article] [PubMed] [Google Scholar]
- 24.McCray AT, Burgun A, Bodenreider O. Aggregating umls semantic types for reducing conceptual complexity. Studies in health technology and informatics, pages 216–220; 2001. [PMC free article] [PubMed] [Google Scholar]
- 25.McInnes BT, Pedersen T, Pakhomov SV. UMLS-Interface and UMLS-Similarity : Open Source Software for Measuring Paths and Semantic Similarity. Proc of the American Medical Informatics Association Symposium; 2009. [PMC free article] [PubMed] [Google Scholar]
- 26.Batet M, Sánchez D, Valls A, Gibert K. Semantic similarity estimation from multiple ontologies. Applied intelligence. 2013;38(1):29–44. [Google Scholar]
- 27.Lesk M. Automatic sense disambiguation using machine readable dictionaries: how to tell a pine cone from an ice cream cone. Proc of the 5th Annual Intl Conference on Systems Documentation, pages 24–26; 1986. [Google Scholar]
- 28.Patwardhan S, Pedersen T. Using WordNet-based Context Vectors to Estimate the Semantic Relatedness of Concepts. Proc of the EACL 2006 Workshop Making Sense of Sense – Bringing Computational Linguistics and Psycholinguistics Together, pages 1–8; 2006. [Google Scholar]
- 29.Dagan I, Lee L, Pereira F. Similarity-based models of word cooccurrence probabilities. Machine Learning. 1999;34(1–3):43–69. [Google Scholar]
- 30.Workman ET, Rosemblat G, Fiszman M, Rindflesch TC. A literature-based assessment of concept pairs as a measure of semantic relatedness. AMIA Annual Symposium Proc, volume 2013, page 1512; American Medical Informatics Association; 2013. [PMC free article] [PubMed] [Google Scholar]
- 31.Pivovarov R, Elhadad N. A hybrid knowledge-based and data-driven approach to identifying semantically similar concepts. Journal of biomedical informatics. 2012;45(3):471–481. doi: 10.1016/j.jbi.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


