Abstract
Statistical text mining and natural language processing have been shown to be effective for extracting useful information from medical documents. However, neither technique is effective at extracting the information stored in semi-structure text elements. A prototype system (TagLine) was developed to extract information from the semi-structured text using machine learning and a rule based annotator. Features for the learning machine were suggested by prior work, and by examining text, and selecting attributes that help distinguish classes of text lines. Classes were derived empirically from text and guided by an ontology developed by the VHA’s Consortium for Health Informatics Research (CHIR). Decision trees were evaluated for class predictions on 15,103 lines of text achieved an overall accuracy of 98.5 percent. The class labels applied to the lines were then used for annotating semi-structured text elements. TagLine achieved F-measure over 0.9 for each of the structures, which included tables, slots and fillers.
Introduction
The analysis of text from the electronic health record (EHR) is an important research activity in medical informatics and particularly in the Veterans Healthcare Administration (VHA) because of its large integrated EHR. A wide variety of methods have been employed to extract information from text. Information retrieval (IR) and information extraction (IE) have been shown to be useful for detecting patterns in patient care1, patient treatment patterns2 and outcomes. IR has been used to identify co-morbidities,3 smoking status,4 as well as detecting fall-related injuries.5 Regular expressions have been used to extract blood pressure values from progress notes.6 Natural language processing (NLP) has been used to extract medical information such as principal diagnosis4 and medication use7 from clinical narratives. This work has led to a better understanding of the conditions patients face and how to treat them.8
Raw medical text passages are voluminous and heterogeneous as is their structure.9 Some of the information is in free-text form, written as full sentences or phrases, but much of it is in the form of semi-structured data, or templates.10 Semi-structured data is defined as data that has some structure but is inconsistent or does not adhere to any rigorous format.11 While some work has been done extracting information from semi-structured data, most of that research focused on extracting data from web pages or research articles and is not easily adapted to the medical domain.12–15 Part-of-speech parsers in off-the-shelf NLP programs do not perform well on semi-structured data because it does not adhere to grammatical rules. If the structures within documents could first be accurately identified, then extraction methods that do not depend on English grammar could be developed to extract the information in these structures.
The goal of this study was to evaluate a method of processing information in semi-structured text in medical progress notes by first, classifying each line of text using machine learning, then using the line classifications in a rules-based parser, annotate the semi-structured text elements. This will allow the information in these structures to be further processed or stored in structured form. To achieve this goal, a prototype system “TagLine” was developed. We exploit non-grammatical features derived from the text in progress notes to apply a class label to each line of text and use these labels in a rule-based annotator to identify semi-structure text elements in the text. Our system combines methods already familiar in IE, such as concept look-ups, regular expressions, rules, and machine learning on features from the text to accurately identify information contained in semi-structured text elements.
Background
Information Extraction
IE is defined as the extraction of predefined types of information from text.16 There are four primary methods available to implement an information extraction system, including NLP, pattern matching, rules, and machine learning. The primary means of performing IE is NLP. NLP research focuses on developing computational models for understanding natural language.17 The use of rules and pattern-matching exploits basic patterns over a variety of structures, such as text strings, part-of-speech tags, semantic pairs, and dictionary entries.18 Regular expressions are effective when the structure of the text and the tokens are consistent, but tend to be one-off methods tailored to the extraction task. Hand coding of complex regular expressions can be a very time consuming effort that requires a priori knowledge of all possible patterns that represent the concept being sought. Machine learning techniques can be an effective method for IE through automated knowledge acquisition.19 Features extracted from the text, such as parts of speech and sentence length, are fed into a learning machine to assist in tasks downstream like word sense disambiguation. The primary disadvantage of using machine learning is that it requires a labeled dataset for training a model.
Semi-Structured Data
Well-structured data, as found in a typical database, conforms to a schema or data model and can be queried using a structured query language to answer questions. Semi-structured data is data that has some structure but is inconsistent or does not adhere to any rigorous format,36 and is very difficult to query. In semi-structured data, information normally associated with a schema is contained within the data, which is sometimes called “self-describing.”11 Semi-structured data can break the conventions for structured data in a number of ways. The structure is often irregular, implicit, or partial.
Efforts to perform IE on semi-structured data are well developed for web pages but less so on research articles from peer-reviewed journals and notes for the electronic medical record. Table 1 shows a summary of the published methods previously used for extracting semi-structured data. Pages on the World Wide Web are written in HTML, which is a standard and provides a healthy measure of reliable structure that these studies used in developing extraction routines. Research papers also adhere to a certain amount of structure. There is an order to the flow of the paper and very specific formatting conventions with journals for section headers, graph labels and tables. For IE on semi-structured data in the electronic medical record, the work is limited and it has been noted that locating the data is difficult, since no standard way to enter the data in the EHR system is reinforced. Furthermore, there are no built-in edit checks available to facilitate data entry.
Table 1.
Previous work on semi-structured data
| Method Used | Document Type |
|---|---|
| Finite state machines20 | HTML |
| Rule generation within specified constraints21 | HTML |
| Example guided object decomposition22 | HTML |
| Example guided structure induction23 | HTML |
| Machine learning on object exchange models24 | HTML |
| Schemas and wrappers on document structures25 | HTML |
| Path expressions26 | HTML |
| Labeled ordered trees on tag structure27 | HTML |
| Ontology and graph based modeling28,29 | HTML |
| Descriptive logics30 | Journal |
| Graph modeling on schemas31–33 | Journal |
| Link grammars connect features with numbers | EHR |
| ID3 trees on NLP features34 | EHR |
| Standard data cleansing techniques35 | EHR |
The Electronic Health Record at the VA
The VHA EHR, VistA, records information regarding a patient’s clinical encounters, in both structured data tables and text. Each line of 80 characters or less is stored as a string associated with a specific document, such as a medical progress note. This preserves the formatting of the document to make it easier to read in the Computerized Patient Record System (CPRS),37 the user interface for VistA. It also provides an artifact useful in text processing. Each note can be separated into a group of individual lines of text. Because progress notes are written for a variety of purposes, notes are assigned descriptive names. Users can create their own custom designed notes using the Progress Notes Construction Set38 and design their own templates for the notes. This causes tremendous variation in the way notes are structured, which means developing extraction routines using techniques like regular expressions and handcrafted rules are typically useful only as one-off solutions.
Document element ontology
This study is guided by an ontology developed by investigators in the Consortium for Health Informatics Research (CHIR) to define the text elements to be targeted. The ontology described here is the result of an error analysis from the 2010 i2b2 challenge submission and a CHIR Information Extraction Methods (IEM) initiative.39 A document is made up from a set of document elements. Sections, slot-value pairs, paragraphs, sentences, phrases, content, questions, lists, tables, and address blocks are examples of document elements. Figure 1 graphically depicts a sample of the structures and their component parts. For purposes of this study, we selected the text elements tables and slot-values. Table 2 presents a sample of the line-of-text classifications being used, along with the parent “is-a” class from the text element ontology and the larger structural (part-of) class. It gives the relationship of the line type to structure type in “is a” or “is part of” relationships. There were a total of 75 distinct class labels derived from the text and ontology.
Figure 1.
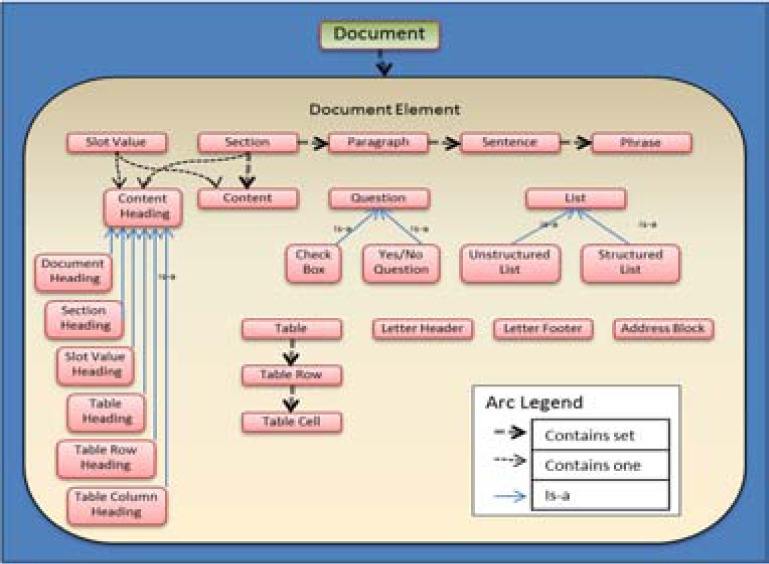
Document element ontology
The Line of Text as the Unit of Analysis
Progress notes in VistA are stored as a series of strings at a length of 80 characters or less. These text strings can be assigned recognizable roles as defined in the text element ontology. Some of these elements stand on their own as atomic text elements, while others are part of larger more complex text element structures. Both tables and slot values can be defined by their constituent parts. Slot values consist of a content heading and content separated by a delimiter and can be contained within a single line of text. Slot value content is a short value, typically a word, numeric value or phrase. The elements of a table can be identified by their parts at the line level. A table, as defined by the document element ontology, is a collection of related items arranged in columns and rows. Tables may have column labels and row labels as well as a caption for the subject of the table. The first line in Table 5, “---- CBC PROFILE ----” stands complete as the table header (THE). The second and third lines are used to label the information in the columns (CLA). The remaining lines are each identifiable table items (TBI) in the table.
Table 5.
Slot Filler Examples
| Class | Line of Text |
|---|---|
| SLF | CURRENT LEVEL OF PAIN: 5 |
| DSV | LMP:11-26-06 PMP:221 |
| TSV | Grava: 2 Para: 2 Abortion: 0 |
| SLT FRT |
Indicate HOW YOUR PAIN FEELS:Aching |
The basic slot-filler consists of a label and a value separated by a delimiter, usually a colon (see Table 6). In its simplest form the slot-filler appears as shown on a single line. This line would be given the label “SLV” for slot-filler or slot-value pair. However, slot-fillers do not always appear in this form; there may be two or three sets of slot-fillers on the same line. These two examples are then labeled “DSV” and “TSV” respectively, for double slot-filler and triple slot-filler. The two elements, the label and the value may also appear on separate lines. The values that fill the slot are not always present, so there may only be a slot. Since each line tends to stand as a unit, we chose this as our unit of analysis in machine learning. Each of these variations must be handled by a specific set of parsing rules, and the class labels identify which set of rules to employ.
Table 6.
Feature Examples
| Feature | Description |
|---|---|
| AllCaps | All uppercase letters |
| Title | In title case |
| NumCaps | Number of uppercase letters |
| Hyphens | Number of hyphens |
| Spaces | Number of spaces |
| Slashes | Number of slashes |
| DecPos | Offset position of decimal |
| ColPos | Offset position of colon |
| QmPos | Offset position of “?” |
| Bar | Formatting bar |
| YesNo | Ends with “Yes” or “No” |
| Icd | Presence of IDC9 code |
| Bullet | Line is bulleted |
| Numbed | Line is numbered |
Feature selection
The selection of features that were used in machine learning to assign classes to each line was crucial to success. We reviewed the work of other studies in this area20,40–42 and derived and tested a number of additional features to detect structure in lines of text. Table 6 shows a sample of the features we adopted. We looked for similar clues in each of the text elements that help us tell them apart from other text elements. These clues fall into one of several types of text features: formatting features, special character usage, term usage, and document structural features. Examples of formatting features include whether the line was in all uppercase letters or in title case, as well as the number of uppercase letters in the line. Special character features included items such as the total number of spaces, slashes, and hyphens in the line and where the first, second, and third decimal and colon could be found in the line. Examples of term usage-based features included position of a question mark, if the line of text ends with a “Yes” or “No” and if the token “ICD” was found in the line. Finally, document structural features included items such as if a line was numbered, had a text bullet, and if a line had a formatting bar (e.g., **********). In all, there were over 70 features defined to describe the differences between classes.
Data Preparation
Document Selection
A set of 162 notes formed the corpus for this study. These notes are a subset of the 5,048 medical progress notes collected in a separate and unrelated study in the VHA to identify patients who have suffered a fall-related injury5. The notes were randomly selected from note types containing the greatest number and variety of semi-structured text elements. The variety of note types selected represents note types that are most frequently used in the VHA for treatment of falls, primarily notes from emergency room and primary care visits. They included “Primary Care Notes”, “Primary Care Nursing Notes”, “Primary Care H&P Notes”, “Primary Care Home Health Consult Notes,” “Emergency Room Nursing Notes”, “Emergency Room Triage Notes” and “Nursing Discharge Notes.” These note types tend to have more structured elements than those written in free text. To evaluate machine learning on assigning line classes, all lines (n=15,103) from the selected notes were randomly split into sets of 10,000 and 5,103 for training and test respectively. To evaluate the structure annotator on identification of targeted semi-structured data, the 162 notes were randomly split into 115 notes for training and 47 notes for testing.
Labeling Lines of text
To provide data for machine learning each of the 15,103 lines in the 162 notes, classes were assigned to lines of text as an intermediate step to finding and extracting information from specific predefined types of semi-structured text elements. Class labels were determined by membership in or relationship to, the structure types defined in the CHIR ontology, and were derived empirically from the text. The classes may describe a part of a structure, such as a table or contain multiple structural parts. Class determination and text line labeling were iterative simultaneous tasks. First we examined the line for a relationship to a structure type in the ontology. If there was a class that describes this relationship, then we applied that class. If not, we created a new class within the ontology that describes the relationship and apply the class label. Structure types in the ontology can be associated with multiple classes of lines. This is grounded in the fact that any given structure type identified by the ontology may appear in the text in multiple forms as in the slot-fillers example above. Rigorous steps were taken to ensure that the classes were unique and that they related to only one text element in the ontology. Each class was evaluated individually and discussed as it was added to the set. As the number classes grew we evaluated their usefulness them by using them in some preliminary machine learning models. Classes that were misclassified were reexamined and if the label applied by the machine learning model was valid, it was relabeled with the predicted label. If the label was not valid, then we applied a new label and new features were added to the machine learning models to improve performance.
When the labeling was completed a count was made of each occurrence of each class and the distribution of the class frequencies was checked. It was found in initial models developed that classes with fewer than six instances in the dataset achieved an F-Measure of less than 0.6. Classes with less than six occurrences in the dataset were removed and the lines with those labels were re-labeled with the next best class. This required that the new class label be re-defined to include the new instances. There were a total of 75 possible class labels derived from the text and ontology. A total of 13 classes were eliminated from the list, leaving 58 remaining classes for use in machine learning models. None of the removed classes were relevant for the structures examined in this effort. The classes that most frequently appeared in the text were free text (FRT), slot-value (SLV), medication list item (MLI), and table item (TBI). Since the annotator uses the predicted line labels in its parsing routines, the errors in line label predictions are likely to cause errors in the structural annotations. It is therefore crucial to achieve the highest possible prediction accuracy in the first step.
TagLine
To accomplish the goal of this study, the prototype TagLine system was designed and implemented. TagLine consists of a series of interacting software modules written in Python. Shown in Figure 2, the paths in yellow show the flow sequence for training a new model and the blue paths show the sequence for structure annotation.
Figure 2.
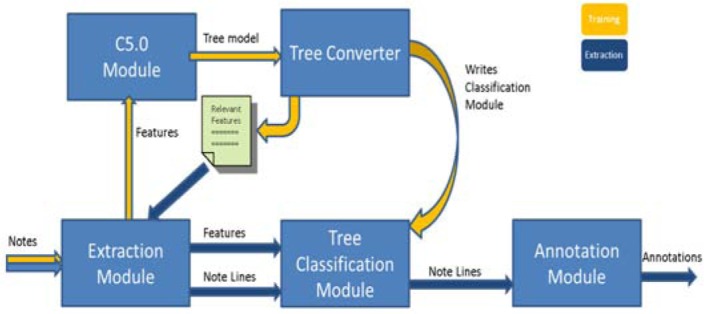
TagLine system architecture
Extraction Module
Notes are converted to a list of text lines and each text line is subjected to a series of functions that extract values for the features to be used in machine learning. When a new model is trained, all features are selected for extraction. The final model determines which features will be extracted when using the system for annotation. The resulting new dataset was used to train the learning machine and make predictions. Each line in the dataset described a line of text in the note. This dataset was sent to the C5.0 module when training a new model or to the next stage for use in classification by the tree classifier.
C5.0 module
Decision trees are a good fit for the work in this study for the following reasons43: They handle high dimensional data well, are computationally efficient, easily interpreted by human beings, and most importantly for this study, can be readily converted to computer executable code. This study uses an updated version of the C4.5 algorithm developed by Ross Quinlan as C5.0.44 The C5.0 module is a Python wrapper for a console application. The C5.0 application was compiled from the GPL C code distributed freely by Ross Quinlan. The C5.0 module develops a decision tree model based on the features provided by the extraction module. The C5.0 algorithm performs winnowing of the attributes before building a tree by evaluating each attributes’ effect on error rate when the attribute is removed. Error-based pruning is also performed to cut back on branches that do not contribute to the models overall efficacy. Finally, this module writes a tree model file out to disk for use later.
Tree converter module
The tree converter parses the original tree model file written by the C5.0 module and writes two new files; a text file and a Python executable classification module. The text file is a list of the features determined by the C5.0 decision tree model found to be useful in prediction. When the new classification module is used, the extractor will only extract those features that are needed.
Tree classification module
The tree classification module is automatically created by the tree converter module by parsing the tree model and writing executable Python code. This module takes the note as a list of text lines and classifies them using a series of if-then rules. Then it passes the classification results along with the lines of text to the annotation module.
Annotation module
The annotation module takes the classification results and the lines of text from the tree classification module and uses them to locate structures in the text based on options submitted by the user. For each type of structure indicated in the options, the annotation module loops through each line of the note looking for the appropriate line labels for the targeted structures. When targeted labels are encountered, a rule-based approach is used to parse and annotate the structure and return values in the form “ElementType|StartOffset|StopOffset.” The annotations can be written to a file for review, recorded in a database for storage and used later as structured data, used as features in another classification task downstream, or sent to an NLP pipeline where annotations can be used for extracting concepts from the text elements using a structured vocabulary. TagLine can also extract the elements and record the notes with the annotated structures removed as a text reduction method.
TagLine Evaluation
TagLine was evaluated in two separate experiments. First, we evaluated the use of decision trees for predicting the classes assigned to the lines of text, then we evaluated the accuracy of our rules based annotator for identifying slot-value pairs and tables based on the classes assigned to the lines.
Results for Line Classification
A decision tree was constructed on the line-level training data and evaluated on unseen test data for prediction accuracy. Winnowing was used for feature reduction before a tree was constructed. Winnowing removes any feature that does add to the models efficacy. The C50 algorithm constructs an initial model on half of the training data and calculates the increase in error rate for each feature when it is left out of the model. Global pruning was performed after tree induction keeping only those branches that had at least one instance associated with it. Error based pruning was also employed; a branch was pruned if its prediction errors exceeded a level of 25 percent. A series of fifty models were tested using graduating numbers of lines in increments of 200 examples starting at 200 and ending with 10,000. The overall prediction accuracy was calculated for each test using a hold out set of 5,103 lines. The results are presented as a learning curve in Figure 4.
Figure 4.

Decision Tree Fragment
With a sample size of 200 lines, the decision tree model was able to achieve an accuracy rating of 80 percent. The accuracy increased to 90 percent at 1,200 lines. An overall accuracy level of 98.5 percent was achieved with a sample set of 10,000 lines, only 3.5 percent higher than at the 3,000-line sample size. The performance results for the five most and least frequent classes in the test set are shown in Table 7. Only five classes did not achieve an overall F-measure of 0.9 or above.
Table 7.
Line Classification Statistics
| Class | N | Precision | Recall | F-Measure |
|---|---|---|---|---|
| Free-text | 934 | 0.9789 | 0.9936 | 0.9862 |
| Tableltem | 900 | 0.9944 | 0.9878 | 0.9911 |
| Slot-filler | 508 | 0.9882 | 0.9882 | 0.9882 |
| Medline | 362 | 0.9564 | 0.9696 | 0.9630 |
| Slot | 273 | 1.0000 | 0.9963 | 0.9982 |
| AphaLine | 6 | 0.8333 | 0.8333 | 0.8333 |
| NumbedQue | 4 | 0.8000 | 1.0000 | 0.8889 |
| NumbMed | 3 | 0.6667 | 0.6667 | 0.6667 |
| LabeledDS | 2 | 0.6667 | 1.0000 | 0.8000 |
| QuestHead | 1 | 0.5000 | 1.0000 | 0.6667 |
Prior to tree construction, the winnowing process eliminated 20 features. The top 10 remaining features are shown below in Table 8 with their respective importance ratings. The importance rating is C5.0’s estimate of the factor by which the true error rate or misclassification cost would increase if that attribute were excluded. The number of colons in a line of text appears to be the most important feature in the model. Colons are important when looking for slot-value pairs, as well as dates and timestamps. The second most predictive feature is the line number for the line of text. Line numbers describe how far into the note a line appears; beginning, middle or end. Interestingly, all of the tests done by the model using the line number feature took place at the top of the note (see figure 4). Question marks, capital letters, and white space gaps also help in distinguishing structured text from un-structured text.
Table 8.
Top 10 Features
| Importance | Feature |
|---|---|
| 835% | Colons |
| 264% | LNum |
| 255% | Slot |
| 179% | LSpc |
| 166% | Bull |
| 150% | Med |
| 148% | Gaps |
| 139% | QM |
| 137% | Caps |
| 127% | Time |
The occurrence of a question mark was the most used feature in the model. This is shown in the tree fragment in Figure 4 where the feature QM is at the top of the decision tree, indicating it is the first attribute the tree splits on.
The tree classification module was used to predict the 5,103 line class labels in the test data. Many classes achieved a perfect score. The poor performers were all low prevalence classes, each occurring less than ten times in the entire test set. The class ALI is alpha list item, or an item in a list that is delineated by an alpha character and some delimiter. One line labeled ALI was misclassified as FRT causing one false negative. The only feature that would distinguish Free-text from an alpha-labeled-line is the use of the delimiter after the delineating character. In most applications a 95 percent accuracy rate would be considered acceptable, so it would not be necessary to use more than 3,000 lines for an acceptable result. However, because the results of the annotation in TagLine is dependent on the accuracy of the line labels it was decided to use the full 10,000 lines for the training set and the remaining for testing in the next section.
Results for Annotating Tables and Slot-Fillers
For this experiment, the data were split into training and test sets segregated at the note level. In the next stage, the parsing routines were tested on two types of structures; slot-value pairs and tables. Since tables are multi-line units, all lines are needed to identify a unit. There were a total of 115 notes and 10,048 lines of text in training set, while the test set had the remaining 47 notes and 5,055 lines of text. A record was constructed noting the number of tables, slots and fillers in the 47 notes. Table 6 shows the occurrences of each structure in the note set. The test set of notes has a total of 96 tables, 770 slots and 566 fillers for the slots. Not all of the slots have fillers associated with them so the numbers of slots and fillers will not match. The complete set of 47 notes was used in a GATE pipeline that called TagLine using a remote procedure call (RPC) server.
As the notes were processed, the server recorded the annotation actions. The extraction results were compiled for evaluation. Table 9 summarizes performance of TagLine on tables, slots, and slot fillers. The TagLine server reads the options, extracts the features from the note, uses the prediction module constructed from the decision tree, applies class labels to the lines, and returns the annotations on the structures found. When tables are targeted, start and stop rules are used to identify the boundaries of the table. The routine sequentially examines the labels applied to each line and when a class is encountered that signals the beginning of a table, a flag is set and all successive lines are included until a line label signaling a stop rule is encountered and the table end boundary is set and the annotation is returned. If a line in the middle of the table is classified as free text, it would prematurely trigger the table’s end and close off the boundaries of the table and erroneously start a new table at the next table line. For this reason, a rule was included to allow for a one-line misclassification gap in the table. While traversing the lines of text after a table beginning has been encountered, if a line is found that is not a table line, the end of table is marked, but held until the next line is checked. If the line after the non-table line is a table line then the table end is cleared and the boundary of the table is extended until there are no more table lines. In the event the next line is a table header or a column labels, then the current table is completed and a new table begins. A constraint enforced by this method is that a table must have at least one table line (TBL) to be annotated as a table.
Table 9.
TagLine Annotation Performance
| Exact Match | Partial Match | ||||||
|---|---|---|---|---|---|---|---|
| Structure | Count | Precision | Recall | F-Measure | Precision | Recall | F-Measure |
| Tables | 96 | 0.9250 | 0.9250 | 0.9250 | 0.9896 | 0.9896 | 0.9896 |
| Slots | 770 | 0.9709 | 0.9974 | 0.9840 | 0.9735 | 1.0000 | 0.9865 |
| Fillers | 566 | 0.9543 | 0.9965 | 0.9749 | 0.9560 | 0.9982 | 0.9767 |
Discussion
TagLine performed well on slots and fillers. There was little difference when evaluating performance on exact versus partial matching criteria. There were 770 slots in the 126 notes used for the test. TagLine found 768 of the slots matching the start and stop end points exactly. The two remaining slots were found but the parsing routine failed to set the offsets properly. One was due to colon placement in the string. The text line in the middle of the note: “(R): 0.7cm × 0.5cm (L): cm × cm” was marked as a DSV, or a double slot value. The “(R):” is the slot, and the filler is “0.7cm × 0.5cm.” TagLine failed to parse this line appropriately for both of the slots and fillers. While they were found, they were not considered exact matches. There were 21 false positives for slots. A total of 19 of the false positives were due to date entries across several notes that were misclassified as slot-values. There were a total of 566 fillers in the 47 test notes. TagLine successfully annotated all but one of the fillers, but there were 26 false positives, 19 associated with the date misclassifications mentioned above, resulting in a lower precision (0.9543) than recall (0.9965). Figure 5 shows the slot-filler annotations highlighted for one of the test notes.
Figure 5.
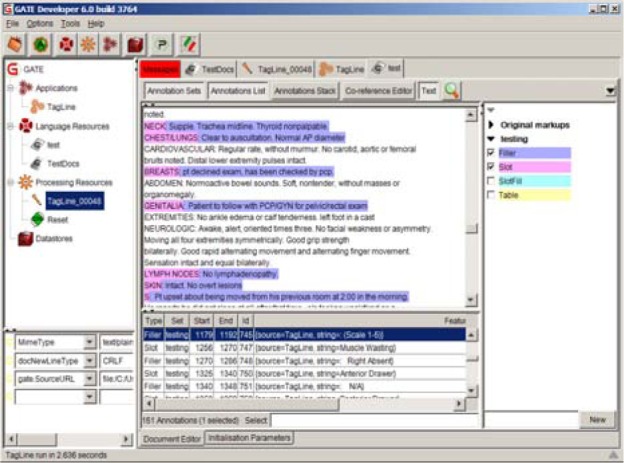
Slot-filler Annotation Example in GATE
There were 96 tables distributed throughout the 47 test notes. TagLine achieved an F-measure of 0.9250 for exact matching, lower than the number achieved for slots and fillers, but encouraging. Of the 96 tables in the test set, 86 were matched exactly according to the start and stop codes. Of the remaining 10 tables, 9 were identified with partial matches and one table was missed completely. Many partial match cases were due to column labels (CLA), or table item (TBI) being misclassified as free text (FRT), causing the parser to miss that portion of the table. In Figure 6, it can be seen that three lines in the table were not annotated. The first of the three is the column label and the next two descriptive entries did not conform to the format of the other table lines and were labeled FRT. Because the lines “color yellow” and “appeara sl cldy” do not conform to the format of the rest of the table, the table was partially captured in two parts. An allowance is made in the case that one line is misclassified, so tables are not broken apart. However, if more than one line is misclassified then the table will only be a partial match. Making allowances for more than one misclassified line causes problems when tables are found stacked directly on top of another table. In these cases, the two tables erroneously become one contiguous table.
Figure 6.

Table Annotation Example in GATE
Conclusions
TagLine was evaluated for parsing and annotating semi-structured text elements both at the macro level, on multi-line structures, and at the micro level, on within-line structures. TagLines’ performance identifying slots and fillers was impressive. The annotator achieved an F-measure of 0.9840 for exact matching criteria for slots and 0.9965 for fillers. The results for tables were not as impressive, achieving an F-measure of 0.925 for exact matches and 0.9896 for partial matches. Since even partial matches are likely to be useful in practice, the results were quite good.
Contributions
In this study, we have shown how using the line of text as the unit of analysis can play an important role in semi-structured elements and that text analysis can be beneficial at this level. TagLine has been shown to be effective for distinguishing specific classes of lines based on their structural roles and for identifying semi-structured text elements in medical progress notes. This will enable researchers to use more of the information in the text than was possible before. The information in the annotations can be stored in a database and used for other analyses as fully structured data. Once identified, semi-structured text elements can also be removed from the document so NLP can focus on the free-text sections which may result in more accurate concept extraction, as well as faster processing times for each note. The counts of these structures could be used as features in a machine learning approach for document classification tasks. TagLine can now be used to do ad hoc concept extraction from the semi-structured text by using rule as implemented in the JAPE module of GATE. Rules such as “if slot = <search term> lookup <filler>.”Only those structures that contained the search terms or concepts would be annotated and returned. Since more information can be extracted from the notes, more complete information is available for patient analysis or document classification. This additional data could enable researchers to explore topics better and perhaps improve the healthcare for veterans easier and faster than before.
Limitations
This study has two primary limitations. First the dataset, while adequate for purposes of providing sufficient examples for developing and evaluating learning machines, is still limited since the sample was taken from an existing study and may not include many note types and their specific challenges. TagLine needs further testing and development to ensure good generalizability. Also, samples of documents from other hospitals should to be included in the corpus to train models for increased generalizability.
Future Work
The parser in TagLine will be expanded to include other types of structures like full templates, questions and checkboxes. The language across note sections tends to differ.45 This feature is often used in NLP systems for “word sense disambiguation,” and is a fertile area for research46. There is a need to improve section identification or section header identification.47,48 We will also extend Tagline to function as a “sectionizer,” which accurately determines which section of the note a line of text belongs to. A sectionizer would be useful in word sense disambiguation and for text reduction, potentially saving significant amounts of time and money on creating annotated data sets. Human annotators would have smaller, more concentrated notes to cover if it is known that certain sections held no interest. This may increase productivity by shortening the time necessary to cover the note and decrease the likelihood of mental fatigue that occurs with longer notes, as well as reducing the amount of time spent on irrelevant documents.
Figure 3.
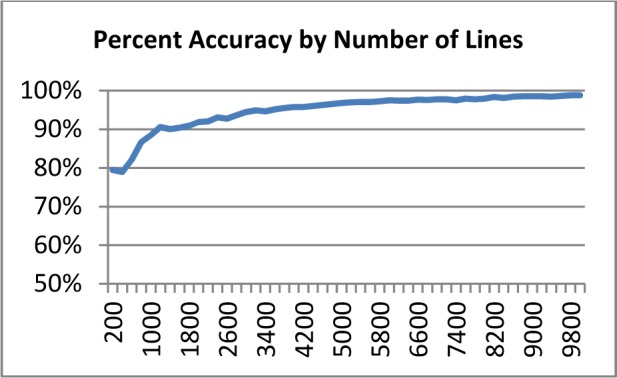
Learning Curve for Machine Learning
Table 3.
Sample line classes and their relationships to document elements.
| Line Class | Is-A | Part-Of |
|---|---|---|
| Double Xed Items | CheckBox | Question |
| Multiple Xed Items | CheckBox | Question |
| Xed item | CheckBox | Question |
| Line list with Header | List | List |
| Comma separated list on a line | List | List |
| List Header | ListHead | List |
| Numbered Item | ListItem | List |
| Medication Footer | MedFooter | Table |
| Medication List Directions | TableRow | Table |
| Med List Item (not numbered) | TableRow | Table |
Table 4.
Table Example
| Class | Line of Text | |||
|---|---|---|---|---|
| THE | ---- CBC PROFILE ---- | |||
| CLA CLA |
BLOOD | 01/19 14:20 |
Units | Reference Ranges |
| TBI | WBC | 5.7 | G/L | 4.2–10.3 |
| TBI | RBC | 4.8 | T/L | 4.2–5.8 |
| TBI | HGB | 14.2 | g/dL | 14–17 |
| TBI | HCT | 43.3 | % | 39–50 |
Acknowledgments
Funding for this work was provided by the Veterans Healthcare Administration Health Services Research & Development grants IIR05-120-3 and SDR HIR 09–002. The views expressed in this paper are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the US government.
References
- 1.Pakhomov S, Bjornsen S, Hanson P, Smith S. Quality performance measurement using the text of electronic medical records. Med Decis Making. 2008 Jul-Aug;28(4):462–470. doi: 10.1177/0272989X08315253. [DOI] [PubMed] [Google Scholar]
- 2.Rao RB, Krishnan S, Niculescu RS. Data mining for improved cardiac care. SIGKDD Explor. Newsl. 2006;8(1):3–10. [Google Scholar]
- 3.Guillen R.Identifying Obesity and Co-morbidities from Medical Records AMIA Symposium November14–182009San Fransisco [Google Scholar]
- 4.Zeng Q, Goryachev S, Weiss S, Sordo M, Murphy S, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Medical Informatics and Decision Making. 2006;6(1):30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCart JA, Berndt DJ, Jarman J, Finch DK, Luther SL. Finding falls in ambulatory care clinical documents using statistical text mining. Journal of the American Medical Informatics Association. 2012 Dec 15;2012 doi: 10.1136/amiajnl-2012-001334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turchin A, Kolatkar NS, Grant RW, Makhni EC, Pendergrass ML, Einbinder JS. Using regular expressions to abstract blood pressure and treatment intensification information from the text of physician notes. Journal of the American Medical Informatics Association. 2006;13(6):691. doi: 10.1197/jamia.M2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H, Shane S, Doan S, Johnson K, Waitman L, JDExtract Medication Information from Clinical Narratives AMIA Symposium November14–182009San Fransisco [Google Scholar]
- 8.Cerrito P. Data and text mining the electronic medical record to improve care and to lower costs. Paper presented at: SUGI-312006; San Fransisco, CA. [Google Scholar]
- 9.Honigman B, Lee J, Rothschild J, et al. Using computerized data to identify adverse drug events in outpatients. Journal of the American Medical Informatics Association. 2001;8(3):254. doi: 10.1136/jamia.2001.0080254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury G. Template Mining for Information Extraction from Digital Documents. Library Trends. 1999;48(1):182–208. [Google Scholar]
- 11.Buneman P. Semistructured data. Paper presented at: Syposium on Principles of Database Systems; Tucson, AZ: 1997. [Google Scholar]
- 12.Singh L, Chen B, Haight R, Scheuermann P. An algorithm for constrained association rule mining in semi-structured data. Lecture Notes in Computer Science. 1999:148–158. [Google Scholar]
- 13.Chang CH, Hsu CN, Lui SC. Automatic information extraction from semi-structured web pages by pattern discovery. Decision Support Systems. 2003;35(1):129–147. [Google Scholar]
- 14.Bratko A, Filipi B. Exploiting structural information for semi-structured document categorization. Information Processing and Management. 2006;42(3):679–694. [Google Scholar]
- 15.Hemnani A, Bressan S. Extracting information from semi-structured Web documents. Advances in Object-Oriented Information Systems. 2002:389–396. [Google Scholar]
- 16.DeJong G. An overview of the FRUMP system. Strategies for natural language processing. 1982:149–176. [Google Scholar]
- 17.Hayes PJ, Carbonell J. Natural Language Understanding. Encyclopedia of Artificial Intelligence. 1987:660–677. [Google Scholar]
- 18.Pakhomov S, Buntrock J, Duffy P. High throughput modularized NLP system for clinical text. 2005 [Google Scholar]
- 19.Lehnert W, Soderland S, Aronow D, Feng F, Shmueli A. Inductive text classification for medical applications. Journal of Experimental and Theoretical Artificial Intelligence. 1995;7:49–49. [Google Scholar]
- 20.Hsu J, Yih W. Template-based information mining from html documents. Paper presented at: National Conference on Artificial Intelligence; 1997. [Google Scholar]
- 21.Singh L, Chen B, Haight R, Scheuermann P, Aoki K. A robust system architecture for mining semi-structured data. Paper presented at: International Conference on Knowledge Discovery and Data Mining; 1998. [Google Scholar]
- 22.Ribeiro-Neto B, Laender A, da Silva A. Top-down extraction of semi-structured data. Paper presented at: String Processing and Information Retrieval Symposium and Internationa; Workshop on Grioupware; 1999. [Google Scholar]
- 23.Ribeiro-Neto B, Laender A, Da Silva A. Extracting semi-structured data through examples. 1999 [Google Scholar]
- 24.Xufa C. Semi-structured Data Extraction and Schema Knowledge Mining. Paper presented at: EUROMICRO; 1999. [Google Scholar]
- 25.Gao X, Sterling L. Semi-structured data extraction from heterogeneous sources. 2000 [Google Scholar]
- 26.Taniguchi K, Sakamoto H, Arimura H, Shimozono S, Arikawa S. Mining semi-structured data by path expressions. Lecture Notes in Computer Science. 2001:378–388. [Google Scholar]
- 27.Kashima H, Koyanagi T. Kernels for semi-structured data. 2002 [Google Scholar]
- 28.Wessman A, Liddle S, Embley D. DW: A generalized framework for an ontology-based data-extraction system. 2005 [Google Scholar]
- 29.Imtiaz H, Darlington J, Zuo L. Ontology Driven Web Extraction from Semi-structured and Unstructured Data for B2B Market Analysis. 2009 [Google Scholar]
- 30.Calvanese D, De Giacomo G, Lenzerini M. What can knowledge representation do for semi-structured data?. Paper presented at: Artificial Intellignce/Innovative applications of Artificial Intelligence; Madison, WI: 1998. [Google Scholar]
- 31.Calvanese D, De Giacomo G, Lenzerini M. Semi-structured data with constraints and incomplete information. Networking and Information Systems Journal. 1999;2:253–273. [Google Scholar]
- 32.Calvanese D, De Giacomo G, Lenzerini M. Queries and constraints on semi-structured data. Vol 1626/2009. Berlin: Springer; 1999. [Google Scholar]
- 33.Calvanese D, De Giacomo G, Lenzerini M. Modeling and querying semi-structured data. Networking and Information Systems Journal. 1999;2:253–273. [Google Scholar]
- 34.Zhou X, Han H, Chankai I, Prestrud A, Brooks A. Converting semi-structured clinical medical records into information and knowledge. Paper presented at: Proceedings of the International Workshop on Biomedical Data Engineering; 2005. [Google Scholar]
- 35.Kristianson K, Ljunggren H, Gustafsson L. Data extraction from a semi-structured electronic medical record system for outpatients: A model to facilitate the access and use of data for quality control and research. Health Informatics Journal. 2009;15(4):305. doi: 10.1177/1460458209345889. [DOI] [PubMed] [Google Scholar]
- 36.Abiteboul S. Querying Semi-Structured Data. 1997 [Google Scholar]
- 37.Fletcher RD, Dayhoff RE, Wu CM, Graves A, Jones RE. Computerized medical records in the Department of Veterans Affairs. Cancer. 2001;91(S8):1603–1606. doi: 10.1002/1097-0142(20010415)91:8+<1603::aid-cncr1173>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 38.Brown S, Hardenbrook S, Herrick L, St Onge J, Bailey K, Elkin P. Usability evaluation of the progress note construction set. 2001. [PMC free article] [PubMed] [Google Scholar]
- 39.Divita GA. Medical Document Text Element Ontology. Paper presented at: 2011 AMIA Annual Symposium; Wash DC: 2011. [Google Scholar]
- 40.Chieu HL, Ng HT. A maximum entropy approach to information extraction from semi-structured and free text. 2002 [Google Scholar]
- 41.Pakhomov SV, Ruggieri A, Chute CG. Maximum entropy modeling for mining patient medication status from free text. Proc AMIA Symp. 2002:587–591. [PMC free article] [PubMed] [Google Scholar]
- 42.Rao BR, Sandilya S, Niculescu R, Germond C, Goel A. Mining time-dependent patient outcomes from hospital patient records. Proc AMIA Symp. 2002:632–636. [PMC free article] [PubMed] [Google Scholar]
- 43.Korting TS. Image Processing Division, National. Institute for Space Research–INPE Sao Jose dos Campos; SP, Brazil: C4. 5 algorithm and Multivariate Decision Trees. [Google Scholar]
- 44.Quinlan JR. Rulequest Free Software Downloads. [ http://www.rulequest.com/download.html]. 2013 Accessed August 1, 2011.
- 45.Zeng Q, Redd D, Divita G, Jarad S, Brandt C. Characterizing Clinical Text and Sublanguage: A Case Study of the VA Clinical Notes. J Health Med Informat S. 2011;3:2. [Google Scholar]
- 46.Patterson O, Igo S, Hurdle JF. Automatic Acquisition of Sublanguage Semantic Schema: Towards the Word Sense Disambiguation of Clinical Narratives. 2010 [PMC free article] [PubMed] [Google Scholar]
- 47.Denny JC, Miller RA, Johnson KB, Spickard A., III Development and evaluation of a clinical note section header terminology. 2008 [PMC free article] [PubMed] [Google Scholar]
- 48.Denny J, Spickard A, III, Johnson K, Peterson N, Peterson J, Miller R. Evaluation of a Method to Identify and Categorize Section Headers in Clinical Documents. Journal of the American Medical Informatics Association. 2009:M3037v3031. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]


