Abstract
Small numbers of patients are a special challenge for rare diseases research. Electronic health record (EHR) data can facilitate research if patients with rare diseases can be reliably identified. We estimate the coverage of the names of a set of 6,519 rare diseases. Using the UMLS, 697 (11%) diseases were matched to ICD-9-CM, 1,386 (21%) to ICD-10-CM and 2,848 (44%) to SNOMED CT. Using published mappings from SNOMED CT to ICD, we further estimate additional broader matches of 2,569 (39%) rare diseases to ICD-9-CM and 1,635 (25%) to ICD-10-CM. The number of codes that match one and only one disease are 1,081 (62%) for ICD-9-CM, 1,403 (73%) for ICD-10-CM, and 3,311 (85%) for SNOMED CT. Our findings confirm that SNOMED CT has the greatest coverage and specificity needed to identify patients with a rare disease from EHR-data, and can facilitate research and evidence-based care.
Introduction
Rare diseases are defined in the US as conditions that affect less than 200,000 Americans and in the European Union as those with a prevalence of 5 per 10,000 or less. They are largely, but not exclusively, genetic disorders. Because of the variation in definition, there is no globally authoritative list of rare diseases, but the number of recognized rare diseases is between 6–7,000 diseases.1–4 Although each condition is uncommon, collectively rare diseases are more common. The National Organization for Rare Disorders estimates that up to 30 million (or 1 in 10) Americans are affected by a rare disease.3 Consequently, rare diseases have emerged as priority research topics in both the US and the EU. Advances in genetic testing and the emergence of personalized medicine increase the number of subtypes of common diseases, further motivating development of research methods for rare diseases.
To identify sufficient numbers of rare diseases patients for research, multiple clinical sites and countries are often required and electronic health record (EHR) data can facilitate the identification of rare disease patients across multiple sites. Phenotype definitions that leverage widely adopted administrative terminologies (such as ICD-9-CM and ICD-10-CM) can potentially enable the consistent identification of patients with rare diseases from different providers and organizations. Our collective national capacity for rare diseases research, therefore, depends upon adequate coverage of rare diseases in these administrative terminologies. On the other hand, according to the “meaningful use” incentive program for the use of EHRs, clinical terminologies (such as SNOMED CT) are required for the encoding of clinical information in the EHR.5 The encoded clinical information will in turn drive the EHR-embedded information and decision support (e.g., InfoButtons6, clinical practice guidelines) and consumer health information (e.g., MedlinePlus Connect) functionalities. To comprehend the national capacity for rare diseases research, EHR-enabled clinical decision support and patient education, we estimate the coverage of rare diseases in ICD-9-CM, ICD-10-CM, and SNOMED CT for a set of 6,519 rare diseases, and explore the granularity of rare disease terms in these terminologies. We examine in detail the matches found for a set of rare diseases that are being studied in the national Patient Centered Outcomes Research network (PCORnet), recently established this year to create a national infrastructure for observational and clinical research in diverse and distributed healthcare organizations. 7
Background
Rare diseases have become an increasingly important topic in health care research and policy contexts. Rare diseases are explicitly represented in important federally-funded research initiatives, such as the Rare Diseases Clinical Research Network8 and the Clinical and Translational Science Awards (CTSA) program. Despite differences in disease etiology and affected populations, there are common logistical challenges for research that can be addressed in part with data standards and informatics expertise and tools.9–12 Generalizable research methods that address issues specific to rare diseases can impact the investigation of thousands of rare conditions and hundreds of thousands of Americans. With increased adoption and meaningful use of EHRs, there is renewed effort in leveraging EHRs for research. The Patient-Centered Outcomes Research Institute (PCORI) was funded from the Affordable Care Act to examine real-world treatment decisions.13 PCORI network is specifically tasked to conduct observational and interventional research on the comparative effectiveness of various treatments using distributed and heterogeneous healthcare organizations and various EHR systems. PCORI currently supports the research for approximately 50 rare diseases (see appendix). The motivation for this paper was to explore the coverage of rare diseases in standard terminologies in order to characterize the current capacity for EHR-based research on those diseases, and to suggest strategies that will increase the national research capacity for all rare diseases.
There are several initiatives that have complete or partial inventories of rare disease names and terms. The Office of Rare Diseases Research (ORDR) of the National Center for Advancing Translational Sciences (NCATS) in the U.S. and Orphanet in the E.U. recognize 6 -7,000 disorders as rare diseases and support various efforts to link these disease names to standard terminologies. The ORDR also supports the Genetic and Rare Diseases Information Center (GARD), a web-based information resource for the public on more than 6,000 rare diseases.14 The Genetics Home Reference, maintained by the National Library of Medicine, includes a smaller set of approximately 800 genetic diseases, most of which are rare. The NLM has identified and validated SNOMED CT codes for these 800 diseases to supports public retrieval of information. Orphanet, an EU-wide advocacy and information organization funded by national and European public institutions and patient organizations, foundations and corporations, provides information to the public on approximately 7,000 rare disorders on its web-based Portal for Rare Diseases and Orphan Drugs.1 Orphanet also sponsors OrphaData, which provides the scientific community with data and tools to support the identification, quantification, and research of rare disorders. As part of this effort, Orphanet recently developed a rare disease ontology (ORDO) which serves as an inventory and classification of rare diseases, cross-referenced with OMIM, ICD-10, and SNOMED-CT and with genes in HGNC, OMIM, UniProtKB and Genatlas.15, 16
Existing standard terminologies, such as ICD and SNOMED CT are important components for EHRs and rare diseases research. Over 3,000 distinct concepts (including diagnoses, findings, treatments and procedures) from 4 medical centers were used to evaluate the content coverage of these and other clinical coding systems.17 Although no coding system captured all concepts, SNOMED was the most complete. The authors concluded that both ICD-9-CM and ICD-10 fail to capture substantial clinical content, and warned that analytic conclusions that depend on these coding systems may be suspect. ICD-10 is critical for global surveillance of rare diseases. The Clinical Modifications (CM), e.g., ICD-9-CM and ICD-10-CM are critical for billing and reimbursement in the U.S. SNOMED CT is becoming increasingly adopted as a supporting clinical terminology in EHR systems worldwide18, and gaining attention in the US since being named as a reporting standard for problem lists.5 Previous studies have shown significant inclusion of rare diseases in SNOMED CT19, 20, and anecdotally the IHTSDO (International Health Terminology Standards Development Organisation) is committing to increasing this coverage. SNOMED CT plays an important role in context-aware knowledge retrieval applications (i.e., InfoButtons) and the identification of patient-directed consumer information from the various information resources such as the Genetic Home Reference and MedlinePlus.
There are different approaches to identifying rare disease names in standard terminologies. Rare disease names from the Office of Rare Diseases have been mapped to the Unified Medical Language System (UMLS) to facilitate coding in other systems such as Medical Subject Headings (MeSH) for medical literature, ICD for public health surveillance, and SNOMED CT for use in clinical records documentation and clinical decision support.19 In 2010, the NLM mapped 8,435 rare disease names (collected from ORDR, Orphanet, and the National Organization for Rare Disorders, a patient advocacy and voluntary health organization in the US) to the UMLS, and found different levels of coverage for Medical Subject Headings (MeSH) (5,663 ; 67%), Online Mendelian Inheritance in Man (OMIM) (3,802 ; 45%), SNOMEDCT (4,192 ; 50%), and ICD-10 (1,029 ;12%).20
In this investigation, we re-examine the current coverage of rare disease names in standard coding systems to support two use cases: 1) the identification of rare disease patients from EHR data for research, and 2) the identification of appropriate rare diseases information, including published medical literature, clinical practice guidelines for providers and authoritative consumer-directed information for patients, using coded data from EHRs.
Further, we explore differences in granularity between various terminologies, all in the context of how EHRs can support the consistent and reliable identification of rare disease patients, to enable evidence-based care and multi-site research.
Methods
Estimating the Coverage of Rare Diseases
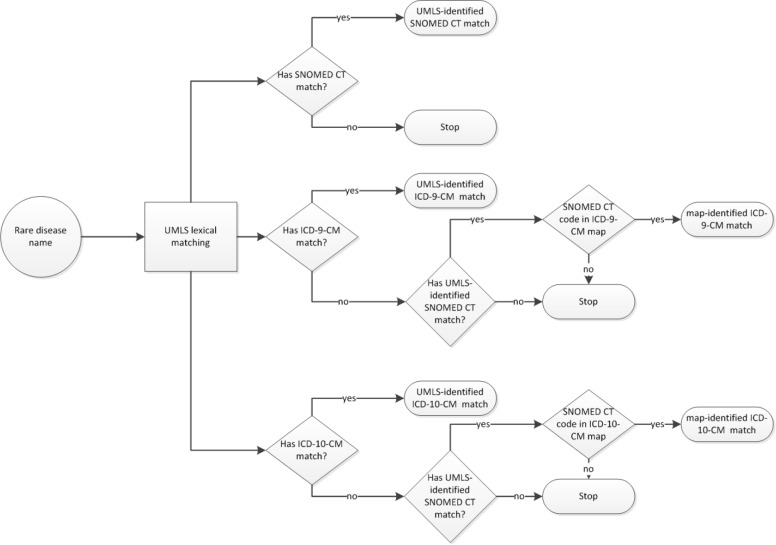
We estimated the coverage of rare diseases in the three terminologies: ICD-9-CM, ICD-10-CM and SNOMED CT. To do this, we used two resources: the UMLS and the published maps from SNOMED CT to ICD-9-CM (developed by IHTSDO) and ICD-10-CM (developed by NLM). We first matched the 6,519 ORDR rare diseases by their names to the UMLS using lexical matching, utilizing both exact and normalized string matches, followed by semantic group validation (with restriction to the Semantic Group Disorders). Through the UMLS concept structure, we identified matches to SNOMED CT, ICD-9-CM and ICD-10-CM codes (we call these UMLS-identified matches). We anticipated that the UMLS-identified codes were mostly equivalent matches, since the UMLS concept structure is based on synonymy (i.e., not broader or narrower matches). For ORDR rare diseases with UMLS-identified SNOMED CT match but no ICD match, we further used the SNOMED CT to ICD-9-CM and ICD-10-CM published maps as an alternative path to match to ICD-9-CM and ICD-10-CM codes (we call these map-identified matches). (Figure 1) The published maps enabled us to identify matches other than equivalent matches, since the ICD map targets could be broader (often) or narrower (seldom) than the SNOMED CT concept. Since the published maps did not cover all of SNOMED CT, we extrapolated the results to estimate the number of map-identified matches that we could potentially find if all SNOMED CT concepts were included in the published maps.
Figure 1.
Overview of matching methods.
Estimating the Granularity of Matches for Rare Diseases
To study the impact of lack of equivalent matches for rare diseases, we looked at the ability of a specific code in the three terminologies to identify a specific disease. Using the matches identified above, we calculated the extent to which multiple rare diseases were included in a single code in the three terminologies.
Manually Validating a Sample of Matches
We reviewed a small set of rare diseases to validate our matching methods. We identified 46 rare disease categories under study in the PCORnet, specifically those diseases listed on applications for the 11 funded Clinical Data Research Networks (CDRNs) and 15 funded Patient Powered Research Networks (PPRNs).7 We exploded some disease categories like ‘vasculitis’ and ‘primary immunodeficiency diseases’ to include specific diseases, such as Churg-Strauss Syndrome and Severe Combined Immunodeficiency.
The matches in the three terminologies found for the PCORnet rare diseases were reviewed by two authors (RR, medical informatician; KWF, physician) familiar with medical terminologies. Specifically, each assessed whether the match for the PCORnet diseases was an equivalent, narrower (more precise), broader (less precise) or related match. Where there was discrepancy between reviewers, consensus was reached through discussion.
As a general reference for comparison, we also calculated the coverage and granularity for the Orphanet’s ORDO, based on the accompanying SNOMED CT mappings in the ontology.
Results
Estimating Rare Diseases Coverage
Using the names and synonyms of the 6,519 ORDR rare diseases for lexical matching in the UMLS, 697 (11%), 1,386 (21%) and 2,848 (44%) diseases were matched to ICD-9-CM, ICD-10-CM and SNOMED CT respectively. These were the UMLS-identified matches.
Among the 5,822 rare diseases with no UMLS-identified match to ICD-9-CM, 2,783 SNOMED CT matches were found, of which 80% (2,448 SNOMED CT codes) were included in the ICD-9-CM published map. This yielded map-identified matches for 2,055 (32%) diseases to ICD-9-CM. If all SNOMED CT concepts were included in the published ICD-9-CM map, the projected map-identified matches for ICD-9-CM would be 2,569 (39%) diseases. Similarly, among the 5,133 rare diseases with no UMLS-identified ICD-10-CM match, 1,841 SNOMED CT matches were identified, of which 56% (1,035 SNOMED CT codes) were included in the ICD-10-CM published map, which yielded map-identified matches for 919 (14%) diseases. The projected map-identified match for ICD-10-CM was 1,635 (25%) diseases. (Table 1)
Table 1.
Coverage of rare diseases in the three terminologies.
| UMLS-identified match | Map-identified match (found) | Map-identified match (projected) | |
|---|---|---|---|
| ICD-9-CM | 697 (11%) | 2055 (32%) | 2569 (39%) |
| ICD-10-CM | 1386 (21%) | 919 (14%) | 1635 (25%) |
| SNOMED CT | 2848 (44%) | n/a | n/a |
As a comparison, Orphanet’s rare disease ontology (ORDO) contained 6,750 diseases, among them 1,446 (21%) diseases were accompanied by matches to SNOMED CT.
Estimating the Granularity of Matches for Rare Diseases
Using the matches identified above, we calculated the extent to which multiple diseases were included in a single code in the three terminologies. (Table 2) We define a unique match as a code that matches to only one rare disease, and a multiple match as a code that matches to more than one rare disease. The number and proportion of unique matches were 1,081 (62%) for ICD-9-CM, 1,403 (73%) for ICD-10-CM, and 3,311 (85%) for SNOMED CT. Overall, 672 (38%) of the matched ICD-9-CM codes were multiple matches, which was lower for ICD-10-CM (n=526, 27%) and lowest for SNOMED CT (n=598, 15%). As for the cardinality of the broader matches, the maximum number of diseases matched to a SNOMED CT code was 5 diseases. The highest number of diseases matched to a single code was 208 for ICD-9-CM and 23 for ICD-10-CM. There were 117 ICD-9-CM and 40 ICD-10-CM codes matching to more than 5 diseases.
Table 2.
Number of Rare Diseases Included in Matched Codes from Source Terminologies.
| # rare diseases matching to a code | # ICD-9-CM codes (% of total codes) | # ICD-10-CM codes (% of total codes) | # SNOMED CT codes (% of total codes) |
|---|---|---|---|
| 1 (unique match) | 1081 (62%) | 1403 (73%) | 3311 (85%) |
| 2 | 319 | 328 | 478 |
| 3 | 125 | 88 | 84 |
| 4 | 68 | 45 | 33 |
| 5 | 43 | 25 | 3 |
| > 5 | 117 | 40 | 0 |
| # codes matching to > 1 disease (% of total codes) (multiple match) | 672 (38%) | 526 (27%) | 598 (15%) |
| Examples | 208 rare diseases matched to 759.89 Other specified congenital anomalies | 22 rare diseases matched to Q82.8 Other specified congenital malformations of skin | 5 rare diseases matched to 28835009 Retinitis pigmentosa |
In Orphanet’s ORDO, 1,446 rare diseases were matched to 1,748 SNOMED CT codes. Most of the SNOMED CT codes (1,735, 99%) were matched to a single disease, and 13 SNOMED CT codes were matched to two diseases.
Manual Review of Matches
Among the UMLS-identified matches, the proportions of equivalent matches were 93%, 68% and 74% for ICD-9-CM, ICD-10-CM and SNOMED CT respectively. Among the map-identified matches, 87% of the ICD-9-CM matches and 64% of the ICD-10-CM matches were broader matches. Overall, 8 (15%) of the 53 diseases could not be matched to any of the three terminologies. Among the 45 diseases that could be matched, an equivalent match could be found for 15 (28%), 22 (42%), 43 (81%) diseases for ICD-9-CM, ICD-10-CM and SNOMED CT respectively. (Table 3)
Table 3.
Manual review of PCORnet rare diseases matches.
| ICD-9-CM | ICD-10-CM | SNOMED CT | ||
|---|---|---|---|---|
| UMLS-identified matches (% of all UMLS-identified matches) | equivalent | 14 (93%) | 13 (68%) | 46 (74%) |
| broader | 0 (0%) | 3 (16%) | 2 (3%) | |
| narrower | 1 (7%) | 2 (11%) | 9 (15%) | |
| related | 0 (0%) | 1 (5%) | 5 (8%) | |
| total | 15 (100%) | 19 (100%) | 62 (100%) | |
| Map-identified matches (% of all map-identified matches) | equivalent | 1 (2%) | 9 (25%) | n/a |
| broader | 45 (87%) | 23 (64%) | n/a | |
| narrower | 4 (8%) | 3 (8%) | n/a | |
| related | 2 (4%) | 1 (3%) | n/a | |
| total | 52 (100%) | 36 (100%) | n/a | |
| # diseases with equivalent match (% of total diseases) | 15 (28%) | 22 (42%) | 43 (81%) | |
| # diseases with no equivalent match | 30 (57%) | 23 (43%) | 2 (4%) | |
| # diseases with no match | 8 (15%) | 8 (15%) | 8 (15%) | |
| Total # diseases | 53 (100%) | 53 (100%) | 53 (100%) | |
Discussion
We first estimated the coverage of ORDR rare disease names in ICD-9-CM, ICD-10-CM, and SNOMED CT by lexical mapping to the UMLS. As expected, we found increasing coverage from ICD-9-CM (697; 13%) to ICD-10-CM (1,386; 26%), with the highest coverage for SNOMED CT (2,848; 53%). This is consistent with previous findings on the higher general clinical coverage of SNOMED CT. 17 The coverage of rare disease names in SNOMED CT is slightly lower than the 50% coverage seen in 2010, 20 although the earlier study used a larger set of rare disease names from multiple sources. As shown by the manual review, most of the UMLS-identified matches are equivalent matches.
This study differs from earlier work by Pasceri 20 in that we did not stop at lexical matching by the UMLS. In view of the low coverage of rare disease names in ICD-9-CM and ICD-10-CM, we explored new ways to match to these terminologies. We made use of the published maps from SNOMED CT to ICD-9-CM and ICD-10-CM to provide a cross-walk to the ICDs via SNOMED CT. This could considerably increase the matching rates to ICD-9-CM (from 11% to 50%) and ICD-10-CM (from 21% to 46%). However, most of these additional matches were not equivalent matches, and were either broader (majority) or narrower/related (minority) matches, as confirmed by the manual review. While these non-equivalent matches may still be useful in some use cases, e.g., to narrow down a large cohort of patients to those who may be suffering from a rare disease, they may not be precise enough to support direct patient care e.g. offering specific advice on the treatment of a particular disease.
Our work specifically supports use cases related to the use of EHRs to support the consistent and reliable identification of rare disease patients to enable evidence-based care and multi-site research. When searching health system data for rare disease patients, fine-grained and specific codes are preferable. Our data show that SNOMED CT has more codes (than ICD-9-CM or ICD-10-CM) that relate to one and only one disease. Due to the need to support statistical analysis in ICD-9-CM and ICD-10-CM, grouper concepts are more prevalent. This will cause problem when a code is required to identify a specific rare disease, such as linking an entry in the EHR to some disease-specific information. Our analysis shows that a higher percentage of ICD-9-CM and ICD-10-CM codes (38% and 27% respectively) lead to more than one rare disease, compared to 15% for SNOMED CT. One ICD code can lead to hundreds of diseases while the number is much smaller in SNOMED CT.
Although the PCORnet rare diseases that we explored in detail are not necessarily representative of all the rare diseases, these conditions are important in that they are being studied now. The proportion of diseases with equivalent match for the PCORnet list is considerably higher than what we saw for the 6,519 rare diseases overall. It is possible that the PCORnet funded research addresses more well-known or important diseases, so that more of them make their way into standard terminologies. Even if the small set of rare diseases that we used for validation are not representative of other rare diseases, their association with the PCORnet national research network that is actively exploring the use of EHR data in observational and interventional research will make them exemplars for refining strategies to increase the national capacity for rare diseases research and evidence-based care.
Limitations of our study include the following. We only focused on one source of rare disease names (ORDR). We did not do a comprehensive review of all the matches found in the three terminologies. The PCORnet rare diseases that we reviewed might not be representative of all rare diseases. Using the published maps to cross-walk to ICD-9-CM and ICD-10-CM was only possible for those diseases with UMLS-identified SNOMED CT matches.
The Way Forward
ICD and SNOMED CT are designed for very different purposes, and the rare disease community can benefit from knowing this distinction. As a classification system, ICD by definition includes categories that are designed to be exhaustive and mutually exclusive. For statistical purposes (in epidemiology and billing use cases) the idea of “multiple counting” (e.g., classifying a disease in two different hierarchies) is discouraged, and residual categories (e.g., some diseases ‘not elsewhere classified’) can be meaningful. In fact, one reason for the residual categories is to avoid the need to assign codes to diseases with very low prevalence (i.e. rare diseases) and to maintain statistical balance of the coding categories. On the other hand, SNOMED CT as a clinical terminology is designed to support the representation of any concept to be stated about the patient. In the context of a terminology, “multiple counting” is desirable, and residual categories are meaningless. For example, for a patient suffering from Laurence-Moon syndrome, the diagnostic label “Other specified congenital malformation syndromes, not elsewhere classified” will not be useful in finding disease-specific patient education information or clinical practice guidelines. Another advantage of SNOMED CT is its shorter update cycle of 6 months, compared to yearly updates for ICD.
There is great potential benefit in the use of SNOMED CT as a source terminology in EHRs. Using a robust and fine-grained terminology such as SNOMED CT, clinicians can document patient data once at the point of care with fidelity to the clinical situation, at the appropriate level of granularity and certainty or uncertainty. These data, encoded in SNOMED CT, could be re-used for automated decision support and accessing customized information at the point of care. Others have shown that coarse disease classifications, such as ICD, are insufficient for these purposes, and our data indicate that SNOMED CT has higher proportion of fine-grained, or highly specific, codes for each disease. The use of SNOMED CT in the EHR can enable clinicians to record data only once at the point of care. Then the mappings of SNOMED CT to ICD classifications can be used to leverage these data for epidemiologic purposes, health system management, and billing. This would avoid duplicate effort of double-coding and potentially avoid the skewing of clinical data for billing purposes. To see how this will work for ICD-10-CM take a look at the I-MAGIC demo tool at NLM’s website. 21
Rare disease advocates should work to include rare diseases specifically in SNOMED CT. In future, a tighter integration between SNOMED CT and ICD-11 is anticipated. Concepts in SNOMED CT will then find a natural path into ICD, avoiding the risk of code translation or mapping errors. This is in line with Orphanet’s exhortation for inclusion of more rare diseases in ICD.
Conclusions
To support patient care, patient education and research in rare diseases, adequate coverage of rare diseases in standard terminologies is essential. Existing coverage in SNOMED CT is higher than ICD-9-CM and ICD-10-CM, and with higher precision. More work is needed to improve coverage.
Acknowledgments
This work was partly supported by the Intramural Research Program of the National Institutes of Health and the National Library of Medicine.
Appendix. List of rare disease categories (46) studied in PCORI networks and grant awards.
| Adrenoleukodystrophy | Granulomatosis with Polyangiitis | Phelan-McDermid Syndrome |
| Aicardi Syndrome | Hepatitis | Primary Immunodeficiency Diseases |
| alpha-1 antitrypsin deficiency | Hypoplastic left heart syndrome | Primary Nephrotic Syndrome (Focal Segmental Glomerulosclerosis) |
| Alström syndrome | Hypothalamic Hamartoma | Pseudoxanthoma elasticum |
| Amyotrophic Lateral Sclerosis | Inflammatory breast cancer | Psoriasis |
| Becker muscular dystrophy | Joubert syndrome | Pulmonary fibrosis |
| Chronic Granulomatous Disease | Juvenile Rheumatic Disease | Rare Cancers |
| Churg-Strauss Syndrome | Kawasaki Disease | Selective IgA Deficiency |
| Co-infection with HIV and hepatitis C virus | Klinefelter syndrome and associated conditions | Severe Combined Immunodeficiency |
| Common Variable Immunodeficiency | Lennox-Gastaut Syndrome | Severe Congenital Heart Disease |
| Cystic fibrosis | Membranous Nephropathy | Sickle Cell Disease |
| DiGeorge Syndrome | Metachromatic leukodystrophy | Sickle cell disease; Recurrent C. Difficile colitis |
| Dravet Syndrome | Microscopic Polyangiitis | Tuberous Sclerosis |
| Duchenne muscular dystrophy | Minimal Change Disease | X-Linked Agammaglobulinemia |
| Dyskeratosis congenital | Multiple Sclerosis | |
| Gaucher disease | Pediatric Transverse Myelitis |
Contributor Information
Kin Wah Fung, Email: kwfung@nlm.nih.gov.
Rachel Richesson, Email: rachel.richesson@dm.duke.edu.
Olivier Bodenreider, Email: obodenreider@mail.nih.gov.
References
- 1.Orphanet The portal for rare diseases and orphan drugs. 2014. [cited 2014 July 21]; Available from: http://www.orpha.net/consor/cgi-bin/index.php.
- 2.NIH Office of Rare Diseases Research (ORDR) Brochure. 2009. [cited 2014 July 21]; Available from: http://rarediseases.info.nih.gov/asp/resources/ord_brochure.html.
- 3.NORD Rare Disease Information. 2014. [cited 2014 July 21]; Available from: http://www.rarediseases.org/rare-disease-information.
- 4.European Commission Public Health Policy Rare Diseases. [cited 2014 July 21]; Available from: http://ec.europa.eu/health/rare_diseases/policy/index_en.htm.
- 5.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med. 2010;363(6):501–4. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 6.Strasberg HR, Del Fiol G, Cimino JJ. Terminology challenges implementing the HL7 context-aware knowledge retrieval (‘Infobutton’) standard. J Am Med Inform Assoc. 2013;20(2):218–23. doi: 10.1136/amiajnl-2012-001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.PCORI PCORnet: The National Patient-Centered Clinical Research Network+2014[cited 2014 July 21]; Available from: http://www.pcori.org/funding-opportunities/pcornet-national-patient-centered-clinical-research-network/ [DOI] [PMC free article] [PubMed]
- 8.Hampton T. Rare Disease Research Gets Boost. JAMA. 2006;295:2836–2838. doi: 10.1001/jama.295.24.2836. [DOI] [PubMed] [Google Scholar]
- 9.Griggs RC, et al. Clinical research for rare disease: opportunities, challenges, and solutions. Mol Genet Metab. 2009;96(1):20–6. doi: 10.1016/j.ymgme.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luisetti M, et al. The problems of clinical trials and registries in rare diseases. Respir Med. 2010;104(Suppl 1):S42–4. doi: 10.1016/j.rmed.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 11.Seminara J, et al. Establishing a consortium for the study of rare diseases: The Urea Cycle Disorders Consortium. Mol Genet Metab. 2010;100(Suppl 1):S97–105. doi: 10.1016/j.ymgme.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckfeldt JH. Statement on the International Rare Diseases Research Consortium. Research and Innovation - Health. 2011. [cited 2014 July 21]; Available from: http://ec.europa.eu/research/health/news-07_en.html.
- 13.Selby JV, Beal AC, Frank L. The Patient-Centered Outcomes Research Institute (PCORI) national priorities for research and initial research agenda. JAMA. 2012;307(15):1583–4. doi: 10.1001/jama.2012.500. [DOI] [PubMed] [Google Scholar]
- 14.NIH Rare Diseases and Related Terms. [cited 2014 July 21]; Available from: http://rarediseases.info.nih.gov/RareDiseaseList.aspx.
- 15.Orphanet Orphanet Rare Disease Ontology (ORDO) 2014. [cited 2014 July 21]; Available from: http://www.orphadata.org/cgi-bin/inc/ordo_orphanet.inc.php.
- 16.Rath A, et al. Representation of rare diseases in health information systems: the Orphanet approach to serve a wide range of end users. Hum Mutat. 2012;33(5):803–8. doi: 10.1002/humu.22078. [DOI] [PubMed] [Google Scholar]
- 17.Chute CG, et al. The content coverage of clinical classifications. For The Computer-Based Patient Record Institute’s Work Group on Codes & Structures. J Am Med Inform Assoc. 1996;3(3):224–33. doi: 10.1136/jamia.1996.96310636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee D, et al. Literature review of SNOMED CT use. J Am Med Inform Assoc. 2014;21(e1):e11–9. doi: 10.1136/amiajnl-2013-001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rance B, et al. Leveraging terminological resources for mapping between rare disease information sources. Stud Health Technol Inform (Proc Medinfo) 2013:529–533. [PMC free article] [PubMed] [Google Scholar]
- 20.Pasceri E. Analyzing rare diseases terms in biomedical terminologies. (LHNCB Medical Informatics Training Program Final Report; Dr. Olivier Bodenreider, Mentor) 2010. [cited 2014 July 21]; Available from: http://mor.nlm.nih.gov/pubs/alum/2010-pasceri.pdf.
- 21.NLM SNOMED CT to ICD-10-CM Map. [cited 2014 July 21]; Available from: http://www.nlm.nih.gov/research/umls/mapping_projects/snomedct_to_icd10cm.html.