Abstract
Use of deceased subject Electronic Health Records can be an important piloting platform for informatics or biomedical research. Existing legal framework allows such research under less strict de-identification criteria; however, privacy of non-decedent must be protected. We report on creation of the decease subject Integrated Data Repository (dsIDR) at National Institutes of Health, Clinical Center and a pilot methodology to remove secondary protected health information or identifiable information (secondary PxI; information about persons other than the primary patient). We characterize available structured coded data in dsIDR and report the estimated frequencies of secondary PxI, ranging from 12.9% (sensitive token presence) to 1.1% (using stricter criteria). Federating decedent EHR data from multiple institutions can address sample size limitations and our pilot study provides lessons learned and methodology that can be adopted by other institutions.
Introduction
The use of electronic health records (EHR) for research is becoming commonplace. Patient privacy and ethical oversight by institutional review boards (IRBs) present practical and administrative challenges for scientists who need to access Big Data.1 De-identification (elimination of the protected sections of health information) can allow EHR records to be considered “not human research subject data”, thus reducing regulation.2 Another approach is to restrict research to the use of deceased patients’ records.3 Although data analyses that use only deceased patient records may have inadequate power due to small sample size, they can provide pilot results whether particular EHR-based data extraction or analysis is feasible. Data analyses in multi centric clinical studies that use data inputs from varying EHR software vendors or varying user groups may benefit from an administratively simpler pilot experiment first. Such a pilot study on deceased subjects’ records can prevent situations in which an investigator tediously obtains IRB approval for a multi-site study that uses more complete record sets (such as de-identified or even identified data) only to discover that the records lack the data necessary for the study.
In a previous perspective paper,3 we outlined several challenges to establishing a deceased subject integrated data repository (dsIDR), such as proving death status, involvement of next of kin (surviving spouse or children), possible deletion of deceased patient data by the organization (to limit the data warehouse size after the minimum record-keeping period expired), and patients’ perceptions of the value of dsIDRs. Creation of dsIDRs using only terminology-coded information, such as structured medication, laboratory results and diagnostic or procedural data that do not require de-identification is relatively easy to accomplish. However, additional challenges are encountered when a dsIDR contains narrative text clinical documents. In such a repository, personal identifiers defined in the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule (Safe Harbor Method) as well as “personally identifiable information” (PII) defined in the Privacy Act must be detected and redacted properly.
One particular challenge with narrative text document inclusion in the dsIDR, identified in our prior analysis, was handling the PHI of people other than the patient. Because research use of private health data of living and deceased is regulated differently, we need to distinguish two types of PHI/PII: primary (about the primary patient in question) and secondary (about individuals other than the primary patient, such as relatives or caregivers). We use the term secondary PxI to refer to both secondary PHI and secondary PII. We also use the term PHI to denote removal of the 18 specified identifiers (e.g., primary or secondary phone number), rather than removal of health data. The regulatory provisions for decedent EHR research do not mandate specific de-identification method (as opposed to the clear definition of 18 PHI identifiers in the Safe Harbor Method). The relaxed approach to de-identification in a dsIDR does not extend, however, to secondary PxI of living relatives, which must be entirely removed. This removal may be more difficult since the typical approach of using the primary patient’s name may not be helpful in identifying secondary names. The process is further complicated because primary PHI that would ordinarily be allowed in a dsIDR must be removed if it helps reveal identity of secondary persons mentioned in the record (e.g., if a primary patient last name remains in the record, it may also provide a last name for a living relative identified in the EHR only by a first name). Based on our own legal analysis, we hypothesize that mention of dates and locations (e.g., city or town) that would have to be scrubbed in traditional de-identification for whole population IDR, may stay in dsIDR.
In this paper, we report on efforts to establish a dsIDR at the National Institutes of Health’s Clinical Center (NIH CC) and our investigation of the rate of secondary PxI in deceased patients’ EHR.
Background
Institutional context
The NIH Clinical Center is a 240-bed hospital dedicated to research. Since 2008, it has established an integrated data repository called the Biomedical Translational Research Information System (BTRIS). BTRIS contains data on over 500,000 research subjects seen at the NIH CC since 1976. BTRIS contains data from the current EHR (Sunrise Clinical Manager v5.5, Allscripts, Chicago, IL), a prior EHR (Medical Information System, Technicon Data Systems, Tarrytown, NY) and several ancillary clinical and research systems.4 BTRIS offers two modes of data export: identified data extracts available to study investigators and de-identified data extracts available to any NIH investigator. Currently, de-identified data exports are limited to structured coded data and a limited number of narrative text documents (e.g., pathology reports).
Design Principles of NIH CC dsIDR (dsBTRIS)
With our dsIDR dataset and framework, we hope to offer interested researchers yet another platform for generating hypotheses and piloting informatics methods. We also hope to learn important lessons about dsIDR design and actual use from creating a single institution dsIDR. These findings can inform development of a federated dsIDR of deceased patient EHR data from multiple institutions. The design principles for our dsIDR include the following elements:
Access is limited to researchers who obtain approval (with exemption from IRB review) from the NIH Office of Human Subject Research Protection (OHSRP)
Researchers must certify that they will not try to identify deceased subjects or other persons in the provided data
- The EHR of deceased patients undergoes the following transformations
- Structured data: direct patient identifiers (e.g., structured name, address or employer field) are removed since they have none or very limited value to research
- Unstructured data: The current dsIDR implementation pilot does not contain any unstructured data; selection of the best de-identification approach is one of the goals of this study; we plan to remove primary patient name from narrative text documents (e.g., progress note and laboratory text comments); dates and locations (e.g., “Suburban Hospital”) would remain in the documents; secondary PxI would also be removed
Legal framework for deceased subject research
A legal framework for use of EHR data post mortem has been explored in greater detail in our earlier perspective paper.3 The most important legal factor is that deceased subject research does not require traditional consent and full IRB review. Decedent research is listed in Title 45 of the Code of Federal Regulation under section §164.512, titled “Uses and disclosures for which an authorization or opportunity to agree or object is not required” [45 CFR 164.512(i)(1)(iii)].5 The enabling statue for this regulation is the Health Insurance Portability and Accountability Act (HIPAA; Public Law 104-191).
The regulation allows disclosure of PHI of decedents without the need to seek full IRB approval and has to adhere to the following conditions:6
use is sought solely for research on the PHI of decedents;
the researcher can provide, on request, documentation of the death for individuals used in the study; and
the PHI is necessary for the research.
Throughout this article, we refer to this HIPAA provision as “decedent research clause”.
When information is private
Of note is also the fact that health related information may become secondary PHI only after it is combined with a clear designation of a secondary person (such as full name or phone number). If a record contains the sentence “Her mother and daughter both have symptoms of multiple sclerosis”, it only becomes secondary PHI when associated with a living person’s full name. How unique and identifying is the context information provided about the secondary person is of significance (quasi-identifiers). This masking phenomenon is well described by the concept of k-anonymity (each dataset record is indistinguishable from at least k-1 other records given a group of identifying attributes) and the concept of l-diversity (each group of identifying attributes is immune to probabilistic inference attack and has at least l well represented value).7 There is no clear established boundary; however, the HIPAA ZIP code rules offer one potential precedence: HIPAA zip code rule (45 CFR 164.514) permits revealing 3-digit ZIP codes as long as the 3-digit ZIP code covers an area populated by more than 20,000 people, as this is considered to be sufficient “masking” of the individual. The masking principle is important in redacting or preserving a sentence, such as, “Patient has a 9-year-old daughter” in a document that otherwise contains unmodified dates and locations, but does not contain the primary patient name.
Fall back policy
In addition to the dsIDR use policy that forbids subject re-identification, NIH CC has a general institutional policy for situations when identifiers are discovered in a dataset that has been thought to be de-identified. The existing NIH CC process generating de-identified data starts with production of an initial de-identified data set. An NIH employee not affiliated with the research (an “honest broker”) reviews any BTRIS-derived dataset and determines whether it is free of PHI. If the initial data set is certified as sufficient, it is provided to the researcher. If PHI is found, the honest broker creates additional rules and data transformations to produce a revised de-identified data set. Ultimately, the researcher receives the initial or revised de-identified data. If the researcher encounters PHI that were missed by the honest broker, NIH OHSRP policy requires him to discontinue the use of the data and obtain IRB approval (typically, with waiver of consent) in order to continue the research. This process has two weak points. First, the review by the honest broker is lengthy, labor intensive, and has to be repeated for each project. Second, the researcher runs the risk of project interruptions and the requirement for additional review if PHI is discovered during the data analysis. The goal of the dsIDR is to produce a resource that does would not require additional human review and could be re-used for multiple projects.
Methods
Establishing the dsIDR
We used existing BTRIS death status data to establish the size of the dsIDR and explored the proportion of structured and narrative-text data available on deceased subjects. We looked at how many data rows are excluded by existing PHI filters (developed for the purpose of current de-identified BTRIS exports). We compared the size of the dsIDR with the full BTRIS repository in terms of number of patients, laboratory results, medication data, diagnoses and clinical documents.
Analysis of narrative text clinical documents
We used National Library of Medicine’s (NLM) Scrubber software to parse dsIDR narrative documents.8 Due to data size, requirements for computing time, and focus on recent and active documents, we limited our analysis to documents authored between October 1st, 2011 and October 1st, 2013). We also limited the analysis to the most frequent documents that accounted for 99% of all documents, ignoring rarely used document types. We obtained ethical approval for our deceased records research from NIH OHSRP office.
In order to correctly detect and remove sensitive patient information, we define the term a priori PHI as patient identifiers that have limited or no research value, that are known prior to the initiation of the PHI scrubbing process, and that definitely have be removed from clinical documents. In other words, our term “a priori PHI” refers to primary patient identifiers that are known from patient demographic data entered in structured form within an EHR. A priori PHI elements in our study were first, middle and last name and medical records number (MRN) of the primary patient. Of note is that patient identifiers do not equate Protected Health Information. Patient identifiers are Personally Identifiable Information, which meet the definition of PHI for living patients but not technically PHI for deceased patients.
Our work also relies on the assumption that any document containing secondary private information must indicate by at least one token who that person is (e.g., husband). We refer to this as the “token presence assumption”. We defined a set of secondary person tokens that may indicate presence of secondary PxI. We used a regular expression search with the following word elements (including plural versions, combinations with applicable prefixes and suffixes, and other variations): mother, father, spouse, parent, wife, husband, daughter, son, sister, brother, children, child, mom, mommy, mama, dad, daddy, papa, sibling, aunt, uncle, “grand-*”, “*-in-law”, “in[-]law”, friend, supervisor, employer, employee, girlfriend, boyfriend, roommate, partner, boss, collaborator. We sought to discover the frequency of secondary person tokens in clinical documents.
In addition to general occurrence, we used the document type (e.g., discharge summary vs. pathology report) as the key document parameter to investigate the occurrence of secondary PxI. We assumed that patterns of secondary PxI occurrence would be similar within a given document type; hence, our analysis is structured by document type. However, we also understand that this assumption may not be fully relied upon, as users may differ in their choice of document type and may differ in how they use those types. For each document type, we measured the frequency of occurrence (in dsIDR), and the frequency with which they contained a priori PHI, any personal name or a secondary person token.
We used NLM Scrubber to detect human names, alphanumeric identifiers (IDs) and addresses. Personal names are detected by multiple methods with the most discriminative method being the computation of the likelihood ratio of name to non-name of a given word. To identify sensitive alphanumeric identifiers, NLM Scrubber uses a two phase process where it first tries to identify laboratory values (that should be preserved) based on a set of hard coded patterns and in the subsequent second phase marks the remaining alphanumeric strings as sensitive tokens. Addresses are recognized mostly via “shapes” component of a specialized part-of-speech tagger (called dTagger) that looks for address-like patterns. For each analyzed document, NLM Scrubber outputs a set of tagged tokens classified by the identified type (e.g., address or personal name) appended to the end of the document or stored within a database. Marked up documents can also be browsed and edited in an associated editor called Visual Tagging Tool (VTT). Because of our focus on secondary PxI, we did not use other features of NLM Scrubber such as de-identification of ages (over age 89) and dates. On the other hand, we added new features, such as the ability to detect a set of secondary person tokens and the ability to analyze sensitive tokens that are in proximity to a secondary person token (e.g., address near “daughter”). We used NLM Scrubber’s token-level output to detect frequency of various identifiers by tag. A full description of NLM Scrubber is available in a prior published study on name detection8 and in an internal NLM report (non-name tokens).9
Manual review of secondary PxI instances
In addition to software-assisted analysis, we employed manual review of a subset of clinical documents to improve our understanding of different types of secondary PxI, to improve our definition of a true positive secondary PxI, and to inform our final scrubbing methodology. Due to a large volume of documents, we picked a single frequent secondary person token and a single frequent document type. We then reviewed all those document instances in a single calendar year (2013) and classified the secondary PxI into classes that could be later used in PxI removal efforts.
Results
Our analysis resulted in the creation of a pilot deceased subject data repository. NIH researchers interested in using the dsIDR can seek ethics approval using an NIH OHSRP form that has pre-filled elements applicable to dsIDR research projects. This form (with pre-filled elements indicating the use of dsIDR platform) is available as an online appendix A at http://dx.doi.org/10.6084/m9.figshare.924793). This form is relatively short and dsIDR projects require less detailed description compared to a full IRB submission.
Size of dsIDR dataset
As of March 2014, the NIH BTRIS data repository contained coded data on 23 962 deceased patients. Table 1 shows side by side comparison of the full BTRIS repository versus deceased subject subset (numbers reflect laboratory test results and medication administrations, not orders). In comparing the number of patients (first row in table 1), we only considered patients with at least one laboratory result due to a significant number of patients that are administratively entered but lack a critical number of clinical diagnostic or treatment data (e.g., trial screening or patient’s relatives). Existing BTRIS de-identification filters excluded 1.4M laboratory test results and 45 644 medication records where associated comments include PHI (patient name or MRN).
Table 1:
Size of structured EHR data (full and decease subject repository)
| Parameter | Full repository (BTRIS) | Deceased subject repository (dsBTRIS) |
|---|---|---|
| # patients (with laboratory data) | 264,885 | 23,962 (9.1%) |
| # laboratory tests results | 213.3 M | 52.0 M (24.4%) |
| # medication administrations | 16.5 M | 5.2 M (31.5%) |
| # diagnoses | 1.9 M | 0.3M (15.8%) |
| # clinical documents | 21.3M | 5.6 M (26.3%) |
A sub-analysis of laboratory data showed that whereas BTRIS contains 13,470 distinct laboratory test types (e.g., plasma folic acid level), dsBTRIS has 8,470 types. However, looking at the most frequent tests10, we found that dsIDR contained 99.85% (1997 test types) of the 2000 most frequent test types (see Figure 1 for cumulative percentage graph).
Figure 1:
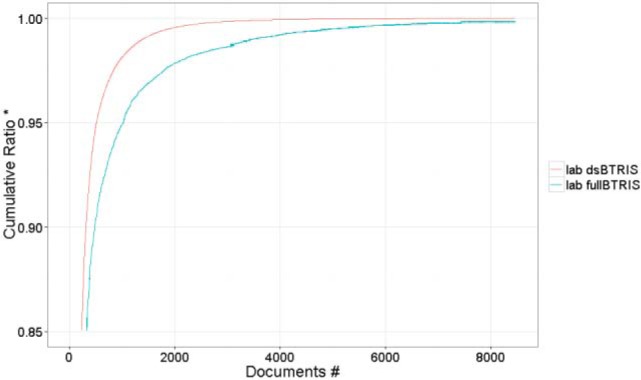
Pareto chart of lab tests for BTRIS and the dsBTRIS. # 8470 dsBTRIS laboratory types; 5000 full BTRIS test types (0.14% of all test instances) are not shown. * Cumulative portion of the respective repository that is accounted for by the test terms on the horizontal axis; the fullBTRIS line stops at 99.86% of all tests covered by all 8470 dsBTRIS tests).
To help researchers seeking recent data within a particular age group, we produced histograms of age of death for different time periods. An example, for deaths occurring during 2009–2013, is shown in Figure 2.
Figure 2:
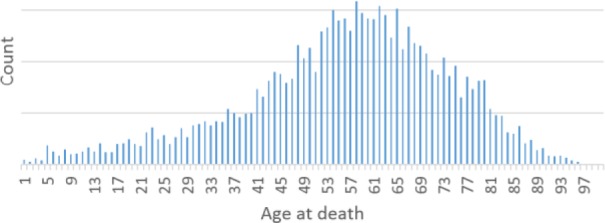
Patient distribution by Age at death (years 2009–2013) (vertical axis legend (count) is not shown to mask exact counts)
Analysis of narrative text clinical documents (by document type)
Within the documents from the last 2 years, we identified over 379 thousand clinical documents in deceased patients that were of 391 distinct document types. Excluding 1% of rare documents, the number of distinct document types reduced to 177 document types (376 051 documents). Each analyzed document type in the final set had at least 80 instances. A small number of document types capture the majority of the data. We found that 28 document types accounted for 80% of all documents and 59 document types accounted for 90% of all documents. An overview of all document types is available as appendix B at http://dx.doi.org/10.6084/m9.figshare.924793.
NLM Scrubber took 105 hours running in 4 parallel processes on a Linux workstation (8 CPU, 16GB RAM) with most computing time spent on part-of-speech tagging. Results were stored in a MySQL database to allow subanalysis by document type, tag or token. NLM Scrubber parsed 112 million tokens overall and marked 1.9 million as sensitive, with 53.4% being tagged as alphanumeric ID, 42.6% as personal name, 2.3% as address, and the remaining 1.7% as a combination of multiple tags. On average, a document contained 5.3 tagged tokens (ranging from 0 to 324) with 5.3% of documents containing no sensitive tokens. No list of local provider names was provided to NLM Scrubber.
Presence of a priori PHI
The presence of a priori PHI discovered by the NLM Scrubber varied widely by document type, from 0% to 100%. Table 2 presents document types that contain a priori PHI in more than 50% of document instances. Six document types (e.g., Speech Language Pathology Document) contain a priori PHI in more than 75% of all instances. The table shows that a priori PHI occurrence also differs by user group (e.g., National Institute of Allergy and Infectious Disease [NIAID] Intramural Research Program). A priori PHI presence data on all document types are available in the online appendix B.
Table 2:
Document types that frequently contain a priori PHI
| Document Type | Total Count | % of instances with a priori PHI | % of instances with secondary person token |
|---|---|---|---|
| NIAID Progress Note - Study Coordinator Document (CC, CRIS) | 154 | 100% | 15% |
| Speech Language Pathology Document (CC, CRIS) | 98 | 99% | 53% |
| Transfusion Medicine Note Document (CC, CRIS) | 88 | 89% | 27% |
| Dermatology Consult (CC, CRIS) | 237 | 86% | 39% |
| SOAP Progress Note Document (CC, CRIS) | 1052 | 83% | 28% |
| Genitourinary (GU) Oncology Free Text (CC, CRIS) | 230 | 80% | 24% |
| Bereavement Intake Progress Note (CC, CRIS) | 121 | 79% | 99% |
| Radiation Oncology HPE Document (CC, CRIS) | 123 | 78% | 96% |
| Radiation Therapy Summary Document (CC, CRIS) | 112 | 75% | 21% |
On the other hand, we found 17 document types (9748 documents in total) that neither contained a priori PHI nor secondary person tokens. Analysis by document type revealed that 9 of these 17 document types always contain the same text telling the clinician to refer to a second document. For example, “Ophthalmology Consult (National Eye Institute)” document type consisted of 350 documents with the text: “NEI Clinical Documentation - Please click the camera icon in the bottom left corner of the window to see this document”. Six document types were very brief forms related to admission, blood transfusion or anesthesia (e.g., Pre-Operative Anesthesia Record, Anesthesia Perioperative Record, and Post Anesthesia Care Unit Record), and often consisted of administrative content. Such “medium value” documents often repeated patient data such as research protocol assignment or patient gender. Only one document type (“Point of Care Testing Document”) contained more variable clinician-entered data, such as glucose bed side test results.
Pooling all documents (regardless of the document type), we found that 4.61% of clinical documents (17,340 out of 376,051) contained a priori PHI.
Presence of a secondary person tokens
Table 3 presents the frequency of secondary person tokens across all documents identified by NLM Scrubber. The most frequent secondary person tokens were: mother, spouse, parent, wife and father. Secondary tokens with less than 0.5% relative occurrence were collapsed into two bottom items “other-family” and “other-non-family” to reduce the size of the table. Data on all tokens are available in a separate sheet of online appendix B.
Table 3:
Absolute and relative occurrences of individual secondary person tokens across all document instances.
| Secondary Person Token | Occurrence | % of All Occurrences | Secondary Person Token | Occurrence | % of All Occurrences |
|---|---|---|---|---|---|
| MOTHER | 17,979 | 14.21% | PARENTS | 3,196 | 2.53% |
| SPOUSE | 16,942 | 13.39% | MOM | 2,695 | 2.13% |
| PARENT | 13,825 | 10.93% | SIBLING | 2,583 | 2.04% |
| WIFE | 11,858 | 9.37% | FRIENDS | 2,540 | 2.01% |
| FATHER | 8,959 | 7.08% | FRIEND | 2,528 | 2.00% |
| DAUGHTER | 6,460 | 5.11% | CHILD | 1,872 | 1.48% |
| HUSBAND | 6,124 | 4.84% | SIBLINGS | 898 | 0.71% |
| SISTER | 5,359 | 4.24% | DAUGHTERS | 754 | 0.60% |
| BROTHER | 5,313 | 4.20% | DAD | 743 | 0.59% |
| SON | 3,867 | 3.06% | OTHER (FAMILY) | 6,477 | 5.12% |
| CHILDREN | 3,742 | 2.96% | OTHER (NON-FAMILY) | 1,780 | 1.41% |
When analyzed by document type, occurrence of secondary person tokens ranges from 0% to 99%. For example, in “Bereavement Intake Progress Note” the occurrence was 99%, while the occurrence in “Tumor Measurements and Response Document” was 0%. Again, online Appendix B also contains data on secondary person token presence for all 177 analyzed document types and Table 2 includes secondary person token column for selected documents.
To produce a conservative estimate and assuming that every document with a secondary person token includes secondary PxI, then NIH CC’s IDR incidence of secondary PxI is 12.94% (48,679 out of 376,051 documents). Using a two-fold condition of simultaneous presence of secondary person token and presence of a priori PHI (primary patient name or MRN), we found that 1.14% of all documents (4,276 out of 376,051) satisfy this stricter condition.
Regarding the detection of personal names, we found that a majority of document types include the names of the authoring clinicians (complete data available in Appendix B). Without prior knowledge about local clinician names or document structure clearly tagging electronic signatures, the detection of personal names was not an optimal signal for detecting instances of secondary PxI due to too many false positives. However, proximity search measures (personal name near a secondary person token) could potentially result in improved detection of true instances of secondary PxI.
Manual review
For manual review, we selected the “daughter” secondary person token since this was the most frequent secondary person token belonging to an offspring which, in turn, is the generation that is likely to be surviving the primary patient. For the reviewed document type, we selected the highly variable document type “Progress Note – Free Text Document”, authored by physicians (rather than nursing staff). This selected document contained a secondary person tokens in 16% of instances. A pilot study of two other pairs of secondary person token and document type did not yield significant findings.
Based on review of all sentences containing the selected token, we classified the secondary PxI instances into three categories listed in Table 4 together with de-identified examples. We found:
Health related secondary information: e.g., secondary person diagnosis. Note that such information technically constitutes protected (in HIPAA sense) health information only if it is explicitly or implicitly combined with a secondary person first name (most often), full name or other identifier in addition to the secondary person token presence. Otherwise, it can be considered de-identified (meets criteria for the Safe Harbor Method).
Non-health secondary information with an identifier (secondary PII) (e.g., secondary name, secondary phone number or secondary age)
Non-health secondary information without a secondary identifier (e.g., description of care situations involving a family member)
Table 4:
Classification of secondary PxI and examples identified during manual review
| Class | Information Domain | Example |
|---|---|---|
| Health related secondary information | Diagnosis | Her mother and daughter both have symptoms of multiple sclerosis. |
| Patient started on antibiotic post exposure prophylaxis for pertusis (daughter tested positive) | ||
| Non-health secondary information with an identifier (secondary PII) | Name | I had a frank 2 hour discussion with Ms Doe and her husband and her daughter Alice. |
| She was accompanied in our meeting by her husband and her daughter Alice | ||
| location | His daughter lives in Richmond. | |
| phone | Mrs. Doe daughter Alice Carol Smith (123-123-1234) contacted research nurse Alice Jones, R.N., and requested that I call her. | |
| I attempted to reach Mrs. Smith (cell 123-123-1234) this morning to convey our condolences on Mr. Smith passing earlier this morning. | ||
| age | He is married and has a 9-year-old daughter. | |
| combination | Mr. Doe young adult daughters (ages 18 and 23) as well as his sister and sister-in-law are anticipated to arrive this evening from Cleveland around 8:30pm. | |
| SH: … Happily married. One son lives in Annandale – active duty Army. Daughter in Pennsylvania. 3 grandkids. | ||
| Mrs. Doe was accompanied by her daughters Alice and Carol who drove her from Pennsylvania for the procedure. | ||
| Non-health secondary information without a secondary identifier | n/a | Pt did ROM exercises in bed with her daughter. |
| We discussed in detail with patient, spouse and daughter the progression of disease based on radiographic findings and worsening symptoms | ||
| The daughter-in-law will be returning with Ms Smith between 10am – 12pm tomorrow for dressing change, supplies, and d/c instructions. (data note: Smith is the primary patient) | ||
| She stated that his daughters did arrive to the hospital but the sons had not made it. |
Discussion
This study is the first to report on an informatics repository and platform created specifically around deceased subjects. The dsIDR contains a smaller number of patients and observations compared with the full IDR; however, the most frequent items are well represented. For example, 99.9% of the top 2000 the full IDR laboratory tests are also present in the dsIDR. Table 1 shows that while the dsIDR consisted of only 9% of patients (24 000 out of all 265 000 IDR patients with any laboratory data), it accounted for 24% of all laboratory tests and 32% of all medication records. This can be explained by the disproportional care provided to older or sicker patients11 and also by the NIH Clinical Center casemix bias due to its research hospital status. In building the dsIDR, we were able to re-use existing processes for structured coded data (e.g., laboratory results) that remove results where comments contain patient name or medical record number. However, due to costs and available resources, robust de-identification techniques for unstructured data (narrative clinical documents) are only now being developed.
Narrative text clinical documents
Since valuable clinical information is often stored in unstructured narrative text clinical documents and the dsIDR’s purpose is to serve as a pilot platform, inclusion of narrative-text clinical documents within the dsIDR is highly desirable. In order to protect the identity of secondary persons, removal of primary patient name is highly advisable. For example, if a record mentions the diagnosis and the first name of a family relative, the presence of the primary patient last name helps reveal the full name of the relative. Although the Privacy Act does not apply to dead people, for non-research use, HIPAA protects health records of decedents for the period of 50 years after death as if they were living.
In our effort to remove a priori PHI (more specifically, primary patient identifiers), we found that their presence in documents differs widely (from 0% to 100%). For document types where all or almost all instances contain primary patient name and medical record number the reason may be due to data integration processes for external ancillary system. If an external system is used, re-entering patient IDs ensures proper data integration of data authored in the external system to the primary system or common data viewing platform. Although some document types do contain with high frequency a priori PHI identifiers (Table 2), the overall rate across all documents is only 4.61%. This implies that in majority of narrative clinical documents, the patient identity is not repeated within the text. Therefore, if a document mentions a family relative by relationship (e.g., daughter), 95% of documents will not provide the context of primary patient full name to identify the secondary person. This is also demonstrated by the considerably lower estimate (1.14%) that uses the two-fold criterion as opposed to mere secondary token presence (12.94%).
The review of alphanumeric identifiers found by NLM Scrubber also showed that date of birth is frequently used as safety check within EHR forms and could potentially be another a priori PHI element type. However, including date of birth in this set (together with name and MRN) may be meaningless because dsIDR already contains structured data on date of birth.
Study limitations
Our pilot exploration of secondary PxI in a deceased subject repository has several limitations. Our goal was not to arrive at a final method capable of removing all secondary PxI. Our set of secondary person tokens, although designed to be broad, may not contain all keywords of importance at other institutions. For example, the presented results reflect an older version of this token set that did not contain the keyword “fiancé”. Also, we have only used a single de-identification tool and data from a single institution.
Our work relies heavily on the secondary person token presence assumption. It was out of scope of this pilot study to formally evaluate and measure, most likely by manual validation, the percentage of excluded documents (based on explicit presence of a secondary person token) truly contain secondary PxI (true positive vs. false positive). Similarly, we did not validate that included narrative documents, lacking secondary person token, are truly free of any implicit secondary PxI (false negative). This evaluation would improve our estimate of secondary PxI presence which should be interpreted as a machine-based upper bound approximation rather than an exact determination.
Lessons learned and future work
During our pilot creation of the dsIDR we found that removing secondary PxI differs significantly from general de-identification tasks that focus on the 18 identifiers specified by the HIPAA Safe Harbor method. While the Safe Harbor research route is well characterized in the regulation, the regulatory guidance for decedent clause research is less precise. We can apply the Safe Harbor Method guidance on any secondary PxI, but it is less clear on how aggressive the scrubbing must be for the primary deceased patient. The classification shown in table 4 (manual review of secondary PxI instances) shows several examples of varying level of sensitive context ranging from family phone number to a simple age fact). In our interpretation, we assumed that all primary patient information can stay, unless it provides context to reveal secondary PxI of a living person. For example, whereas traditional de-identification within a whole population IDR would require removal of locations and dates, they are permitted to stay in the dsIDR (unless related to a secondary person, which is much less frequent). It is also important to note that the recent regulatory change in decedent PHI protection (shortening to 50 years instead of indefinitely) does not apply to the research context; the decent research clause provides research access to decedents’ records immediately after death.
Another useful lesson was about importance of empty (un-filled) EHR forms. We found that many documents are semi-structured and clinicians enter only short entries into this pre-populated structure (e.g., anesthesia type). Some of those semi-structured document fields are even pre-populated by software (e.g., protocol number, gender) and never edited by the user. Documenting this as metadata at the document level (list of all document questions/sections, software field pre-population, editability) provides useful prior knowledge for scrubbing. In our future revisions, we may consider measuring Levenshtein distance, which is a string metric for measuring the difference between two text sequences as number of single-character edits (empty EHR form vs. complete document). Such an approach is suitable for documents with minimal clinician-entered content (e.g., Post Anesthesia Care Unit Record).
Alternatively, for a limited set of non-dictated documents authored within the modern EHR form paradigm, the optimal dsIDR unit of information is a clinical document section or question rather than the full document. In a separate study,12 we analyzed problem list sections in dsIDR as well as full IDR. Due to the OHSRP requirement for de-identification, we performed a manual review to verify that truly only problem list information is entered into the problem list document section. We found that clinicians may occasionally forget to hit <tab> or otherwise advance to the next form field and enter social history or family history into the problem list section resulting in PHI being present in the dataset. Another observed problem related to the issue of empty forms and form structure was the presence of a false positive secondary person token (e.g., children) in the form questions or pre-populated entry (e.g., clinical trial title “Continuing Treatment for Children and Adults in the Center for Cancer Research; clinicaltrials.gov: NCT00001295).
Regarding the detection of phone numbers as the most frequent alphanumeric identifiers, we found that local hotels, local pharmacies and internal department pagers were often detected as false positive secondary PxI identifiers. Scrubbing software could take advantage of a prior list of permissible phone numbers. Detecting the nature of the phone number by an Internet search engine is another strategy that can distinguish publicly known phone number from phone numbers intentionally kept private. If an EHR contains a structured and well used field that provides primary patient’s phone number, it can be used as a priori PHI element to improve the scrubbing accuracy. Similarly, names of investigative drugs could be extracted from clinical trial descriptions and white-listed as allowed alphanumeric identifiers.
We plan to incorporate some of the above mentioned issues (see Table 5 for summary) in the future versions of our overall processing pipeline and NLM Scrubber feature set. Our pilot study and presented experience offer some guidance to other institutions interested in establishing their own dsIDR in terms of how to characterize the size of the dsIDR, narrative document lessons learned and the value of collecting and verifying death status data. NLM Scrubber is planned to be released freely prior to the AMIA Annual Symposium in November 2014 and could provide to interested institutions as a no-cost software tool with potentially additional experimental features for detecting secondary person tokens. Federation of multiple dsIDRs can address the sample size limitations of any single repository. Central Intelligence Agency Factbook estimates that each month about 221 thousand deaths occur in the USA and 66.5% of those deaths occur outside the home (in hospitals, nursing homes or other facilities; according to a 2009 Medicare study).13 Internal records and first-hand primary death data are important because national death registries, such as the Social Security Death Master File contained 0.03% false positive deaths and since 2011 omits records from several US states.3,14
Table 5:
Summary of lessons learned
| • | Lack of clear regulatory guidance for de-identification of decedent records for research use (in contrast to the Safe Harbor Method for general de-identification within a whole population IDR) |
| • | Need for metadata on document structure (empty EHR forms; pre-populated fields, secondary person tokens as part of the form question; plain free-text vs. structured provider e-signatures) |
| • | Phone number as the most sensitive secondary ID and possible disambiguation methods for phone numbers (local hotel or pharmacy vs. secondary person private number) |
| • | Provision of prior information to scrubber about local provider names or permitted alphanumeric IDs, such as experimental drug names. |
| • | Importance of accurate deceased status using local institutional data or external data from State Vital Statistics Administration (research context only) or federal death index data |
Conclusions
The HIPAA decedent clause represents an important and often overlooked alternative to using de-identified EHR records. Use of deceased subject records does not require full IRB review or patient consent. The 18 patient identifiers listed in the HIPAA Safe Harbor Method do not have to be removed in dsIDR; however, in order to protect the privacy of secondary living persons, we argue for removal of those that have limited research value. Removal of only secondary PxI results in less distorted research data compared with general de-identification used in whole population IDR. Our study is the first to suggest a basic method for de-identification of decedent records and provides the first estimate of the rate of secondary PxI in EHR data (12.9% using only the secondary person token presence criterion and 1.1% using stricter criteria). With potential increased use of decedent EHR data, clearer guidelines addressing the issue of secondary PxI and what constitutes acceptable proof of death could facilitate such research. We also recommend modifications of existing de-identification programs (such as NLM-NS) to offer a dsIDR scrubbing mode in addition to existing modes where all 18 PHI identifiers are being targeted. In our exploration of secondary PxI, we found secondary person phone numbers to be most frequent and most sensitive secondary person identifiers. Federated dsIDRs can address the issue of limited sample size and make them even primary research platform for certain hypotheses.
Acknowledgments
This work has been supported by intramural research funds from the NIH Clinical Center and the National Library of Medicine. The opinions expressed in this article are authors’ own and do not reflect the view of the National Institutes of Health, or the Department of Health and Human Services.
Appendix A
Appendix B
References
- 1.Kushida CA, Nichols DA, Jadrnicek R, Miller R, Walsh JK, Griffin K. Strategies for de-identification and anonymization of electronic health record data for use in multicenter research studies. Med Care. 2012;50(Suppl):S82–101. doi: 10.1097/MLR.0b013e3182585355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meystre SM, Friedlin FJ, South BR, Shen S, Samore MH. Automatic de-identification of textual documents in the electronic health record: a review of recent research. BMC medical research methodology. 2010;10:70. doi: 10.1186/1471-2288-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huser V, Cimino JJ. Don’t take your EHR to heaven, donate it to science: legal and research policies for EHR post mortem. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2013-002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy EC, Ferris FL, 3rd, O’Donnell WR. An electronic medical records system for clinical research and the EMR EDC interface. Investigative ophthalmology & visual science. 2007;48:4383–9. doi: 10.1167/iovs.07-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Electronic Code of Federal Regulations: Title 45: §164.512 Uses and disclosures for which an authorization or opportunity to agree or object is not required. http://www.ecfr.gov/cgi-bin/retrieveECFR?SID=0c83756f3a487d6d70dd3232e084c0a0&n=45y1.0.1.3.78.5&r=SUBPART&ty=HTML#45:1.0.1.3.78.5.27.8.
- 6.HIPAA Privacy Rule and Its Impact on Research. 2013. http://privacyruleandresearch.nih.gov/pr_08.asp.
- 7.Ninghui L, Tiancheng L, Venkatasubramanian S. t-Closeness: Privacy Beyond k-Anonymity and l-Diversity. Data Engineering, 2007 ICDE 2007 IEEE 23rd International Conference on; 2007 15–20 April 2007; 2007. pp. 106–15. [Google Scholar]
- 8.Kayaalp M, Browne AC, Callaghan FM, Dodd ZA, Divita G, Ozturk S, et al. The pattern of name tokens in narrative clinical text and a comparison of five systems for redacting them. J Am Med Inform Assoc. 2013 doi: 10.1136/amiajnl-2013-001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M K, AC B, Z D, P S, CJ M. Technical Report to the LHNCBC Board of Scientific Counselors: Clinical Text De-Identification Research. 2013 [Google Scholar]
- 10.LOINC Top 2000+ Lab Observations (US version) 1.1. 2014. http://loinc.org/usage/obs/loinc-top-2000-plus-loinc-lab-observations-us.csv/view.
- 11.Gielen B, Remacle A, Mertens R. Patterns of health care use and expenditure during the last 6 months of life in Belgium: differences between age categories in cancer and non-cancer patients. Health policy. 2010;97:53–61. doi: 10.1016/j.healthpol.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Huser V, Fung KW, Cimino JJ. Natural Language Processing of Free-text Problem List Sections in Structured Clinical Documents: a Case Study at NIH Clinical Center. AMIA Summits Transl Sci Proc (accepted) 2014 [Google Scholar]
- 13.Teno JM, Gozalo PL, Bynum JP, Leland NE, Miller SC, Morden NE, et al. Change in end-of-life care for Medicare beneficiaries: site of death, place of care, and health care transitions in 2000, 2005, and 2009. JAMA. 2013;309:470–7. doi: 10.1001/jama.2012.207624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Death Master File: Important Notice. 2011. http://www.ntis.gov/pdf/import-change-dmf.pdf.


