Abstract
An opportunity exists for meaningful concept extraction and indexing from large corpora of clinical notes in the Veterans Affairs (VA) electronic medical record. Currently available tools such as MetaMap, cTAKES and HITex do not scale up to address this big data need. Sophia, a rapid UMLS concept extraction annotator was developed to fulfill a mandate and address extraction where high throughput is needed while preserving performance. We report on the development, testing and benchmarking of Sophia against MetaMap and cTAKEs. Sophia demonstrated improved performance on recall as compared to cTAKES and MetaMap (0.71 vs 0.66 and 0.38). The overall f-score was similar to cTAKES and an improvement over MetaMap (0.53 vs 0.57 and 0.43). With regard to speed of processing records, we noted Sophia to be several fold faster than cTAKES and the scaled-out MetaMap service. Sophia offers a viable alternative for high-throughput information extraction tasks.
Introduction
There is a pressing need for clinical concept extraction and concept indexing to unlock currently obscured information from large corpora holding clinical narratives. Natural language processing (NLP) tools such as MetaMap1, cTAKES2 and HITex3 have traditionally been used for concept extraction and have performed well in the clinical domain. However, these have not been scaled-up to handle big data while preserving processing speeds. Large health care systems such as Kaiser Permanente, Mayo, Vanderbilt and the US Department of Veterans Affairs (VA) would have need for scaling up their concept (information) extraction tasks. As an example, the VA maintains a fast growing corpora of 2.6 billion clinical notes through a secure research environment (Veterans Informatics and Computing Infrastructure, VINCI4). Using currently available tools running on several multi-core servers, we estimated that it would take multiple years to create concept indexes for the notes available in VINCI notes to facilitate further information extraction and retrieval.
For clinical, health services and genomic research, there is a critical and ongoing need for NLP tools to mine the free text of medical records to supplement structured data queries to identify patient cohorts and phenotypes. In developing these tools, researchers consider several criteria: usability, maintenance, efficacy (in terms of recall/precision/f-score), ability to incorporate and use local lexica (or terminology), high throughput performance and adoption within the NLP community. While no currently available tools satisfy all criteria, we set out to develop a tool which would be useful for high throughput while maintaining efficacy.
We report the development of Sophia which is a UIMA-AS5 based UMLS6 concept extraction annotator. Sophia is now a key component of the v3NLP Framework used by VINCI for information extraction tasks. Sophia shares some methodologies found in MetaMap and cTAKES, but includes some attributes that cTAKES does not, and also excludes some functionality that MetaMap has. More importantly, Sophia is designed for fast processing, while most prior efforts emphasize extraction accuracy.
State of the Art in Extracting UMLS Concepts
There are a number of open source NLP tools and techniques specifically developed to extract UMLS concepts from clinical text. Among them, cTAKES and HITex are well represented in the field. MetaMap and SAPHIRE7 were tools initially designed for UMLS concept extraction within the bio-literature domain that have been adapted for use with clinical text by several organizations. There are a number of non-open source successful efforts to extract UMLS concepts within clinical text include MedLEE8, MedKAT9 and KnowledgeMap10.
Many of these efforts are built upon two frameworks adopted or developed for use within the NLP field: GATE11 and Apache-UIMA. A relevant component common to these two frameworks is the notion of a pipeline composed of a sequence, or end-to-end chaining, of atomic modules often referred to as annotators. An annotator adds stand-off highlights, mark-ups, labels or annotations related to the original text. The annotations from one module are used as input to a downstream annotator. A phrase chunker annotator, for example, depends upon part-of-speech annotations added via an upstream annotator in a pipeline. Efforts built upon the UIMA platform have the potential for being scaled out through the replication of pipeline instances via a related framework: Apache UIMA’s Asynchronous Scale-out12 (UIMA-AS). The UIMA-AS framework provides for pipeline component replication in addition to the full pipeline instance replication to address bottleneck annotators.
Methods
Sophia Annotator Defined
The Sophia annotator identifies UMLS Concepts using a lookup algorithm to match longest spanning matches to an index of known UMLS concepts. A conscious decision was made to find longest spanning matches rather than shortest spanning or by including all possible matches. Longest spanning matches reduce the ambiguity issue by finding the most specific match, for instance finding chest pain rather than chest and pain. While including the constituent components such as pain and chest might be useful for building google-like search indexes to aid retrieval techniques, the first iteration of Sophia does not include this capability because such a capability was not part of the motivating use cases.
The lookup algorithm relies on exact match retrieval to keys in the index, rather than uninflected or stemmed key retrieval. An exact match retrieval looks up the words as they appear in the sentence to find keys in an index. Within the sentence the patients were transferred, exact match retrieval would look up the words patients, were, and transferred within a dictionary. Within an uninflected lookup algorithm, each of the words within the sentence would be transformed into the uninflected keys: patient is transfer. These uninflected keys are what would be looked up within an index that holds the uninflected forms. The index includes all possible fruitful variants13 for a given UMLS concept to insure that valid matches will be found. Fruitful variants for a given term includes spelling variants, inflections, synonyms, acronyms and abbreviations, acronym and abbreviation expansions, derivations and combinations of these transformations such as the spelling variants of synonyms. The burden of computation to make a match is shifted from the cost of normalizing words in the text to be looked up, to having a larger index where the variant expansion cost was taken up at index creation time. An early MetaMap paper14 showed that this technique increases match precision or accuracy over stemming normalization techniques.
The lookup algorithm works on a window that initially includes all the tokens of a sentence as the longest span to find. Subsequent lookups drop successive tokens from the beginning of the sentence until a match is made. The algorithm does not rely on phrasal boundaries on the grounds that there are important UMLS terms that include multiple phrases, particularly those that include multiple prepositional phrases (of, with, without, with/without). Techniques that rely on phrasal barriers to determine the window size sometimes miss the longer, less ambiguous, more specific matches. Both MetaMap and cTAKES have post phrase identification to re-join specific kinds of prepositional phrases to the adjoining noun phrases to partially ameliorate this condition.
No phrasal boundaries are necessary for Sophia’s lookup technique. As a consequence, no part-of-speech tagger is necessary to identify phrasal markers, eliminating two common up-stream annotators commonly found in other concept extraction systems.
The Sophia lookup algorithm evolved from the SPECIALIST Text Tools15. The SPECIALIST Text Tools lookup dropped tokens from the beginning side of the sentence where-as the Sophia algorithm drops tokens from the ending side of the sentence. While neither version is perfect, dropping tokens from the ending side of the sentence favors having the head of a term as the last token matched. For example, the prior version would have matched heavy chain and smoking from the sequence heavy chain smoking whereas the current version would match heavy and chain smoking, given the situation where the index includes heavy chain, chain smoking, and heavy as keys (and not heavy chain smoking).
The index entry key creation is important to the overall Sophia scheme. Each UMLS string has a set of lexical variants generated to create keys in the Sophia index. These lexical variants include spelling variants, inflections, un-inflections, synonymy, derivations, acronym or abbreviation expansions and acronyms and abbreviations. Fruitful combinations of each of the above mentioned variants are also generated including derivations of spelling variants, derivations of synonyms, and derivations of derivations. These variants are generated from a configuration of the LVG tool16 distributed by the National Library of Medicine called the fruitful variants flow. This tool over-generates variants for Sophia’s purposes. Variants that are generated from terms that, themselves are acronym/abbreviation are most likely fallacious. For instance, generating spelling variants to the acronym A.I.D.S. generates the term AIDS, applying (un) inflections to that term creates AID (already fallacious), and applying either inflectional or derivational suffixes such as ing will lead to additional fallacious terms including aiding.
A post-processing filter is applied to prune out any variant generation combination that includes acronym/abbreviation or acronym/abbreviation expansion plus any additional mutation. Long sequences of synonyms or derivations are likewise pruned out.
The Sophia Pipeline
To evaluate Sophia against MetaMap and cTAKES, we created a pipeline that includes the prerequisite upstream annotators, the Sophia annotator, and needed downstream annotators (the Sophia Pipeline). The pipeline is built upon the UIMA-AS framework and is expected to be configured to utilize the resources of well-endowed production hardware to provide big-data concept extraction across the Veteran Administration’s 2.6 billion clinical records.
The Sophia annotator works on the tokens within sentence or utterance boundaries. As such, the Sophia annotator requires upstream token and sentence annotators to identify words and sentences. Sophia was developed with the v3NLP framework, which includes token and sentence boundary annotators. V3NLP’s sentence boundary annotator takes advantage of an annotator that identifies slot:value structures (they have a special kind of sentence grammar), and an annotator that identifies section headings. Since many clinical notes include question and checkbox boiler-plated sections, an annotator was added to recognize questions and their related checkbox structures to correctly handle concept assertions within these entities. The Sophia Annotator will blindly find UMLS concept mentions. The conText17 assertion annotator is run downstream of the Sophia annotator to provide assertion attributes to the concepts found. Figure 1 shows a skeuomorphic representation of the Sophia pipeline with a medical problems identification annotator at the tail end of the pipeline. The problems identification component was added here to extract medical problems from clinical text as an extrinsic evaluation.
Figure 1.
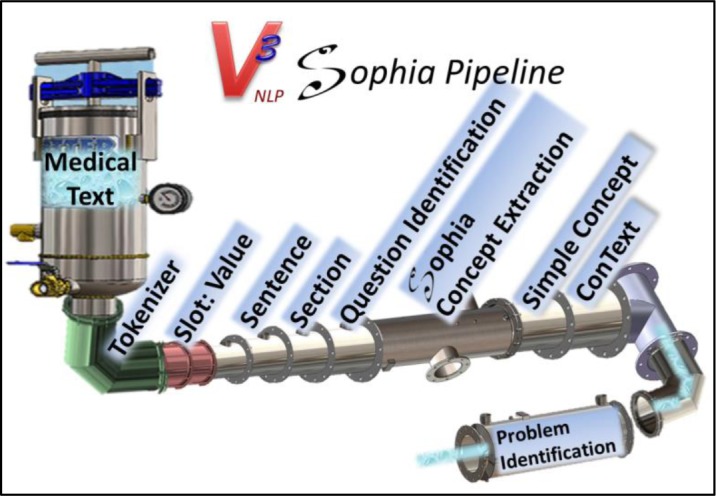
v3NLP Sophia pipeline
Evaluation
The Sophia Pipeline, MetaMap Pipeline and cTAKES were evaluated in an extrinsic task to identify medically significant problems mentioned in clinical text. The evaluation includes a span comparison compared to a human reference set and the throughput performance, i.e., how many records per second were processed. This paper provides the basis for baseline efficacy and performance metrics of the software devoid of the deployment environment.
Span Evaluation
A gold standard reference corpus with span level clinically significant utterances was developed through a Consortium for Healthcare Informatics Research18 (CHIR) Information Extraction and Modeling task from the Veterans Administration. Human annotators highlighted problems (as defined by the i2b2/VA 2010 Challenge19) in 145 clinical records in a corpus chosen at random from the 100 most frequent VA document types by the annotation team that annotated the i2b2/VA 2010 Challenge. Assertion attributes of negation, conditional, and subject were also annotated for these problems.
A MetaMap pipeline and a Sophia pipeline were created for this effort. Each of the pipelines included the problem identification annotator after the concept extraction annotation. The problem annotator filtered to concepts that were of the proscribed problem semantic type. The conText annotator was also applied to add assertion attributions. The conText annotator marked concepts with asserted, negated, conditional, applies-to the-patient, and historical attributions.
Figure 2 shows the MetaMap pipeline used for this evaluation. The cTAKES part-of-speech annotator and a phrase annotator were added and the Sophia annotator is replaced with the MetaMap annotator. The MetaMap annotator is a wrapper around a client that goes out to a MetaMap service running on external machines. This annotator gathers and uniques the phrases for a given record, then makes one request out to the MetaMap service. The MetaMap service runs MetaMap in the term processing mode. The MetaMap service is a restful service that includes 60 instances of MetaMap. The service treats each incoming term as a new request to the next available MetaMap process. Even with this environment, 86% of the processing time taken within the MetaMap pipeline is taken within this one annotator when analyzed via the UIMA CPE tool.
Figure 2.
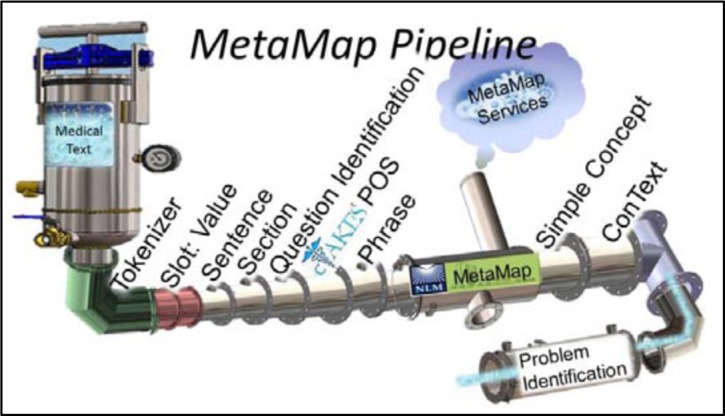
MetaMap pipeline
The cTAKES application was run separately on the 145 records. The output was fed into a post processing v3NLP pipeline that converted the cTAKES UMLS concept annotations to the CHIR model’s CodedEntries. The same problem annotator was applied. cTAKES includes assertion attributions.
The span level comparison was done with overlapping matching spans compared to the reference standard on asserted problems associated with the patient.
Both the MetaMap server and Sophia indexed using the 2011AA Level0+9 configuration. cTAKES uses the 2011AA SNOMED concepts. The evaluation used an f-score computed as
where
Efficacy Results
Table 1 shows MetaMap, cTAKES, and Sophia compared to the 145 record reference standard. The evaluation was a span-only evaluation, where credit was given for partial matches. cTAKES has the overall better F-Score at 0.568, followed closely by Sophia at 0.531. MetaMap had an overall f-score of 0.431. Sophia performed better at recall with a metric of 0.71 followed by cTAKES at 0.66. MetaMap had a recall metric of 0.38. cTAKES and MetaMap had a precision metric of 0.5 vs Sophia’s .422. Sophia’s precision was noticeably lower because of a plethora of false positives for this task. Those false positives from Sophia that were reviewed indicated that many could have been considered medical problems associated with the patient but the annotators chose not to mark them as such.
Table 1.
Problem span comparison
| TP | FP | FN | Recall | Precision | F-Score | |
|---|---|---|---|---|---|---|
| MetaMap | 436 | 436 | 717 | 0.380 | 0.500 | 0.431 |
| cTAKES | 757 | 760 | 391 | 0.660 | 0.500 | 0.568 |
| Sophia | 823 | 1125 | 325 | 0.717 | 0.422 | 0.531 |
Differences between Sophia, MetaMap and cTAKES
There was a large overlap between the systems. It cannot be construed that those concepts unique to one system or another are fallacious. Of interest is how well each system identified multi-word spans. The more tokens involved in the match, the less ambiguity is left for downstream processors to deal with. Table 2 shows the Sophia pipeline compared to MetaMap and cTAKES, using MetaMap and cTAKES as reference standards.
Table 2.
Problem span comparison using MetaMap and using cTAKES as the reference standard
| TP | FP | FN | Recall | Precision | F-Score | |
|---|---|---|---|---|---|---|
| Sophia compared to MetaMap reference standard | 1000 | 1238 | 169 | 0.855 | 0.45 | 0.587 |
| Sophia compared to cTAKES reference standard | 1496 | 1117 | 562 | 0.727 | 0.57 | 0.641 |
Many of the differences found included how each system chunked phrases, with no clear indication of whether either system did better or not. Table 3 shows instances where MetaMap picked up multi-word concepts but Sophia chunked them into separate concepts and instances of where Sophia picked up multi-word terms that include phrasal barrier markers.
Table 3.
Multiword matching differences between MetaMap, Sophia and cTAKES
| MetaMap | Sophia | cTAKES |
|---|---|---|
| right sided facial weakness | facial weakness | facial weakness |
| multiple old infarcts | infarcts | Infarcts |
| hard of hearing | ||
| change in bowel habits | ||
| lives with family | ||
| unable to sit | ||
| Sensitive to touch |
The largest category of differences between Sophia and cTAKES was that CTAKES annotated terms found in section headings that Sophia did not. A concept mention within a section heading would not indicate that the mention is related to the patient. For instance, a section heading Pain Management would not automatically indicate the patient has or does not have pain; only that there is a section in the document that includes a section with pain in the name. The reference standard did not include annotations from within section headings. The Sophia pipeline includes a sectionizer that marks section headings to be ignored. Table 3 shows multi-word terms that Sophia suggested that were missed by cTAKES.
Time Performance
The Sophia pipeline, the MetaMap pipeline and the cTAKES assertion aggregate annotator were run against two corpora on a development virtual machine (VM) provided by the VA to securely process clinical records. The CHIR reference standard has a shorter average character length than other available corpora. The i2b2 2010/VA Corpusi provided an additional benchmark to a corpus with known attributes within the NLP community. The fastest of 3 runs are reported here (Table 4). The throughput numbers are meant to be interpreted as a means to rank the relative performance between the three systems. The performance time of these systems on well- endowed production servers are vastly different than the VM’s provided for development purposes or current desktop machines. cTAKES and Sophia performance time on the i2b2 corpus on the a core i7 desktop was 4 times faster than the development virtual machine, and the MetaMap performance time was 2 times faster on the same corpus on the core-i7 using a less endowed MetaMap server.
Table 4.
Time performance in milliseconds to run Sophia, MetaMap and cTAKES on two corpora of records
| # of Records | Sophia | MetaMap | cTAKES | |
|---|---|---|---|---|
| i2b2 2010 V A Corpus | 349 | 1,395,271 (23.24 min) |
4,804,951 (80.08 min) |
24,524,827 (408 min) |
| Problem Reference Standard Corpus | 145 | 343,384 (5.7 min) |
478,824 (8 min) |
3,060,000 (51 min) |
Sophia has a significant initialization cost to load all the keys into an in-memory hash. This initialization is the same whether kicking off one instance or 100 due to the way the hash is shared across server threads. The impact of this initialization becomes less as more records are processed. Table 5 shows the initialization cost and the average per-record cost with and without taking into account the initialization cost. The initialization cost with MetaMap is hidden behind the running MetaMap service that was employed. CTAKES does have an initialization time of 36 seconds vs Sophia’s 40 seconds observed on a desktop core i7 with solid state drives.
Table 5.
Time performance to run Sophia on two corpora of records, reported in milliseconds
| # of records | Initialization in milliseconds | Average milliseconds per Record | Average milliseconds per record w/out initialization | Total milliseconds | |
|---|---|---|---|---|---|
| i2b2 2010 VA Corp us | 349 | 187,013 | 70,271 | 69,735 | 24,524,827 |
| Problem Reference Standard Corpus | 145 | 185,664 | 2,351 | 1,079 | 343,384 |
This time evaluation is not perfect. The number of external CPU’s and threads employed by the MetaMap services makes it difficult to replicate the same MetaMap pipeline performance if moved to an environment that does not employ the VA’s MetaMap services. Even with these constraints, the single threaded Sophia annotator out-performs the MetaMap annotator by a factor of 7 and out-performs cTAKES by a factor of 18.
A pipeline performance analysis was performed to analyze the time contribution for each of the components within the Sophia, MetaMap, and cTAKES pipelines. The UIMA Component Processing Engine (CPE) was employed to break down each of the component times. See the pie charts in figures 3–5 of the relative amount of time each component consumed. It is the assertion component that takes up the most time (70%) within the Sophia pipeline, and the second highest amount time (27%) the cTAKES pipeline, yet it is a mere 9% within the MetaMap pipeline. Within Metamap and cTAKES, other components consume much more processing relative to the assertion module. Efficiencies to conText should be explored before improving performance elsewhere for the Sophia pipeline.
Figure 3.
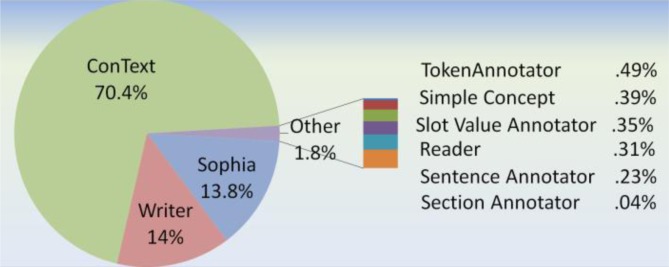
Sophia pipeline proportion of each annotator’s processing
Figure 5.
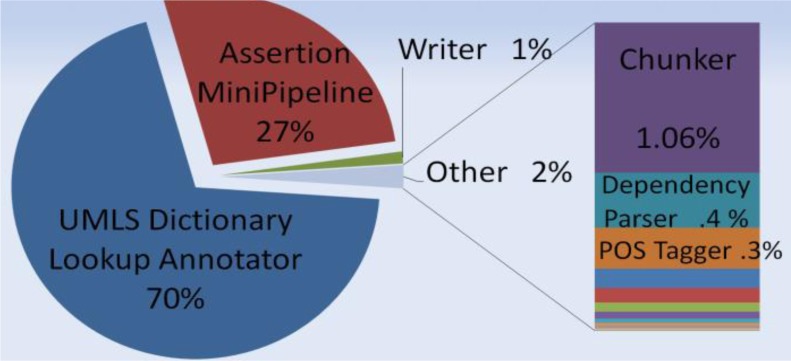
cTAKES pipeline proportion of each annotator’s processing
Some efficiency had been built into Sophia’s conTEXT wrapper, by spawning off a pool of threads to handle the conTEXT processing. This change contributed a 30% performance improvement compared to using no additional threads.
Discussion
Inspired by currently available tools and with the objective of improving total throughput performance in NLP tasks, we developed Sophia as an expedient UMLS concept extraction annotator. The Sophia pipeline, as configured as a single end-to-end UIMA application for evaluation purposes significantly out performs both MetaMap and cTAKES in throughput. Components of each of the pipelines were examined to further elucidate the bottleneck components. For the Sophia Pipeline, assertion is the most time consuming component, even with some efficiency built around the ConText methods. Evaluation using the extrinsic task of finding clinical problems showed that Sophia has a similar over-all f-score to cTAKES, and out performs MetaMap. Furthermore, Sophia had a better recall than both cTAKES and MetaMap on this task.
The techniques within Sophia are an evolution of the techniques embedded within MetaMap. Sophia borrowed heavily from NLM’s SPECIALIST Text Tools™, which were included in the first Java implementation of MetaMap Technology Transfer (MMTx). The Text Tools included the lexical lookup, the part-of-speech tagger and the phrase identification components of MetaMap. Sophia’s lexical lookup is a direct descendant to the Text Tools™. Sophia differs from MetaMap in that it does not do the brute force mapping that was included in MMTx; it keeps only the longest spanning matches from the variant table, that is, it does not compute partial matches; it does not do part-of-speech tagging or phrase identification; and it combines the concept information within the variant and lookup table, rather than relying on tables to do the lexical lookup, then lookup in tables to find the variants, followed by lookup in tables to find the concept information for each match.
MetaMap’s strength is in the evaluation and ranking it achieves once candidate concepts are pulled from the index. It is in this evaluation where MetaMap churns away. It is the most computationally expensive part of the algorithm, by far. Neither Sophia nor cTAKES includes such an evaluation component. This evaluation component allows MetaMap to retrieve and rank quality near matches that don’t quite cover, or cover too much (partial matches, concept gaps and over-matches) from the corpus text. Neither cTAKES nor Sophia retrieves partial matches, concept gaps or over-matches. This increases coverage for information retrieval tasks. If one limits to exact matches (those that have 1000 as the final mapping score within MetaMap), results, in theory, should be equivalent. MetaMap’s ranking takes into account the cognitive distance it took between seen text and a UMLS Concept. Sophia retains the cognitive distance but does not use it. Even with this ranking mechanism, MetaMap still returns ambiguous concepts when the ambiguity is at the lexical level. Embedded within MetaMap are techniques to limit ambiguity where it can without having to call upon the services of Word Sense Disambiguation (WSD). Such techniques include stop word filters, the ability to filter by semantic type, truncating by frequency hit cut offs and the like. MetaMap has an add-on WSD service to help ameliorate this facet as well. The Sophia pipeline considers all its ambiguous retrieval results to be a WSD issue that should be addressed properly in a downstream process or annotator, where both local and global context can be utilized.
In comparing Sophia, MetaMap and cTAKES methodology, the Sophia annotator shares many attributes with the cTAKES dictionary lookup annotator, and the Dictionary Lookup Annotator UMLS aggregate engine. Both are UIMA based, both include similar windowed lookup techniques. Whereas cTAKES uses LVG’s normalization to a normalized index of UMLS strings, Sophia looks up unadulterated tokens in Sophia indexes that are generated via LVG’s fruitful variants flow using UMLS Strings as its input. This algorithm was developed as part of MetaMap1, and had become an LVG function in the early 2000’s. Sophia relies on a post filtering of this flow to prune off unnecessarily aggressive or likely to be fallacious variants.
CTAKES matches to the SNOMED vocabulary subset of the UMLS. Sophia indexes to the level 0 + 9 UMLS terminologies which include MeSH and SNOMED. Both the cTAKES and Sophia pipelines were designed for use within the clinical setting, and as such, utilize tokenizer, sentence and section annotators and downstream annotators to add negation, conditional, hypothetical, or not-relating-to-the-patient context.
Sophia relies on sentence annotations created from upstream annotators within a UIMA pipeline. In this way, Sophia is similar to the cTAKES Lookup annotator functionality. Sophia adds Clinical Statements filled with CodedEntries to each annotated document. Clinical Statements are roughly equivalent to cTAKES EntityMentions and EventMentions, and even more roughly equivalent to MetaMap’s final mappings. A CodedEntry is equivalent to cTAKES’ UMLSConcept, and roughly equivalent to MetaMap’s Candidate Concept.
Whereas MetaMap and cTAKES formulate candidate phrases for lookup using similar techniques, Sophia does not. MetaMap and cTAKES break text into phrases before concept lookup by first tokenizing into sentences, then doing part-of-speech annotation, followed by phrase detection prior to phrase-to-candidate concept lookup. Sophia, in contrast, relies on upstream annotators to label sentences. Sophia looks up longest matching terms within the sentence, similar to MetaMap’s lexical lookup algorithm. Like MetaMap’s lexical lookup, Sophia’s term lookup uses a longest spanning match, which is an evolution of the algorithm embedded in the SPECIALIST Text Tools, which was embedded in MMTx, the java implementation of MetaMap. It should be noted that MetaMap has the ability to do both longest and shortest spanning matches. Sophia’s lookup mechanism has two new attributes not found in the SPECIALIST Text Tools. First, it starts its matches from left to right, using an index where the token keys are reversed. This is done to favor picking up right headed noun phrases. Second, UMLS Concept information is embedded within the indexes, so further lookup is not needed.
Whereas MetaMap first looks up terms within the SPECIALIST Lexicon, then uses those terms for phrase barrier determination, then looks up the phrase tokens to find UMLS concepts from an index of UMLS Concept variants, Sophia looks up terms in the UMLS Concept variant table directly without need for part-of-speech or phrasal boundaries.
An early UMLS principle was to keep knowledge resources like the SPECIALIST Lexicon and the UMLS Metathesaurus separate to keep semantic components out of the SPECIALIST Lexicon and to keep syntactic components out of the Metathesaurus. This allowed maintenance cycles for these resources to be de-coupled. This principle carried forth to continue to decouple the syntactic processing from the semantic processing via first finding terms, then phrases, then concepts within those phrases, as MetaMap, and to some extent, cTAKES does. Finding multi-word terms, particularly if they come from the SPECIALIST Lexicon, and particularly if they decrease ambiguity, greatly helps phrasal boundary detection from part-of-speech taggers that tag at the single token level of granularity. MetaMap uses the MedPost part-of-speech tagger, which was trained using a corpus that had sparse coverage of the majority of multi-words found in the SPECIALIST lexicon, and from the UMLS Metathesaurus. Term lookup followed by part-of-speech tagging on those words and terms within MMTx is used to make phrasal barrier decisions.
Sophia does away with the need for phrases and consequently, parts-of-speech. The term indexes and the UMLS Concept information are folded into one, indexed off the same key. That’s not to say that other annotators shouldn’t be run to keep around both part-of-speech and phrasal information. A consequence of ignoring phrasal boundaries within Sophia, longest matching terms that span across phrasal boundaries are retrievable within Sophia, but would be missed via MetaMap and cTAKES.
Although not incorporated here, the Sophia Pipeline, in practice, is often augmented with a local concept annotator combined with a file of local terms and their categories to address tasks where the UMLS lacks coverage. This is a capability included within the v3NLP framework that is not easily replicated within MetaMap or cTAKES.
Future Work
Future versions of Sophia will be integrated into v3NLP’s scaled-out architecture, where the slower annotators are replicated as multiple instances behind services, and called via wrappers around clients to these services. The next version of Sophia will be updated to the latest version of the UMLS.
The next version of the Sophia pipeline will include annotators to filter out non-salient false positives including the units of measure, dates, and the like that MetaMap effectively filters out. Further analysis will be spent to understand those multi-word instances that Sophia missed, and vice versa.
Assertion attribution will be looked at further to choose what assertion modules perform the best in respect to time and efficacy.
There is on-going interoperability work to enable v3NLP annotators, Sophia being one, with cTAKES to enable the use of cTAKES annotators within v3NLP and vise-versa.
Availability
Sophia is available via an Apache license, and is distributed from the http://v3nlp.utah.edu/sophia. End users are required to validate their own UMLS license via an application that validates UMLS licenses through the National Library of Medicine’s UMLS Terminology Services (UTS) before unlocking the content of indexes that contain UMLS derivative content within this distribution.
Conclusions
Sophia has been developed as an expedient UMLS concept annotator. The Sophia pipeline out performs both cTAKES and MetaMap in recall and has an f-score that is only 0.04 different than cTAKES. The pipeline runs 18 times faster than cTAKES and 7 times faster than the scaled-out MetaMap services. For those information extraction applications where fast throughput is needed and/or recall is favored over precision, the Sophia pipeline is an acceptable solution.
Figure 4.
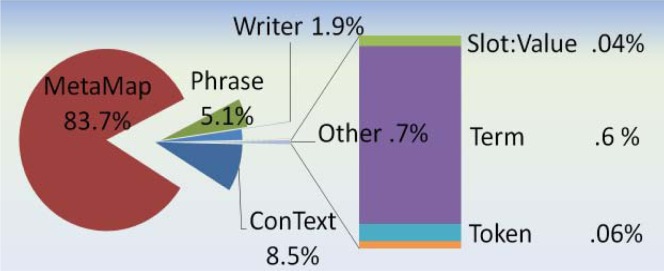
MetaMap pipeline proportion of each annotator’s processing
Acknowledgments
This work is funded by US Department of Veterans Affairs, Office of Research and Development, Health Services Research and Development grants VINCI HIR-08-204, CHIR HIR 08-374, ProWATCH grants HIR-10-001 and HIR 10-002. We would like to express our gratitude to the administration and staff of the VA Informatics and Computing Infrastructure (VINCI) for their support of our project. A special thanks to Shuying Shen for her efforts in providing the reference annotations. We also acknowledge the staff, resources and facilities of the VA Salt Lake City IDEAS Center for providing a rich and stimulating environment for NLP research.
Footnotes
Parts of the Sophia pipeline were used within an entry in the i2b2 2010 VA Challenge. The whole corpus was used for additional training to improve pipeline components after the challenge. This training invalidates any efficacy evaluation to this corpus.
References
- 1.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings / AMIA Annual Symposium AMIA Symposium; 2001. pp. 17–21. Epub 2002/02/05. [PMC free article] [PubMed] [Google Scholar]
- 2.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association : JAMIA. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. Epub 2010/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC medical informatics and decision making. 2006;6:30. doi: 10.1186/1472-6947-6-30. Epub 2006/07/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.VA Informatics and Computing Infrastructure (VINCI) 2012. [cited 2013]; Available from: http://www.hsrd.research.va.gov/for_researchers/vinci/
- 5.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Lang Eng. 2004;10(3–4):327–48. [Google Scholar]
- 6.Lindberg C. The Unified Medical Language System (UMLS) of the National Library of Medicine. J Am Med Rec Assoc. 1990;61(5):40–2. Epub 1990/04/09. [PubMed] [Google Scholar]
- 7.Hersh W, Hickam D. Information retrieval in medicine: the SAPHIRE experience. Medinfo MEDINFO. 1995;(8 Pt 2):1433–7. [PubMed] [Google Scholar]
- 8.Friedman C. A broad-coverage natural language processing system. Proceedings /AMIA Annual Symposium AMIA Symposium; 2000. pp. 270–4. [PMC free article] [PubMed] [Google Scholar]
- 9.MedKAT/p. http://ohnlp.org/index.php/MedKAT/p.
- 10.Denny JC, Peterson JF, Choma NN, Xu H, Miller RA, Bastarache L, et al. Development of a natural language processing system to identify timing and status of colonoscopy testing in electronic medical records. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2009. p. 141. [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham H. GATE, a general architecture for text engineering. Computers and the Humanities. 2002;36(2):223–54. [Google Scholar]
- 12.Apache UIMA-AS http://uima.apache.org/doc-uimaas-what.html.
- 13.Lexical Variant Generation Documentation: Fruitful Variants. http://lexsrv3.nlm.nih.gov/LexSysGroup/Projects/lvg/current/docs/designDoc/UDF/flow/fG.html.
- 14.Aronson AR. The effect of textual variation on concept based information retrieval. Proceedings : a conference of the American Medical Informatics Association / AMIA Annual Fall Symposium AMIA Fall Symposium; 1996. pp. 373–7. [PMC free article] [PubMed] [Google Scholar]
- 15.Brown AC, Divita G. The SPECIALIST Text Tools. http://lexsrv3.nlm.nih.gov/LexSysGroup/Projects/textTools/
- 16.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proceedings / the Annual Symposium on Computer Application [sic] in Medical Care Symposium on Computer Applications in Medical Care; 1994. pp. 235–9. [PMC free article] [PubMed] [Google Scholar]
- 17.Chapman WW, Chu D, Dowling JN. ConText: an algorithm for identifying contextual features from clinical text. Proceedings of the Workshop on BioNLP 2007: Biological, Translational, and Clinical Language Processing; Prague, Czech Republic. 1572408: Association for Computational Linguistics; 2007. pp. 81–8. [Google Scholar]
- 18.Collaboration between VINCI and CHIR. 2012. [cited 2013]; Available from: http://www.hsrd.research.va.gov/for_researchers/vinci/chir.cfm.
- 19.Uzuner O, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. Journal of the American Medical Informatics Association : JAMIA. 2011;18(5):552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]


