Abstract
The title compound, C6H12O6, a C-3 position epimer of fructose, was crystallized from an aqueous solution of equimolar mixture of d- and l-psicose (1,3,4,5,6-pentahydroxyhexan-2-one, ribo-2-hexulose, allulose), and it was confirmed that d-psicose (or l-psicose) formed β-pyranose with a 2 C 5 (or 5 C 2) conformation. In the crystal, an O—H⋯O hydrogen bond between the hydroxy groups at the C-3 and C-2 positions connects homochiral molecules into a column along the b axis. The columns are linked by other O—H⋯O hydrogen bonds between d- and l-psicose molecules, forming a three-dimensional network. An intramolecular O—H⋯O hydrogen bond is also observed. The cell volume of racemic β-d,l-psicose [763.21 (6) Å3] is almost the same as that of chiral β-d-psicose [753.06 Å3].
Keywords: crystal structure, hydrogen bonding, racemic compound, rare sugar
Related literature
For the crystal structure of the chiral β-d-psicose, see: Kwiecien et al. (2008 ▸); Fukada et al. (2010 ▸). For the synthesis of the chiral d-psicose, see: Itoh et al. (1995 ▸); Takeshita et al. (2000 ▸). For the synthesis of the chiral l-psicose, see: Takeshita et al. (1996 ▸).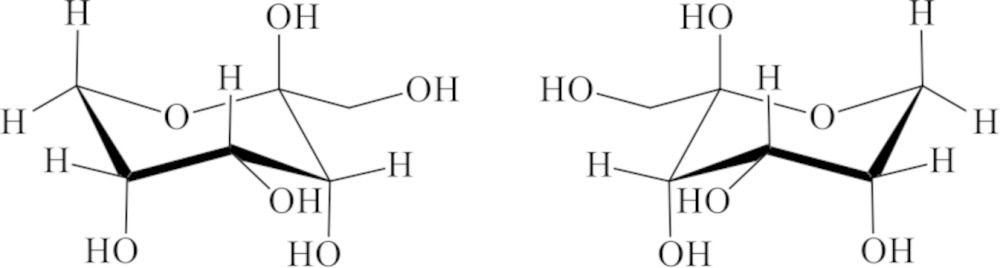
Experimental
Crystal data
C6H12O6
M r = 180.16
Orthorhombic,
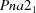
a = 11.2629 (5) Å
b = 5.3552 (3) Å
c = 12.6538 (6) Å
V = 763.21 (6) Å3
Z = 4
Cu Kα radiation
μ = 1.25 mm−1
T = 296 K
0.10 × 0.10 × 0.10 mm
Data collection
Rigaku R-AXIS RAPID diffractometer
Absorption correction: multi-scan (ABSCOR; Higashi, 1995 ▸) T min = 0.442, T max = 0.883
12119 measured reflections
1400 independent reflections
1295 reflections with F 2 > 2σ(F 2)
R int = 0.139
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.102
S = 1.04
1400 reflections
116 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.25 e Å−3
Δρmin = −0.23 e Å−3
Absolute structure: Flack (1983 ▸), 666 Friedel pairs
Absolute structure parameter: 0.1 (4)
Data collection: RAPID-AUTO (Rigaku, 2009 ▸); cell refinement: RAPID-AUTO; data reduction: RAPID-AUTO; program(s) used to solve structure: SIR2011 (Burla et al., 2012 ▸); program(s) used to refine structure: SHELXL2013 (Sheldrick, 2015 ▸); molecular graphics: CrystalStructure (Rigaku, 2014 ▸); software used to prepare material for publication: CrystalStructure.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989015006623/is5394sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015006623/is5394Isup2.hkl
ORTEP . DOI: 10.1107/S2056989015006623/is5394fig1.tif
ORTEP view of the title compound with the atom-labeling scheme. The thermal ellipsoids of all non-hydrogen atoms are drawn at the 50% probability level. H atoms are shown as small spheres of arbitrary radius.
b . DOI: 10.1107/S2056989015006623/is5394fig2.tif
Part of the crystal structure of the title compound with hydrogen-bonding network represented as green solid lines, viewed down the b-axis. The hydrogen atoms are omitted for clarity.
d et al. . DOI: 10.1107/S2056989015006623/is5394fig3.tif
Part of the crystal structure of the chiral β-d-psicose (Fukada et al., 2010) with hydrogen-bonding network represented as green solid lines. The hydrogen atoms are omitted for clarity.
CCDC reference: 1057484
Additional supporting information: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (, ).
| DHA | DH | HA | D A | DHA |
|---|---|---|---|---|
| O1H1AO3i | 0.82 | 1.91 | 2.715(3) | 168 |
| O2H2AO4ii | 0.82 | 1.92 | 2.724(3) | 166 |
| O3H3AO2iii | 0.82 | 2.20 | 2.874(3) | 140 |
| O3H3AO5 | 0.82 | 2.36 | 2.822(4) | 117 |
| O4H4AO6iv | 0.82 | 2.14 | 2.829(3) | 141 |
| O5H5AO1v | 0.82 | 1.94 | 2.746(4) | 169 |
Symmetry codes: (i) 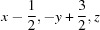 ; (ii)
; (ii) 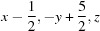 ; (iii)
; (iii)  ; (iv)
; (iv) 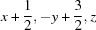 ; (v)
; (v) 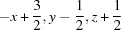 .
.
Acknowledgments
The authors are grateful to Grants-in-Aid for Rare Sugar Research of Kagawa University.
supplementary crystallographic information
S1. Comment
In the crystal of the title compound, the D- and L-molecules are located alternatively in a-c plane, so that the main hydrogen bonding networks can be created between D- and L-molecules. An additional hydrogen bonding between two D-molecules (and two L-molecules) are observed along to the b-axis (O3—H3A···O2iii; Table 1). The molecular structure of D-psicose (or L-psicose) is β-pyranose form with a 2C5 (or 5C2) conformation. Orientations of two OH groups at C-3 and C-5 positions are axial, therefore an intramolecular hydrogen bonding can be observed (O3—H3A···O5; 2.36 Å) [hereafter, (O3···O5)]. The intramolecular hydrogen bonding unit (O3···O5) shown in the racemic D,L-crystal has also observed in a chiral D-crystal (Fukada et al., 2010). In the chiral one, one-dimensional hydrogen bonding chain, that is (O3···O5) -> (O3···O5) -> (O3···O5) ->···, can be observed by connecting through an another hydrogen bonding between two D-molecule units (O5—H5···O3). On the other hand in the case of the racemic one, the L-molecule (or D-molecule) plays as a role of a bridging between two adjacent intramolecular hydrogen bonding in D-molecule (or L-molecule) (O3···O5) units, that is (D O3···O5) -> (L O1) -> (D O3···O5) -> (L O1) -> ··· (or, (L O3···O5) -> (D O1) -> (L O3···O5) -> (D O1) -> ···). Concerning the intermolecular hydrogen bonding, there are four kinds of bondings are also observed between D- and L- psicose molecules (O1—H1A···O3 (a-axis), O2—H2A···O4, O4—H4A···O6 (a-axis), and O5—H5A···O1 (c-axis),). The cell volume of racemic β-D,L-psicose [763.21 (6) Å3 at r.t.] is almost the same as that of chiral β-D-psicose [753.06 Å3 at r.t.].
S2. Experimental
D-Psicose was prepared from D-fructose by enzymatic epimerization using D-tagatose 3-epimerase (Itoh et al., 1995; Takeshita et al., 2000). L-Psicose was prepared from allitol by microbial oxidation using Gluconobacter frateurii IFO 3254 (Takeshita et al., 1996). D-Psicose and L-psicose were mixed in equal amount and dissolved in hot water to give 60, 65, 70, 75, and 80 wt% solution. And these samples were kept at 10, 20, and 30 °C. After one day, small crystals were obtained in 65, 70, 75, and 80 wt% solution at 10, 20, and 30 °C.
S3. Refinement
H atoms bounded to methine-type C (H3B, H4B, H5B) were positioned geometrically and refined using a riding model with C—H = 0.98 Å and Uiso(H) = 1.2Ueq(C). H atoms bounded to methylene-type C (H1B, H1C, H6A, H6B) were positioned geometrically and refined using a riding model with C—H = 0.97 Å and Uiso(H) = 1.2Ueq(C). H atoms bounded to O (H1A, H2A, H3A, H4A, H5A) were positioned geometrically and refined using a riding model with O—H = 0.82 Å and Uiso(H) = 1.2Ueq(O), allowing for free rotation of the OH groups.
Figures
Fig. 1.
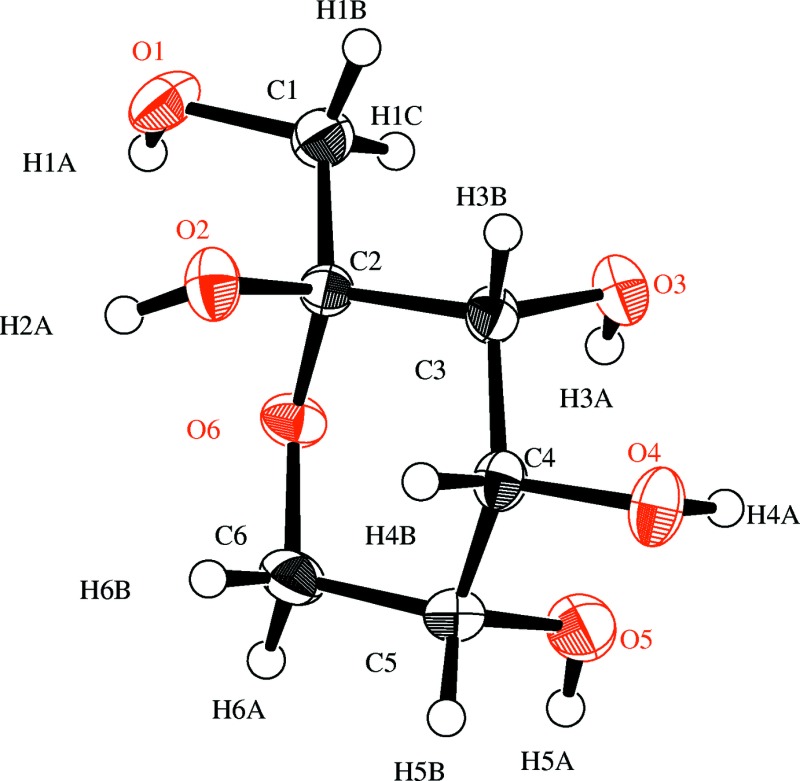
ORTEP view of the title compound with the atom-labeling scheme. The thermal ellipsoids of all non-hydrogen atoms are drawn at the 50% probability level. H atoms are shown as small spheres of arbitrary radius.
Fig. 2.
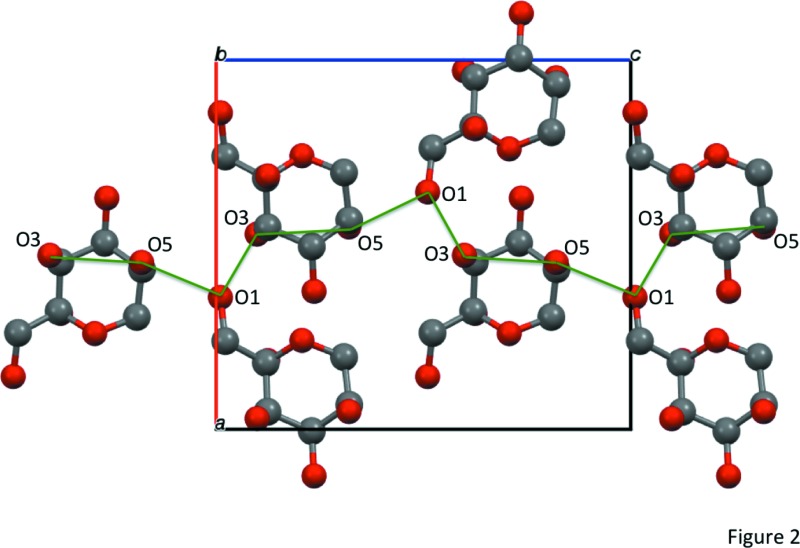
Part of the crystal structure of the title compound with hydrogen-bonding network represented as green solid lines, viewed down the b-axis. The hydrogen atoms are omitted for clarity.
Fig. 3.
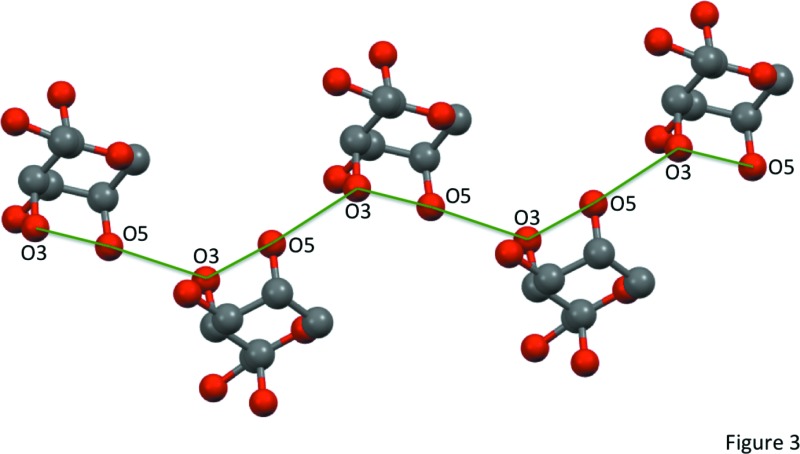
Part of the crystal structure of the chiral β-D-psicose (Fukada et al., 2010) with hydrogen-bonding network represented as green solid lines. The hydrogen atoms are omitted for clarity.
Crystal data
| C6H12O6 | Dx = 1.568 Mg m−3 |
| Mr = 180.16 | Cu Kα radiation, λ = 1.54187 Å |
| Orthorhombic, Pna21 | Cell parameters from 5584 reflections |
| a = 11.2629 (5) Å | θ = 3.5–68.5° |
| b = 5.3552 (3) Å | µ = 1.25 mm−1 |
| c = 12.6538 (6) Å | T = 296 K |
| V = 763.21 (6) Å3 | Block, colorless |
| Z = 4 | 0.10 × 0.10 × 0.10 mm |
| F(000) = 384.00 |
Data collection
| Rigaku R-AXIS RAPID diffractometer | 1295 reflections with F2 > 2σ(F2) |
| Detector resolution: 10.000 pixels mm-1 | Rint = 0.139 |
| ω scans | θmax = 68.2°, θmin = 7.0° |
| Absorption correction: multi-scan (ABSCOR; Higashi, 1995) | h = −13→13 |
| Tmin = 0.442, Tmax = 0.883 | k = −6→6 |
| 12119 measured reflections | l = −15→15 |
| 1400 independent reflections |
Refinement
| Refinement on F2 | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.048 | w = 1/[σ2(Fo2) + (0.0359P)2] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.102 | (Δ/σ)max < 0.001 |
| S = 1.04 | Δρmax = 0.25 e Å−3 |
| 1400 reflections | Δρmin = −0.23 e Å−3 |
| 116 parameters | Extinction correction: SHELXL |
| 1 restraint | Extinction coefficient: 0.039 (3) |
| Primary atom site location: structure-invariant direct methods | Absolute structure: Flack (1983), 666 Friedel pairs |
| Secondary atom site location: difference Fourier map | Absolute structure parameter: 0.1 (4) |
| Hydrogen site location: inferred from neighbouring sites |
Special details
| Geometry. ENTER SPECIAL DETAILS OF THE MOLECULAR GEOMETRY |
| Refinement. Refinement was performed using all reflections. The weighted R-factor (wR) and goodness of fit (S) are based on F2. R-factor (gt) are based on F. The threshold expression of F2 > 2.0 σ(F2) is used only for calculating R-factor (gt). |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.6428 (2) | 1.0475 (5) | 0.0101 (2) | 0.0321 (7) | |
| O2 | 0.8138 (2) | 1.3225 (4) | 0.1244 (2) | 0.0283 (6) | |
| O3 | 0.9712 (2) | 0.7407 (4) | 0.0986 (2) | 0.0278 (6) | |
| O4 | 1.12635 (19) | 0.9941 (5) | 0.2360 (2) | 0.0306 (7) | |
| O5 | 0.9520 (3) | 0.6941 (5) | 0.3201 (2) | 0.0368 (7) | |
| O6 | 0.75926 (19) | 0.9487 (5) | 0.2068 (2) | 0.0243 (6) | |
| C1 | 0.7610 (3) | 0.9600 (8) | 0.0218 (3) | 0.0275 (8) | |
| C2 | 0.8199 (3) | 1.0614 (6) | 0.1199 (3) | 0.0211 (7) | |
| C3 | 0.9525 (3) | 0.9956 (6) | 0.1238 (3) | 0.0223 (7) | |
| C4 | 1.0049 (3) | 1.0665 (7) | 0.2306 (2) | 0.0233 (8) | |
| C5 | 0.9337 (3) | 0.9564 (7) | 0.3210 (3) | 0.0266 (8) | |
| C6 | 0.8044 (3) | 1.0258 (7) | 0.3083 (3) | 0.0289 (8) | |
| H1B | 0.80695 | 1.00892 | −0.03968 | 0.0330* | |
| H1C | 0.76042 | 0.77904 | 0.02497 | 0.0330* | |
| H1A | 0.59772 | 0.95756 | 0.04384 | 0.0386* | |
| H2A | 0.75043 | 1.36467 | 0.15074 | 0.0340* | |
| H3A | 0.92832 | 0.65324 | 0.13561 | 0.0333* | |
| H3B | 0.99273 | 1.0959 | 0.06985 | 0.0268* | |
| H4B | 1.00147 | 1.24872 | 0.23691 | 0.0280* | |
| H4A | 1.1313 | 0.84167 | 0.23198 | 0.0367* | |
| H5A | 0.92192 | 0.63218 | 0.37294 | 0.0442* | |
| H5B | 0.96322 | 1.0246 | 0.38792 | 0.0319* | |
| H6A | 0.75828 | 0.94707 | 0.36378 | 0.0347* | |
| H6B | 0.79566 | 1.20525 | 0.31544 | 0.0347* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0232 (14) | 0.0431 (18) | 0.0301 (13) | −0.0019 (11) | −0.0058 (11) | 0.0126 (12) |
| O2 | 0.0238 (12) | 0.0228 (13) | 0.0384 (14) | 0.0010 (9) | 0.0057 (11) | 0.0025 (13) |
| O3 | 0.0252 (12) | 0.0240 (13) | 0.0340 (14) | 0.0013 (10) | 0.0068 (10) | −0.0020 (11) |
| O4 | 0.0193 (12) | 0.0265 (14) | 0.0459 (17) | 0.0004 (10) | −0.0022 (10) | 0.0013 (12) |
| O5 | 0.0475 (17) | 0.0309 (15) | 0.0322 (15) | 0.0064 (12) | 0.0101 (11) | 0.0101 (12) |
| O6 | 0.0230 (12) | 0.0297 (14) | 0.0203 (11) | −0.0057 (10) | 0.0027 (10) | 0.0002 (12) |
| C1 | 0.025 (2) | 0.032 (2) | 0.0254 (19) | −0.0001 (15) | −0.0001 (14) | 0.0024 (18) |
| C2 | 0.0211 (17) | 0.0228 (17) | 0.0195 (16) | 0.0012 (12) | 0.0034 (14) | 0.0043 (16) |
| C3 | 0.0197 (19) | 0.0233 (17) | 0.0239 (17) | 0.0009 (12) | 0.0038 (14) | 0.0042 (14) |
| C4 | 0.0189 (17) | 0.025 (2) | 0.0265 (19) | 0.0009 (13) | 0.0000 (13) | 0.0006 (15) |
| C5 | 0.030 (2) | 0.030 (2) | 0.0201 (17) | 0.0027 (14) | −0.0011 (15) | −0.0001 (15) |
| C6 | 0.0272 (18) | 0.039 (2) | 0.0209 (17) | 0.0006 (15) | 0.0055 (15) | −0.0002 (15) |
Geometric parameters (Å, º)
| O1—C1 | 1.419 (4) | O1—H1A | 0.820 |
| O2—C2 | 1.401 (4) | O2—H2A | 0.820 |
| O3—C3 | 1.418 (4) | O3—H3A | 0.820 |
| O4—C4 | 1.423 (4) | O4—H4A | 0.820 |
| O5—C5 | 1.419 (4) | O5—H5A | 0.820 |
| O6—C2 | 1.428 (4) | C1—H1B | 0.970 |
| O6—C6 | 1.442 (4) | C1—H1C | 0.970 |
| C1—C2 | 1.509 (5) | C3—H3B | 0.980 |
| C2—C3 | 1.535 (5) | C4—H4B | 0.980 |
| C3—C4 | 1.523 (5) | C5—H5B | 0.980 |
| C4—C5 | 1.516 (5) | C6—H6A | 0.970 |
| C5—C6 | 1.512 (5) | C6—H6B | 0.970 |
| C2—O6—C6 | 113.3 (2) | C4—O4—H4A | 109.469 |
| O1—C1—C2 | 112.3 (3) | C5—O5—H5A | 109.469 |
| O2—C2—O6 | 111.6 (3) | O1—C1—H1B | 109.152 |
| O2—C2—C1 | 111.8 (3) | O1—C1—H1C | 109.152 |
| O2—C2—C3 | 106.0 (3) | C2—C1—H1B | 109.145 |
| O6—C2—C1 | 105.7 (3) | C2—C1—H1C | 109.145 |
| O6—C2—C3 | 110.1 (3) | H1B—C1—H1C | 107.867 |
| C1—C2—C3 | 111.8 (3) | O3—C3—H3B | 107.581 |
| O3—C3—C2 | 111.0 (3) | C2—C3—H3B | 107.578 |
| O3—C3—C4 | 112.5 (3) | C4—C3—H3B | 107.580 |
| C2—C3—C4 | 110.4 (3) | O4—C4—H4B | 107.757 |
| O4—C4—C3 | 110.3 (3) | C3—C4—H4B | 107.761 |
| O4—C4—C5 | 111.5 (3) | C5—C4—H4B | 107.767 |
| C3—C4—C5 | 111.6 (3) | O5—C5—H5B | 109.099 |
| O5—C5—C4 | 107.5 (3) | C4—C5—H5B | 109.103 |
| O5—C5—C6 | 112.5 (3) | C6—C5—H5B | 109.093 |
| C4—C5—C6 | 109.5 (3) | O6—C6—H6A | 109.357 |
| O6—C6—C5 | 111.4 (3) | O6—C6—H6B | 109.357 |
| C1—O1—H1A | 109.471 | C5—C6—H6A | 109.358 |
| C2—O2—H2A | 109.471 | C5—C6—H6B | 109.358 |
| C3—O3—H3A | 109.470 | H6A—C6—H6B | 107.992 |
| C2—O6—C6—C5 | −60.7 (3) | C1—C2—C3—C4 | −171.7 (2) |
| C6—O6—C2—O2 | −58.2 (3) | O3—C3—C4—O4 | 52.5 (3) |
| C6—O6—C2—C1 | −179.9 (2) | O3—C3—C4—C5 | −72.0 (3) |
| C6—O6—C2—C3 | 59.2 (3) | C2—C3—C4—O4 | 177.1 (2) |
| O1—C1—C2—O2 | −53.5 (4) | C2—C3—C4—C5 | 52.6 (3) |
| O1—C1—C2—O6 | 68.1 (3) | O4—C4—C5—O5 | −54.2 (3) |
| O1—C1—C2—C3 | −172.1 (2) | O4—C4—C5—C6 | −176.7 (2) |
| O2—C2—C3—O3 | −168.2 (2) | C3—C4—C5—O5 | 69.6 (3) |
| O2—C2—C3—C4 | 66.4 (3) | C3—C4—C5—C6 | −52.9 (3) |
| O6—C2—C3—O3 | 71.0 (3) | O5—C5—C6—O6 | −63.8 (4) |
| O6—C2—C3—C4 | −54.4 (3) | C4—C5—C6—O6 | 55.7 (4) |
| C1—C2—C3—O3 | −46.2 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1A···O3i | 0.82 | 1.91 | 2.715 (3) | 168 |
| O2—H2A···O4ii | 0.82 | 1.92 | 2.724 (3) | 166 |
| O3—H3A···O2iii | 0.82 | 2.20 | 2.874 (3) | 140 |
| O3—H3A···O5 | 0.82 | 2.36 | 2.822 (4) | 117 |
| O4—H4A···O6iv | 0.82 | 2.14 | 2.829 (3) | 141 |
| O5—H5A···O1v | 0.82 | 1.94 | 2.746 (4) | 169 |
Symmetry codes: (i) x−1/2, −y+3/2, z; (ii) x−1/2, −y+5/2, z; (iii) x, y−1, z; (iv) x+1/2, −y+3/2, z; (v) −x+3/2, y−1/2, z+1/2.
Footnotes
Supporting information for this paper is available from the IUCr electronic archives (Reference: IS5394).
References
- Burla, M. C., Caliandro, R., Camalli, M., Carrozzini, B., Cascarano, G. L., Giacovazzo, C., Mallamo, M., Mazzone, A., Polidori, G. & Spagna, R. (2012). J. Appl. Cryst. 45, 357–361.
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Fukada, K., Ishii, T., Tanaka, K., Yamaji, M., Yamaoka, Y., Kobashi, K. & Izumori, K. (2010). Bull. Chem. Soc. Jpn, 83, 1193–1197.
- Higashi, T. (1995). ABSCOR. Rigaku Corporation, Tokyo, Japan.
- Itoh, H., Sato, T. & Izumori, K. (1995). J. Ferment. Bioeng. 80, 101–103.
- Kwiecien, A., Slepokura, K. & Lis, T. (2008). Carbohydr. Res. 343, 2336–2339. [DOI] [PubMed]
- Rigaku (2009). RAPID-AUTO. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2014). CrystalStructure. Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Takeshita, K., Shimonishi, T. & Izumori, K. (1996). J. Ferment. Bioeng. 81, 212–215.
- Takeshita, K., Suga, A., Takada, G. & Izumori, K. (2000). J. Biosci. Bioeng. 90, 453–455. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989015006623/is5394sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989015006623/is5394Isup2.hkl
ORTEP . DOI: 10.1107/S2056989015006623/is5394fig1.tif
ORTEP view of the title compound with the atom-labeling scheme. The thermal ellipsoids of all non-hydrogen atoms are drawn at the 50% probability level. H atoms are shown as small spheres of arbitrary radius.
b . DOI: 10.1107/S2056989015006623/is5394fig2.tif
Part of the crystal structure of the title compound with hydrogen-bonding network represented as green solid lines, viewed down the b-axis. The hydrogen atoms are omitted for clarity.
d et al. . DOI: 10.1107/S2056989015006623/is5394fig3.tif
Part of the crystal structure of the chiral β-d-psicose (Fukada et al., 2010) with hydrogen-bonding network represented as green solid lines. The hydrogen atoms are omitted for clarity.
CCDC reference: 1057484
Additional supporting information: crystallographic information; 3D view; checkCIF report


