Abstract
The large numbers of microorganisms that inhabit mammalian body surfaces have a highly co-evolved relationship with the immune system. Although many of these microbes carry out functions that are critical for host physiology, they nevertheless pose the threat of breach with ensuing pathologies. The mammalian immune system plays an essential role in maintaining homeostasis with resident microbial communities, thus ensuring that the mutualistic nature of the host-microbial relationship is maintained. At the same time, resident bacteria profoundly shape mammalian immunity. Here we review advances in our understanding of the interactions between resident microbes and the immune system and the implications of these findings for human health.
Complex communities of microorganisms, termed the ‘microbiota’, inhabit the body surfaces of virtually all vertebrates. In the lower intestine these organisms reach extraordinary densities and have evolved to degrade a variety of plant polysaccharides and other dietary substances (1). This simultaneously enhances host digestive efficiency and ensures a steady nutrient supply for the microbes. Metabolic efficiency was likely a potent selective force that shaped the evolution of both sides of the host-microbiota relationship. Millions of years of co-evolution, however, have forged pervasive interconnections between the physiologies of microbial communities and their hosts that extend beyond metabolic functions. These interconnections are particularly apparent in the relationship between the microbiota and the immune system.
Despite the symbiotic nature of the intestinal host-microbial relationship, the close association of an abundant bacterial community with intestinal tissues poses immense health challenges. The dense communities of bacteria in the lower intestine (≥1012/cm3 intestinal contents) are separated from body tissues by the epithelial layer (10μm) over a large intestinal surface area (~200m2 in humans). Opportunistic invasion of host tissue by resident bacteria has serious health consequences including inflammation and sepsis. The immune system has thus evolved adaptations that work together to contain the microbiota and preserve the symbiotic relationship between host and microbiota. The evolution of the vertebrate immune system has therefore been driven by the need to protect the host from pathogens and to foster complex microbial communities for their metabolic benefits (2).
In this review we survey the state of our understanding of microbiota-immune system interactions. We also highlight key experimental challenges that must be confronted in order to advance our understanding in this area, and consider how our knowledge of these interactions might be harnessed in order to improve public health.
Tools for analyzing the microbiota-immune system relationship
Much of our current understanding of microbiota-immune system interactions has been acquired from studies of germ-free animals. Such animals are reared in sterile isolators in order to control their exposure to microorganisms, including viruses, bacteria, and eukaryotic parasites. Germ-free animals can be studied in their microbiologically sterile state, or can serve as living test tubes for the establishment of simplified microbial ecosystems composed of a single microbial species or defined species mixtures. The technology has thus come to be known as “gnotobiotics”, a term derived from Greek meaning “known life”. Gnotobiotic animals, particularly rodents, have become critical experimental tools for determining which host immune functions are genetically encoded and which require interactions with microbes.
The current impetus for gnotobiotic experimentation has been driven by several important technical advances. First, because any mouse strain can be derived to germ-free status (3), large numbers of genetically-targeted and wild-type inbred isogenic mouse strains have become available in the germ-free state. The contribution of different immune system constituents to host microbial mutualism can thus be determined by comparing the effects of microbial colonization in genetically-altered and wild-type mice (4, 5).
Second, next-generation sequencing technologies have opened the black box of microbiota complexity. Although advances in ex vivo culturability are still needed, the composition of human and animal microbiotas can be operationally defined from polymorphisms of bacterial genes, especially those encoding the 16S ribosomal RNA sequences. Such analyses have made possible the construction of defined microbiotas, whose distinct effects on host immunity can now be examined (6). Moreover, these advances allow the study of experimental animals that are both isobiotic and, in a defined inbred host, isogenic. A dominant goal of these efforts is to benefit human health (see Review by Blumberg and Powie (7)). With the developing technology, the species differences can be closed using mice with a defined humanized microbiota (8). On the horizon, there is even the prospect of humanized isobiotic mice that also have a humanized immune system (9).
A third advance has been the development of experimental systems that allow the uncoupling of commensal effects on the immune system from microbial colonization. This cannot be achieved by antibiotic treatment alone because a small proportion of the targeted microbes will persist. Deletion strains of bacteria lacking the ability to synthesize prokaryotic-specific amino acids have been developed which can be grown in culture but do not persist in vivo, so the animals become germ-free again. This allows issues of mucosal immune induction, memory and functional protection to be explored without permanent colonization (10).
Finally, important insights about the impact of resident microbial communities on mammalian host biology have been acquired by using high throughput transcriptomic and metabolomic tools to compare germ-free and colonized mice (11, 12). These tools include DNA microarrays, which have led to a detailed understanding of how microbiota shape many aspects of host physiology, including immunity (13, 14) and development (15), as well as mass spectrometry and nuclear magnetic resonance spectroscopy, which have provided important insights into how microbiota influence metabolic signaling in mammalian hosts (12). The application of these new approaches to the older technology of gnotobiotics has revolutionized the study of interactions between the microbiota and the immune system.
Looking inside-out: Immune system control of the microbiota
A major driving force in the evolution of the mammalian immune system has been the need to maintain homeostatic relationships with the microbiota. This encompasses control of microbial interactions with host tissues as well as the composition of microbial consortia. Here we discuss recent insights into how the immune system exerts “inside-out” control over microbiota localization and community composition.
Stratification and compartmentalization of the microbiota
The intestinal immune system faces unique challenges relative to other organs, as it must continuously confront an enormous microbial load. At the same time it is necessary to avoid pathologies arising from innate immune signaling or from microbiota alterations that disturb essential metabolic functions. An important function of the intestinal immune system is to control the exposure of bacteria to host tissues, thereby lessening the potential for pathologic outcomes. This occurs at two distinct levels: first, by minimizing direct contact between intestinal bacteria and the epithelial cell surface (stratification), and, second, by confining penetrant bacteria to intestinal sites and limiting their exposure to the systemic immune compartment (compartmentalization).
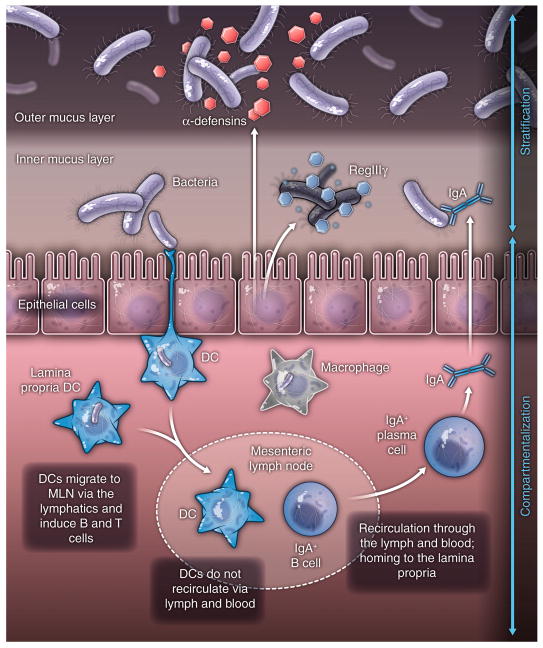
Several immune effectors function together to stratify luminal microbes and to minimize bacterial-epithelial contact (Fig. 1A). Intestinal goblet cells secrete mucin glycoproteins that assemble into a ~150 μm thick viscous coating at the intestinal epithelial cell surface. In the colon, there are two structurally distinct mucus layers. Although the outer mucus layer contains large numbers of bacteria, the inner mucus layer is resistant to bacterial penetration (16). In contrast, the small intestine lacks clearly distinct inner and outer mucus layers (17). Here, compartmentalization depends in part on antibacterial proteins that are secreted by the intestinal epithelium. RegIIIγ is an antibacterial lectin that is expressed in epithelial cells under the control of Toll-like receptors (TLRs) (18–20). RegIIIγ limits bacterial penetration of the small intestinal mucus layer, thus restricting the number of bacteria that contact the epithelial surface (5).
Figure 1. Looking inside-out: immune system control of the microbiota.
(A) Several immune effectors function together to stratify luminal microbes and to minimize bacterial-epithelial contact. This includes the mucus layer produced by goblet cells, epithelial antibacterial proteins, and immunoglobulin A secreted by lamina propria plasma cells.
(B) The zones of containment of the intestinal microbiota by the immune system are illustrated by dashed lines. The red dotted line represents the lower intestinal epithelial cell layer, above which secreted mucus, peptides, proteins and antibodies combine to stratify the intestinal microbiota and limit access to the epithelium. Some microbes are sampled by intestinal dendritic cells (DC), shown as green cells carrying brown bacilli. The loaded DC traffic to the mesenteric lymph nodes through the intestinal lymphatics (yellow), but do not penetrate further into the body. This compartmentalizes live bacteria and induction of immune responses to the mucosal immune system, shown by the blue dashed line. There is recirculation of induced B cells (blue) and some T cell subsets through the lymphatics and the bloodstream to home back to mucosal sites, where B cells differentiate into IgA-secreting plasma cells. The spleen and liver are capable of filtering any live microbes that penetrate the systemic bloodstream – these are eliminated by the biocidal activity of splenic and hepatic phagocytes. The stomach and upper intestine have very low levels of microbial colonization as a result of acid and bile secretion in the stomach and duodenum and constant intestinal motility: this creates the necessary microbial exclusion zone for the host to have the first priority to digest dietary nutrients.
Stratification of intestinal bacteria on the luminal side of the epithelial barrier also depends on secreted immunoglobulin A (IgA). IgA specific for intestinal bacteria is produced with the help of intestinal dendritic cells that sample the small numbers of bacteria that penetrate the overlying epithelium. These bacteria-laden dendritic cells interact with B and T cells in the Peyer’s patches, inducing B cells to produce IgA directed against intestinal bacteria (21). IgA+ B cells home to the intestinal lamina propria and secrete IgA that is transcytosed across the epithelium and deposited on the apical surface. The transcytosed IgAs bind to luminal bacteria, preventing microbial translocation across the epithelial barrier (22).
Mucosal compartmentalization functions to minimize exposure of resident bacteria to the systemic immune system (Fig. 1B). Although bacteria are largely confined to the luminal side of the epithelial barrier, the sheer number of intestinal bacteria makes occasional breach inevitable. Typically, commensal microorganisms that penetrate the intestinal epithelial cell barrier are phagocytosed and eliminated by lamina propria macrophages (23). However, the intestinal immune system samples some of the penetrant bacteria, engendering specific immune responses that are distributed along the length of the intestine (21). Bacteria that penetrate the intestinal barrier are engulfed by dendritic cells (DCs) residing in the lamina propria and are carried alive to the mesenteric lymph nodes. However, these bacteria do not penetrate to systemic secondary lymphoid tissues. Rather, the commensal-bearing DCs induce protective secretory IgAs (21), which are distributed throughout all mucosal surfaces via recirculation of activated B and T cells. Thus, distinctive anatomical adaptations in the mucosal immune system allow immune responses directed against commensals to be distributed widely while still being confined to mucosal tissues.
Other immune cell populations also promote the containment of commensal bacteria to intestinal sites. Innate lymphoid cells reside in the lamina propria and have effector cytokine profiles resembling those of T helper (Th) cells (24). Innate lymphoid cells that produce interleukin (IL)-22 are essential for containment of lymphoid-resident bacteria to the intestine, thus preventing their spread to systemic sites (25).
The compartmentalization of mucosal and systemic immune priming can be severely perturbed in immune deficient mice. For example, mice engineered to lack IgA show priming of serum IgG responses against commensals, indicating that these bacteria have been exposed to the systemic immune system (22). A similar outcome is observed when innate immune sensing is defective. Mice lacking MyD88 or TRIF signaling adaptors for TLR-mediated sensing of bacteria also produce serum IgG responses against commensals (26). This probably results from the fact that in these settings, large numbers of commensals cross the epithelial barrier and phagocytic cells are less able to eliminate the penetrant organisms.
Immune system control of microbiota composition
The development of high-throughput sequencing technologies for microbiota analysis has provided insight into the many factors that determine microbiota composition. For example nutrients, whether derived from the host diet (27) or from endogenous host sources (28), are critically important in shaping the structure of host-associated microbial communities. Recent evidence suggests that the immune system is also likely to be an important contributor to “inside-out” host control over microbiota composition.
Certain secreted antibacterial proteins produced by epithelial cells can shape the composition of intestinal microbial communities. α-defensins are small (2–3 kD) antibacterial peptides secreted by Paneth cells of the small intestinal epithelium. Analysis of the microbiota in mice that were either deficient in functional α-defensins, or that overexpressed human α-defensin-5, showed that although there was no impact on total numbers of colonizing bacteria, there were substantial α-defensin-dependent changes in community composition, with reciprocal differences observed in the two mouse strains (29).
An interesting question is how far secreted innate immune effectors “reach” into the luminal microbial consortia. For example, the impact of human α-defensin-5 on luminal community composition contrasts with the antibacterial lectin RegIIIγ, which limits penetration of bacteria to the epithelial surface but does not alter luminal communities (5). This suggests that some antimicrobial proteins, such as α-defensins, reach into the lumen to shape overall community composition, whereas others, such as RegIIIγ, have restricted effects on surface-associated bacteria and thus control microbiota location relative to host surface tissues. Questions remain as to exactly how α-defensin-5 controls luminal community composition, however. In one scenario, these small antimicrobial peptides diffuse through the mucus layer and directly act on bacteria that inhabit the lumen. Another possibility is that α-defensin-5 exerts its antibacterial activity on bacteria that are trapped in the outer reaches of the mucus layer, with those bacteria acting as reservoirs that seed luminal communities and thus dictate their composition. Answering these questions will require improved tools for fine-mapping microbiota composition and consortia from the surface of the intestine to the interior of the lumen.
The impact of the immune system on microbiota composition is also suggested by several immune deficiencies that alter microbial communities in ways that predispose to disease. For example, Garrett et al. studied mice that lack the transcription factor T-bet (encoded by Tbx21), which governs inflammatory responses in cells of both the innate and the adaptive immune system (30). When Tbx21−/− mice were crossed onto Rag2−/− mice, which lack adaptive immunity, the Tbx21−/−/Rag2−/− progeny developed ulcerative colitis in a microbiota-dependent manner (30). Remarkably, this colitis phenotype was transmissible to wild-type mice by adoptive transfer of the Tbx21−/−/Rag2−/− microbiota. This demonstrated that altered microbiota were sufficient to induce disease and can thus be considered “dysbiotic”. Similarly, mice lacking the bacterial flagellin receptor TLR5 exhibit a syndrome encompassing insulin resistance, hyperlipidemia, and increased fat deposition associated with alterations in microbiota composition (31). These metabolic changes are transferable to wild-type mice that acquire the Tlr5−/− gut microbiota. A third example of immune-driven dysbiosis is seen in mice deficient for epithelial cell expression of the inflammasome component NLRP6. These mice develop an altered microbiota with increased abundance of members of the Bacteroidetes phylum associated with increased intestinal inflammatory cell recruitment and susceptibility to chemically-induced colitis. Again, there is evidence that dysbiosis alone is sufficient to drive the intestinal inflammation, because wild-type mice that acquire the dysbiotic microbiota show similar immunopathology (32).
Together these findings suggest that the immune system affords mammalian hosts some control over the composition of their resident microbial communities. It is also clear that these communities can be perturbed by defects in the host immune system. This leads to the idea of the immune system as a form of ecosystem management that exerts critical control over microbiota composition, diversity and location (see Review by Costello et al. (33)). However, a number of questions remain. First, although it is apparent that the immune system shapes community composition at the species level, it is not yet clear whether the immune system shapes the genetics and physiology of individual microbial species. Second, how much does the immune system combine with gastric acid and intestinal motility to control the longitudinal distribution of microbial species in the gastrointestinal tract? Finally, it will be important to determine the extent to which the immune system also controls microbial community composition and location in other organ systems such as the respiratory tract, urogenital tract, and skin.
Looking outside-in: how microbiota shape immunity
The earliest comparisons of germ-free and colonized mice revealed a profound effect of microbial colonization on the formation of lymphoid tissues and subsequent immune system development. It was thus quickly apparent that the microbiota influence the immune system from “outside-in”. Recent studies have greatly amplified this understanding and have revealed some of the cellular and molecular mediators of these interactions.
The impact of the microbiota on lymphoid structure development and epithelial function
The tissues of the gastrointestinal tract are rich in myeloid and lymphoid cells, many of which reside in organized lymphoid tissues. It has long been appreciated that the gut microbiota have a critical role in the development of organized lymphoid structures and in the function of immune system cells. For example, isolated lymphoid follicles in the small intestine do not develop in germ-free mice, and such mice are also deficient in secretory IgA and CD8αβ intraepithelial lymphocytes. The specific microbial molecules endowed with this inductive function have not yet been described, however.
Sensing of commensal microbiota through the TLR-MyD88 signaling pathway triggers several responses that are critical for maintaining host-microbial homesostasis. The microbiota induce repair of damaged intestinal epithelium through a MyD88-dependent process that can be rescued in microbe-depleted animals by gavage with bacterial lipopolysaccharide (LPS). The innate signals, conveyed largely through myeloid cells, are required to enhance epithelial cell proliferation (34, 35). As discussed above, MyD88-dependent bacterial signals are also required for the induction of epithelial antimicrobial proteins such as RegIIIγ (5, 19). This expression can be induced by LPS (19, 20) or flagellin (36). The flagellin signals are relayed through TLR5 expressed by CD103+CD11b+ dendritic cells in the lamina propria, stimulating production of IL-23 that, in turn, promotes the expression of IL-22 by innate lymphoid cells (37). IL-22 then stimulates production of RegIIIγ, which is also secreted upon direct activation of MyD88 in epithelial cells (5, 20). This is one clear example of the importance of commensals in the induction of host innate responses, but it likely represents a tiny fraction of the multitude of effects of microbiota on the host immune system.
Microbiota shaping of T cell subsets
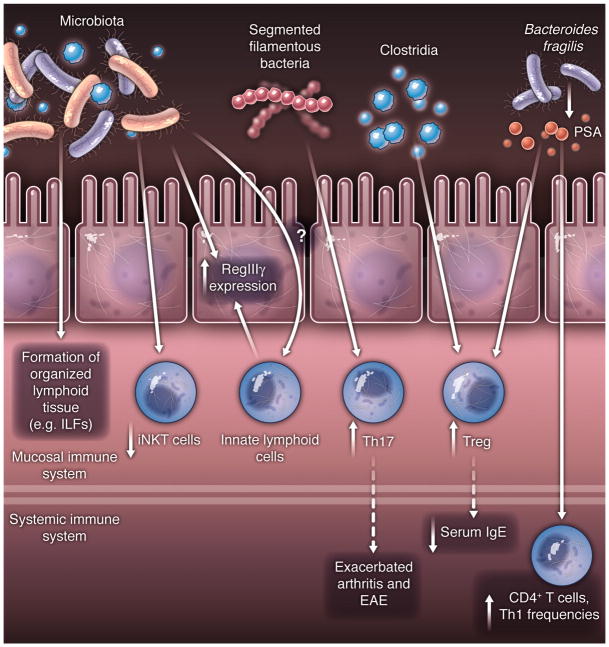
It has recently become evident that individual commensal species influence the makeup of lamina propria T lymphocyte subsets that have distinct effector functions. Homeostasis in the gut mucosa is maintained by a system of checks and balances between potentially pro-inflammatory cells, which include Th1 cells that produce interferon-γ, Th17 cells that produce IL-17a, IL-17f, and IL-22, diverse innate lymphoid cells with cytokine effector features resembling Th2 and Th17 cells, and anti-inflammatory Foxp3+ regulatory T cells (Tregs). Colonization of mice with segmented filamentous bacteria (SFB) results in accumulation of Th17 cells and, to a lesser extent, in an increase in Th1 cells (38, 39). SFB appear able to penetrate the mucus layer overlying the intestinal epithelial cells in the terminal ileum, and they interact closely with the epithelial cells, inducing host cell actin polymerization at the site of interaction and, presumably, signaling events that result in a Th17 polarizing environment within the lamina propria. There is little known about host cell signaling pathways initiated by SFB. It is possible that SFB influence epithelial gene expression, resulting, for example, in expression of antimicrobial proteins such as RegIIIγ and of molecules that participate in Th17 cell polarization. SFB may also act directly on cells of the immune system, either through interactions with myeloid cells that extend processes through the epithelium to the mucus layer or by production of metabolites that act on various receptors expressed by host cells.
Other bacteria have been shown to enhance the anti-inflammatory branches of the adaptive immune system by directing the differentiation of Tregs or by inducing IL-10 expression. For example, colonization of gnotobiotic mice with a complex cocktail of 46 mouse Clostridial strains, originally isolated from mouse feces and belonging mainly to cluster IV and XIVa of the Clostridium genus, results in the expansion of lamina propria and systemic Tregs. These have a phenotype characteristic of Tregs induced in the periphery in response to transforming growth factor (TGF)-β and retinoic acid (in contrast to thymic-derived natural (n) Tregs) (40), and many of these iTregs express IL-10. The exact Clostridial strains within the complex experimental mixture that drive this regulatory response remain to be defined. Furthermore, polysaccharide-A (PSA) of Bacteroides fragilis induces an IL-10 response in intestinal T cells, which prevents the expansion of Th17 cells and potential damage to the mucosal barrier (41). In contrast, mutant B. fragilis lacking PSA has a pro-inflammatory profile and fails to induce IL-10. Production of PSA by B. fragilis has been proposed to be instrumental for the bacterium’s success as a commensal.
Within the intestine, the balance of effector lymphoid cells and Treg cells can have a profound influence on how the mucosa responds to stresses that elicit damage. The relative roles of commensal-regulated T cells differ according to the models used to study inflammation. For example, in mice subjected to chemical or pathogen-induced damage to the mucosa, Th17 cells have a beneficial effect that promotes healing. In contrast, Th1 and Th17 cells, as well as IL-23-dependent innate lymphoid cells, promote colitis in models in which Treg cells are depleted. It is likely that inflammatory bowel diseases in humans can be similarly triggered by commensal-influenced imbalance of lymphoid cell subsets. This is supported by numerous observations, including the strong linkage of IL23R polymorphisms with Crohn’s disease, a serious condition with relapsing intestinal inflammation and a risk of malignancy, and the severe enterocolitis associated with IL10 and IL10R mutations (42, 43).
Microbiota effects on systemic immunity
The influence of commensal bacteria on the balance of T cell subsets is now known to extend well beyond the intestinal lamina propria. Homeostatic T cell proliferation itself is driven through the microbiota or their penetrant molecules (44). Systemic autoimmune diseases have long been suggested to have links to infections, but firm evidence for causality has been lacking. Recent studies in animal models, however, have reinforced the notion that commensal microbiota contribute to systemic autoimmune and allergic diseases at sites distal to the intestinal mucosa. Several mouse models for autoimmunity are dependent on colonization status. Thus, germ-free mice have marked attenuation of disease in models of arthritis and experimental autoimmune encephalomyelitis (EAE), as well as in various colitis models. In models of Th17 cell-dependent arthritis and EAE, monoassociation with SFB is sufficient to induce disease (42, 45, 46). In all of these models, induction of Th17 cells in the intestine has a profound influence on systemic disease. Exacerbation of arthritis and EAE is likely the consequence of an increase in the number of arthritogenic or encephalitogenic Th17 cells that traffic out of the lamina propria. The antigen specificity of such cells remains to be examined.
Induction of iTregs by the cluster IV and XIVa Clostridia also has a systemic effect on inflammatory processes. Colonization of germ-free mice with these bacteria results not only in attenuated disease after chemical damage of the gut epithelium, but also reduces the serum IgE response following immunization with antigen under conditions that favor a Th2 response (40). As with pathogenic Th17 cells, the antigen specificity of the commensal-induced iTregs that execute systemic anti-inflammatory functions is not yet known, although at least some of the Tregs in the gut have T cell receptors with specificity for distinct commensal bacteria (47).
Finally, B. fragilis PSA impacts the development of systemic T cell responses. Colonization of germ-free mice with PSA-producing B. fragilis results in higher numbers of circulating CD4+ T cells as compared to mice colonized with B. fragilis lacking PSA. PSA-producing B. fragilis also elicits higher Th1 cell frequencies in the circulation (48). Together, these findings show that commensal bacteria have a general impact on immunity that reaches well beyond mucosal tissues.
Microbiota influences on invariant T cells and innate lymphoid cells
A recent study extends the role of microbiota to the control of the function invariant natural killer T cells (iNKT cells), which bear an invariant T cell receptor specific for lipid antigens presented by the atypical class I molecule CD1d. Germ-free mice were found to have increased susceptibility to iNKT cell-mediated oxazolone-induced colitis and ovalbumin-induced asthma. Remarkably, this effect could be reversed only if mice were exposed to microbiota in the neonatal period. The regulation of iNKT cell expansion was ascribed to reduced expression of the chemokine CXCL16 in the presence of microbiota. Thus, signals elicited by commensals may repress systemic expression by epithelial cells of a chemokine that interacts with CCR6 that is selectively expressed by iNKT cells (49).
Innate lymphoid cells that produce either IL-17 or IL-22 are protective against damage in an innate model of colitis and during Citrobacter rodentium enteric infection (50, 51). The extent to which innate lymphoid cells are regulated by the microbiota is not yet clear (52–54), but cryptopatches, which are formed by a subset of innate lymphoid cells in the small intestine, differentiate into isolated lymphoid follicles only when commensals are present (55). Thus, it is likely that, even if innate lymphoid cell numbers are not influenced by commensals, their function may be subject to microbiota signals.
Microbiota can trigger inflammation in immunocompromised hosts
The commensal microbiota clearly have important effects on the normal development of immunity. However, commensal bacteria can also trigger inflammatory responses in immunodeficient hosts. For example, defective signaling through the phosphatase SHP-1 causes a microbiota-dependent autoinflammatory syndrome with lesions on the feet, salivary glands and lungs; such inflammation also occurs in mice without B- or T-lymphocytes (56, 57). There are a series of monogenic conditions of the NOD receptor family (58) considered to be ‘autoinflammatory’. One of the best characterized of these is in the NLRP3 inflammasome protein (59). Depending on the exact activating mutation involved, the clinical spectrum in humans encompasses urticaria triggered by the cold, episodic fevers occurring with unknown triggers, and neonatal onset multisystem inflammatory disease (60). Although the exact cause of these pathologies is not yet clear, these outcomes are consistent with studies in mice showing that NLRP3-deficiency can cause dysbiosis of the intestinal microbiota (61), as well as studies showing that TLR ligands can trigger pro-inflammatory IL-1β secretion in the presence of activating NLRP3 mutations (62, 63).
Another NOD family member, NOD2 (CARD15), a receptor for the muramyl dipeptide structural unit of bacterial peptidoglycan, was the first susceptibility gene identified for Crohn’s disease (64, 65). This reinforced early clinical observations of the benefits of surgically diverting the intestinal stream or treating with antibiotics, thus implicating intestinal microbes in the etiology (66, 67). More recent genetic data from genome-wide association studies (GWAS) of human inflammatory bowel disease have revealed a highly polygenic picture, with over 70 loci described for Crohn’s disease alone. These include modulators of the mucosal immune response, proteins functioning in the epithelial stress response, and the IL23R polymorphisms described above (68, 69). However, the sum total of the contributions of these loci to overall disease incidence leaves a considerable gap. It is clear that some cases can be explained by phenotypes from private mutations, such as those affecting IL-10 signaling (43), that are too infrequent to be detected by GWAS but that disrupt host-microbial mutualism in animal models (70).
Microbiota can protect against autoimmune disease
Type 1 diabetes (T1D) results from autoimmune damage to the insulin-secreting islets of Langerhans in the pancreas. This autoimmune condition is also shaped by the interactions between immunity and the microbiota, but unlike EAE and arthritis, where SFB drive autoimmunity, the microbiota can protect from T1D. The non-obese diabetic (NOD) mouse is a good model of T1D with some genetic predispositions similar to those in humans and defined CD4 and CD8 diabetogenic T cell populations (71, 72). The incidence of T1D in an isogenic NOD colony is dependent on the housing conditions, as both the presence of pathogens and microbiota diversity are determining factors (73, 74). In congenic NOD mice with a MyD88 adaptor deficiency (disrupting most TLR signaling), germ-free animals have the same frequency of diabetes as the parent NOD strain. In contrast, when colonized with a microbiota, NOD/Myd88−/− mice, but not NOD animals, are largely protected from diabetes onset (75). MyD88-deficiency has complex effects on host-microbial mutualism, including increased access of intestinal microbes to the epithelial surface, increased penetration of commensals to the systemic immune system, and reduced costimulation in induction of adaptive immunity (5, 26, 36, 76). It is therefore not yet clear whether this is purely a failure of accumulation of autoimmune T cells (77), or whether there is also immune deviation arising from commensal barrage that dilutes the frequency of diabetogenic lymphocytes (26). Either way, it may offer insight into why better hygiene associates with a higher frequency of autoimmune and allergic disease in human populations (78).
Microbiota-immune system interactions and metabolic health
As described in the Review by Nicholson et al. (79), our bodies are bathed with microbial molecules, generating a host-microbial molecular embrace (12). Recent studies have shown that the immune response to these microbial molecules profoundly impacts the metabolic health of mammalian hosts.
Metabolic syndrome is a constellation of abnormalities, including insulin resistance, obesity, dyslipidemia and hypertension, that predisposes affected individuals to cardiovascular disease and diabetes (80). It may seem counterintuitive to blame metabolic syndrome on our intestinal microbiota rather than our modern dissipations of stress, eating too much and taking little exercise, but there is now good evidence that how the immune system responds to the microbiota makes us vulnerable to these diseases. These effects appear to be independent of the microbial contributions to energy harvest. In mice, dysfunctional sensing of microbial molecular patterns can cause low-grade intestinal inflammation. As discussed above, mice lacking the bacterial flagellin sensor exhibit features of metabolic syndrome that are associated with changes in microbiota composition and can be acquired by wild-type mice through microbiota transfer (31). Similarly, mice lacking the inflammosome components NLRP3 or NLRP6 exhibit a low grade intestinal enteropathy dependent on overgrowth of Prevotellaceae and Porphyromonadacae members of the Bacteroidetes (61). This results in TLR agonist influx into the hepatic portal vein, which supplies blood from the gut to the liver. Consequently, TLR4 and 9 signaling increases TNFα expression. In mice and humans, TNFα promotes insulin resistance and accumulation of fat in hepatocytes. The metabolic changes leading to fatty liver are transmissible to wild-type mice upon microbiota transfer (61), although the microbiota alterations may not persist in the absence of innate immune deficiency. Together, these examples show that innate immune system defects can result in dysbiosis of the intestinal microbiota with downstream metabolic consequences for the host.
Future challenges
The challenges for understanding host-microbial immune mutualism are intimately connected with human health. The first question is how much is immune dysfunction a cause or consequence of disease-associated alterations in the microbiota? As outlined above, there is good evidence from animal models that alterations in immunity can cause dysbiosis and conversely, that certain microbial species can trigger immunopathology in the face of immunodeficiency. The idea that immunodeficiency is relatively common among human populations, and often presents in adults with a limited range of opportunistic infections, may extend to weak phenotypes of immune dysfunction exerting long term inflammatory, metabolic or autoimmune effects through microbial dysbiosis. We need a better understanding of the mechanisms whereby altered immunity shapes microbiota composition and determines which microbes are present to embrace us with their metabolites. Alternatively, given the pervasiveness of this metabolic embrace, we need a better understanding of how the metabolic handshake shapes the host immune system in health and disease.
The next issue is how to reproducibly and effectively advance the science of host-microbial mutualism. Defined animal models are clearly essential for determining the relative contributions of host genetics and microbiota composition. Excellent resources are available to study the in vivo effects of host genetic changes, especially in mice, and isogenic mouse models are widely shared between investigators. Such standardization is not yet available for the microbiotas within these isogenic mouse strains. Animals in the vivaria of different institutions and sourced from different commercial breeders have variable microbiota compositions that have likely contributed to conflicting reports with various models of autoimmune disease (39, 81). We therefore urgently need different standardized microbiotas in isobiotic mice that can be shared between institutions for reproducible experimentation. Otherwise we may face confusion from variable results attributable to differences between experimental microbiotas. Creating animals that are realistic models of human disease is an even bigger challenge, requiring us to reduce the species differences through ‘humanized’ immune systems combined with a ‘humanized’ gnotobiotic microbiota. The latter poses additional potential pitfalls, as many microbial species are likely adapted to specific hosts.
Many essential details of the immune response to the microbiota remain to be discovered. For example, how commensal microorganisms influence the differentiation of immune cells, such as Th17 and Treg cells, remains to be unraveled. What microbial molecules drive differentiation of these cells? What is the molecular basis for the differential ability of distinct commensal species to trigger these developmental pathways? Why do some commensal species exert powerful effects on immune cell differentiation whereas other species are essentially ignored, and is there a general mechanism of mucosal adaptation or is it tuned to the microbial species present? Can we explain the hygiene effect of allergy and autoimmunity in terms of the T- and B-cell repertoires being shaped by our microbiota?
Most of our current understanding of microbiota-immune system interactions is derived from studies of bacteria in the gastrointestinal tract. However, other body surfaces, such as the skin, upper respiratory tract, and urogenital tract, harbor rich communities of indigenous microorganisms and thus are also likely to be sites where such interactions influence host health. Do resident microbes shape immune cell differentiation at these sites as they do in the intestine and is there crosstalk among the immune effectors at different sites? At the same time, gut microbiota also include viruses, bacteriophage (82), and eukaryotic organisms such as fungi. These other elements of the microbial community undoubtedly also have highly coevolved relationships with the mammalian immune system and will provide fascinating targets for further exploration of microbe-immune system relationships.
Finally, little is known about the influences of the microbiota in the critical window of immune development in the neonatal period, despite the fact that breastfeeding is one of the most effective known health-promoting measures. Immediate maternal immune effects through lactation are well defined; the longer-term consequences for both the host microbiota and the immune system are less clear. It will be important to determine how we simultaneously transmit our microbiota to our offspring and shape the education of their immune system through lactational effects on health.
Perspectives
It is now clear that the immune system plays a central role in shaping the composition of the microbiota as well as its proximity to host tissues. At the same time, resident microbes provide signals that foster normal immune system development and influence the ensuing immune responses. Disruption of these complex and dynamic interactions can have profound consequences for host health. However, there are still major gaps in our understanding of how the immune system regulates the microbiota, and of how the microbiota shape host immunity. The questions that remain are challenging and will require the innovation of new tools and approaches. Ultimately, these efforts should lead to deeper insight into host-microbial relationships and provide exciting new opportunities to improve human health.
Figure 2. Looking outside-in: how microbiota shape host immunity.
Timelines of development of lymphopoietic tissues, secondary lymphoid structures and leukocyte populations. Events dependent on colonisation with a microbiota are shown in red text and with red arrows. Immune system development and maintenance that is independent of microbiota colonisation is shown in black. En = embryo age (days), birth = E19-21, weaning at approximately postnatal day 20, CSR=class switch recombination.
Contributor Information
Lora V. Hooper, Email: lora.hooper@utsouthwestern.edu.
Dan R. Littman, Email: dan.littman@med.nyu.edu.
Andrew J. Macpherson, Email: andrew.macpherson@insel.ch.
References
- 1.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Nat Rev Micro. 2008;6:776. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McFall-Ngai M. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 3.Macpherson AJ, Geuking MB, Kirundi J, Collins S, McCoy KD. Encyclopedia of Microbiology. Vol. 156. Elsevier; 2009. pp. 237–246. [Google Scholar]
- 4.Geuking MB, et al. Immunity. 2011;34 doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 5.Vaishnava S, et al. Science. 2011;334:255. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Infect Immun. 1999;67:1992. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumberg RS, Powrie F. Sci Transl Med. doi: 10.1126/scitranslmed.3004184. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodman AL, et al. Proc Natl Acad Sci USA. 2011;108:6252. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Legrand N, et al. Cell Host Microbe. 2009;6:5. [Google Scholar]
- 10.Hapfelmeier S, et al. Science. 2010;328:1705. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper LV, et al. Science. 2001;291:881. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 12.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Trends Microbiol. 2011;19:349. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Nat Immunol. 2003;4:269. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 14.Peterson DA, McNulty NP, Guruge JL, Gordon JI. Cell Host Microbe. 2007;2:328. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Stappenbeck TS, Hooper LV, Gordon JI. Proc Natl Acad Sci USA. 2002;99:15451. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johansson MEV, et al. Proc Natl Acad Sci USA. 2008;105:15064. [Google Scholar]
- 17.Johansson MEV, Larsson JMH, Hansson GC. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Science. 2006;313:1126. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brandl K, Plitas G, Schnabl B, DeMatteo RP, Pamer EG. J Exp Med. 2007;204:1891. doi: 10.1084/jem.20070563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Proc Natl Acad Sci USA. 2008;105:20858. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Macpherson AJ, UHR T. Science. 2004;303:1662. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 22.Macpherson AJ, et al. Science. 2000;288:2222. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 23.Kelsall B. Mucosal Immunol. 2008;1:460. doi: 10.1038/mi.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spits H, Di Santo JP. Nat Immunol. 2011;12:21. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- 25.Sonnenberg GF, et al. Science. this issue. [Google Scholar]
- 26.Slack E, et al. Science. 2009;325:617. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnenburg ED, et al. Cell. 2010;141:1241. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonnenburg JL, et al. Science. 2005;307:1955. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 29.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Nature. 2003;422:522. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 30.Garrett WS, et al. Cell. 2007;131:33. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijay-Kumar M, et al. Science. 2010;328:228. [Google Scholar]
- 32.Elinav E, et al. Cell. 2011;145:745. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costello EK, Relman DA. Science. this issue. [Google Scholar]
- 34.Rakoff-Nahoum S, Medzhitov R. Science. 2007;317:124. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 35.Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Proc Natl Acad Sci USA. 2005;102:99. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kinnebrew MA, et al. J Infect Dis. 2010;201:534. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinnebrew MA, et al. Immunity. 2012;36:276. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaboriau-Routhiau V, et al. Immunity. 2009;31:677. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 39.Ivanov II, et al. Cell. 2009;139:485. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Atarashi K, et al. Science. 2011;331:337. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Round JL, et al. Science. 2011;332:974. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abraham C, Cho JH. Annu Rev Med. 2009;60:97. doi: 10.1146/annurev.med.60.051407.123757. [DOI] [PubMed] [Google Scholar]
- 43.Glocker EO, et al. N Engl J Med. 2009;361:2033. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kieper WC, et al. J Immunol. 2005;174:3158. doi: 10.4049/jimmunol.174.6.3158. [DOI] [PubMed] [Google Scholar]
- 45.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4615. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu HJ, et al. Immunity. 2010;32:815. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lathrop SK, et al. Nature. 2011;478:250. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. Cell. 2005;122:107. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 49.Olszak T, et al. Science. 2012;336:489. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buonocore S, et al. Nature. 2010;464:1371. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vonarbourg C, et al. Immunity. 2010;33:736. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satoh-Takayama N, et al. Immunity. 2008;29:958. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Sawa S, et al. Science. 2010;330:665. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 55.Lorenz RG, Chaplin DD, McDonald KG, McDonough JS, Newberry RD. J Immunol. 2003;170:5475. doi: 10.4049/jimmunol.170.11.5475. [DOI] [PubMed] [Google Scholar]
- 56.Croker BA, et al. Proc Natl Acad Sci USA. 2008;105:15028. doi: 10.1073/pnas.0806619105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu CC, et al. J Exp Med. 1996;183:371. doi: 10.1084/jem.183.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inohara N, Nuñez G. Nat Rev Immunol. 2003;3:371. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 59.Brydges SD, et al. Immunity. 2009;30:875. doi: 10.1016/j.immuni.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Henderson C, Goldbach-Mansky R. Curr Opin Rheumatol. 2010;22:567. doi: 10.1097/BOR.0b013e32833ceff4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henao-Mejia J, et al. Nature. 2012;482:179. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tschopp J, Schroder K. Nat Rev Immunol. 2010;10:210. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 63.Meng G, Zhang F, Fuss I, Kitani A, Strober W. Immunity. 2009;30:860. doi: 10.1016/j.immuni.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ogura Y, et al. Nature. 2001;411:603. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 65.Hugot JP, et al. Nature. 2001;411:599. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 66.Rutgeerts P, et al. Lancet. 1991;338:771. doi: 10.1016/0140-6736(91)90663-a. [DOI] [PubMed] [Google Scholar]
- 67.Cummings JH, Kong SC. Novartis Found Symp. 263:99–111. discussion 111–4, 211 8 2004. [PubMed] [Google Scholar]
- 68.Franke A, et al. Nat Genet. 2010;42:1118. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGovern DPB, et al. Nat Genet. 2010;42:332. doi: 10.1038/ng.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Cell. 1993;75:263. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 71.Verdaguer J, et al. J Exp Med. 1997;186:1663. doi: 10.1084/jem.186.10.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmidt D, Amrani A, Verdaguer J, Bou S, Santamaria P. J Immunol. 1999;162:4627. [PubMed] [Google Scholar]
- 73.Ohsugi T, Kurosawa T. Lab Anim Sci. 1994;44:386. [PubMed] [Google Scholar]
- 74.Leiter E. In: The role of microorganisms in non-infectious disease. de Vries RRP, Cohen IR, van Rood JJ, editors. Springer Verlag; Berlin: 1990. pp. 39–55. [Google Scholar]
- 75.Wen L, et al. Nature. 2008;455:1109. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carvalho FA, Aitken JD, Vijay-Kumar M, Gewirtz AT. Annu Rev Physiol. 2012;74:177. doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Y, et al. J Immunol. 2010;184:5645. doi: 10.4049/jimmunol.0901814. [DOI] [PubMed] [Google Scholar]
- 78.Bach JF. N Engl J Med. 2002;347:911. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 79.Nicholson JK, et al. Science. this issue. [Google Scholar]
- 80.Moller DE, Kaufman KD. Annu Rev Med. 2005;56:45. doi: 10.1146/annurev.med.56.082103.104751. [DOI] [PubMed] [Google Scholar]
- 81.Ivanov II, et al. Cell Host Microbe. 2008;4:337. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reyes A, et al. Nature. 2010;466:334. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]