Abstract
A recent history of failed clinical trials suggests that waiting until even the early stages of onset of Alzheimer’s disease may be too late for effective treatment, pointing to the importance of early intervention in young people. Early intervention will require markers of Alzheimer’s risk that track with genotype but are capable of responding to treatment. Here, we sought to identify a functional MRI signature of combined Alzheimer’s risk imparted by two genetic risk factors. We used a task of executive attention during fMRI in participants genotyped for two Alzheimer’s risk alleles: APOE-ε4 and CLU-C. Executive attention is a sensitive indicator of the progression of Alzheimer’s even in the early stages of mild cognitive impairment, but has not yet been investigated as a marker of Alzheimer’s risk in young adults. Functional MRI revealed that APOE-ε4 and CLU-C had an additive effect on brain activity such that increased combined genetic risk was associated with decreased brain activity during executive attention, including in the medial temporal lobe, a brain area affected early in Alzheimer’s pathogenesis.
1. Introduction
Developing the empirical groundwork for preventive interventions against Alzheimer’s disease (AD) is a priority because the recent history of failed clinical trials suggests that waiting until even the early stages of frank disease onset may be too late for effective treatment (Zahs & Ashe, 2010). Identifying neurocognitive markers of genetic risk for AD in young people is an important component of this groundwork (Goldberg & Weinberger, 2004; Green, Fugelsang, Kraemer, & Dunbar, 2008; Meyer-Lindenberg & Weinberger, 2006; Tan, Callicott, & Weinberger, 2008). Genetic association studies of AD have repeatedly confirmed that the ε4 allele of the apolipoprotein E (APOE) gene is by far the strongest common genetic risk factor for late onset AD (e.g., (Bertram, McQueen, Mullin, Blacker, & Tanzi, 2007; Harold, Abraham, Hollingworth, Sims, Gerrish, Hamshere, Pahwa, Moskvina, Dowzell, Williams, Jones, Thomas, Stretton, Morgan, Lovestone, Powell, Proitsi, Lupton, Brayne, Rubinsztein, Gill, Lawlor, Lynch, Morgan, Brown, Passmore, Craig, McGuinness, Todd, Holmes, Mann, Smith, Love, Kehoe, Hardy, Mead, Fox, Rossor, Collinge, Maier, Jessen, Schurmann, van den Bussche, et al., 2009; Lambert, et al., 2009). Inheritance of one copy of APOE-ε4 markedly increases the risk of AD and decreases the average age of onset (Farrer, et al., 1997). The mechanism by which APOE affects AD risk is still unclear, although mouse studies show that in normal brain, APOE-ε4 is associated with alterations in synaptic components (Dumanis, DiBattista, Miessau, Moussa, & Rebeck, 2013; Dumanis, et al., 2009) and activity (Hunter, et al., 2012). These effects may lead to earlier amyloid deposition observed in mouse models of AD (Kim, Basak, & Holtzman, 2009). Studies in humans suggest that, prior to clinical symptoms, APOE genotype affects medial temporal lobe (MTL) activity (S. Bookheimer & Burggren, 2009; S. Y. Bookheimer, et al., 2000) and distributed network connectivity (Pena-Gomez, et al., 2012), and that APOE-ε4 increases the risk of converting from mild cognitive impairment (MCI) to AD (Fei & Jianhua, 2012). These data suggest that APOE-ε4 is associated with an increased susceptibility of the hippocampus and surrounding medial temporal regions to damage that occurs early in the development of AD (Liu, Kanekiyo, Xu, & Bu, 2013).
While alleles of APOE have the strongest known effects on genetic risk for AD, genome-wide association studies have identified polymorphisms in other genes that have small but significant effects on the risk of AD (Harold, Abraham, Hollingworth, Sims, Gerrish, Hamshere, Pahwa, Moskvina, Dowzell, Williams, Jones, Thomas, Stretton, Morgan, Lovestone, Powell, Proitsi, Lupton, Brayne, Rubinsztein, Gill, Lawlor, Lynch, Morgan, Brown, Passmore, Craig, McGuinness, Todd, Holmes, Mann, Smith, Love, Kehoe, Hardy, Mead, Fox, Rossor, Collinge, Maier, Jessen, Schurmann, van den Bussche, et al., 2009; Lambert, et al., 2009). Together, these genes identify several potential pathways that could affect the risk of AD, including neuroinflammation, cholesterol homeostasis, and endocytic regulation (Bertram, et al., 2007). Individual genes may affect common pathways to AD pathogenesis, or entirely independent pathways. One of these genetic risk factors is CLU, the gene for clusterin (or apolipoprotein J). Extant evidence indicates that the CLU-C polymorphism is associated with a slightly higher risk of Alzheimer’s disease. Large-scale meta analyses have replicated that the CLU-C allele is associated with elevated AD risk (Odds Ratio = 1.22; p = 8.6 × 10−5) (Carrasquillo, et al., 2010; Harold, Abraham, Hollingworth, Sims, Gerrish, Hamshere, Pahwa, Moskvina, Dowzell, Williams, Jones, Thomas, Stretton, Morgan, Lovestone, Powell, Proitsi, Lupton, Brayne, Rubinsztein, Gill, Lawlor, Lynch, Morgan, Brown, Passmore, Craig, McGuinness, Todd, Holmes, Mann, Smith, Love, Kehoe, Hardy, Mead, Fox, Rossor, Collinge, Maier, Jessen, Schurmann, Heun, et al., 2009)
To the authors’ knowledge, there are no reports demonstrating an interaction effect between APOE and CLU to increase Alzheimer’s disease risk. However, the apoE and apoJ proteins share a number of important characteristics: they are among very few proteins associated with brain lipoproteins (Elliott, Weickert, & Garner, 2010; Koch, et al., 2001); they interact with a shared set of cell surface receptors (Kounnas, et al., 1995); they promote neurite outgrowth (Kang, et al., 2005; Nathan, et al., 1994); and elimination of apoE or apoJ in an AD mouse model caused similar effects on accumulation of Aβ, a component of amyloid plaques associated with AD neuropathology (DeMattos, et al., 2004). Due to these biological connections, APOE and CLU polymorphisms may affect similar pathways leading to the development of AD (Wu, Yu, Li, & Tan, 2012).
The neural effects of CLU genotype in young humans have not yet been well characterized. Extant studies indicate that the risk-associated CLU-C allele alters structural and functional connectivity as well as memory-related neuronal activity (Braskie, et al., 2011; Erk, et al., 2011; Lancaster, et al., 2011) in ways that may ultimately contribute to disordered blood flow (Thambisetty, et al., 2013) and atrophy (Thambisetty, et al., 2012). To our knowledge, no prior research has investigated combined neural effects of CLU and APOE in young people.
The most conspicuous neurocognitive deficit associated with AD is memory impairment, but the disease also has dramatic effects on a set of complex thinking skills referred to as executive function (Kane & Engle, 2003; Silveri, Reali, Jenner, & Puopolo, 2007). One of these skills, executive attention, or the ability to maintain appropriate focus despite the presence of salient but irrelevant stimuli, appears to be a sensitive indicator of the progression of AD even in the early stages of mild cognitive impairment, yielding effects on both behavioral (Saunders & Summers, 2011; Wylie, Ridderinkhof, Eckerle, & Manning, 2007) and brain-based (Neufang, et al., 2013; Schroeter, et al., 2012) measures. Executive attention relies most strongly on prefrontal and cingulate regions associated with top-down response inhibition and selection (Kane & Engle, 2003), though MTL has also been implicated to a lesser extent (Banich, et al., 2009; Casey, Thomas, Davidson, Kunz, & Franzen, 2002; Epstein, Harris, Stanley, & Kanwisher, 1999; Preston & Gabrieli, 2008; Ryan, Lin, Ketcham, & Nadel, 2010). The few brain-imaging studies carried out thus far on the effects of the APOE-ε4 allele in young healthy individuals have focused on memory tasks and have not yet examined executive attention (Borghesani, et al., 2008; Bunce, Anstey, Burns, Christensen, & Easteal, 2011; Dennis, et al., 2010; Filippini, et al., 2009; Mondadori, et al., 2007; Scarmeas, et al., 2005).
Here, we investigated a task of executive attention as a brain-imaging marker for AD risk in a cohort of healthy young adults, testing for combined effects of APOE and CLU genotype. Based on their likely involvement in shared molecular biological pathways, we hypothesized that possession of the CLU-C risk allele would exacerbate neural effects of the APOE-ε4 allele. Because of the early involvement of MTL in AD pathogenesis, we focused on combined genetic risk effects in this region in our young cohort.
2. Methods
2.1 Participants
Participants were selected from a superset of 160 healthy, right-handed native English speakers (131 male, mean age = 23.7 years) who were undergraduate students and community members with no history of mental illness, brain injury, or psychoactive medication, providing informed consent for fMRI. Several exclusions were made to maximally match groups on genetic variables (except for those we sought to directly contrast), and to make group sizes proximate. Table 1 displays demographic data for the individuals included in our analyses. Carriers of the APOE-ε2 allele were excluded due to the potential confound of protective effects conferred by the ε2 allele (Farrer, et al., 1997) (Bertram, et al., 2007). APOE-ε4ε4 (N = 4) were excluded because the impact of putative exacerbating effect of ε4 homozygosity (Bertram, et al., 2007; Farrer, et al., 1997) could not be meaningfully assessed given the small group size. Because there were far more APOE-ε3ε3/CLU-C individuals (N = 83) than APOE-ε3ε3/CLU-NonC individuals (N = 16) or APOE-ε4ε3/CLU-C individuals (N = 23), sixteen APOE-ε3ε3 were randomly selected for analysis by taking the first sixteen in a randomly assigned order using the “RAND” function in Microsoft Excel (2011). This enabled proximate group sizes for contrasts between combined APOE and CLU genotype groups and between all of the included ε4-positive (N = 26) vs. ε4-negative (N = 32) individuals. The randomly selected group of sixteen APOE-ε3ε3/CLU-C individuals did not differ from the full group of eighty-three APOE-ε3ε3/CLU-C individuals with respect to IQ, MSIT performance, or MSIT executive attention-related activity within an a priori region of interest in bilateral medial temporal lobe (all p > .1). In the two groups of CLU-C-positive individuals (i.e., APOE-ε3ε3/CLU-C and APOE-ε4ε3/CLU-C), the proportion of individuals who were heterozygous (CC) vs. homozygous (CT) did not differ, c2(1, N = 43) = 0.26, p = .61). The four genotype groups selected for analysis did not differ on age (F(3, 54) = 1.14, p = .34; all between-group p > .1) or IQ (F(3, 54) = .90. p = .45; all between-group p > .1). Additionally, selected participants did not differ by Age or IQ when grouped as APOE-ε4-positive vs. APOE-ε4-negative or as CLU-C-positive vs. CLU-C-negative (all p > .25). Our study was composed predominantly of men. The effects of APOE genotype on AD risk appear to be similar for men and women in the broader population (Farrer, et al., 1997; Ghebremedhin, et al., 2001). However, reports of sex differences in the effects of APOE on brain biomarkers (Damoiseaux, et al., 2012; Lehmann, et al., 2006) motivated a test of the effect of sex within our sample, which we report below. Our study was predominantly Caucasian. Only 4 of the 55 participants in the selected genotype groups were not Caucasian, with both the ε4-positive and ε4-negative groups including one participant identifying as Black and one identifying as Asian. Thus, it is highly unlikely that our data are substantially affected by potential confounds related to ethnic stratification.
Table 1.
Demographic Information
| Genotype | Sex | Age | IQ |
|---|---|---|---|
| APOE-E3 E3/CLU-NonC | M: 15; F: 1 | 26 ± 6 | 127.56 ± 8.21 |
| APOE- E3 E3/CLU-C (CC: 4; CT: 12) | M: 15; F: 1 | 25.25 ± 5.87 | 121.47 ± 13.97 |
| APOE- E3 E3/CLU-NonC | M: 3; F: 0 | 20 ± 1.73 | 119.5 ± 11.32 |
| APOE- E4 E3/CLU-C (CC: 5; CT: 18) | M: 19; F: 4 | 24 ± 5.49 | 123.22 ± 12.30 |
Values for age and IQ represent the mean ± standard deviation.
2.2 Genotyping
Human APOE genotypes were determined using TaqMan® SNP Genotyping Assays per manufacturer’s protocol (Applied Biosystems, Inc.). Briefly, extracted DNA samples were amplified using the standard Allelic Discrimination Protocol on an ABI 7900HT system and SDS software using either the rs429358 (codon 112) or rs7412 (codon 158) primer/probe sets for APOE and rs11136000 primer/probe set for CLU. For APOE genotyping, human DNA of known APOE genotypes (ε2ε2, ε2ε3, ε2ε4, ε3ε3, ε4ε3, ε4ε4) obtained from the National Cell Repository for Alzheimer’s Disease (NCRAD, Indiana University) were run on reaction plates as standards. Both APOE and CLU genotype runs included negative controls lacking DNA template. We obtained 100% correct calls using the APOE standards for this gene and for both genes we obtained ≥95% quality value on all calls and 100% recall on ~20% of samples that were rerun for quality control purposes.
2.3 Experimental Procedure
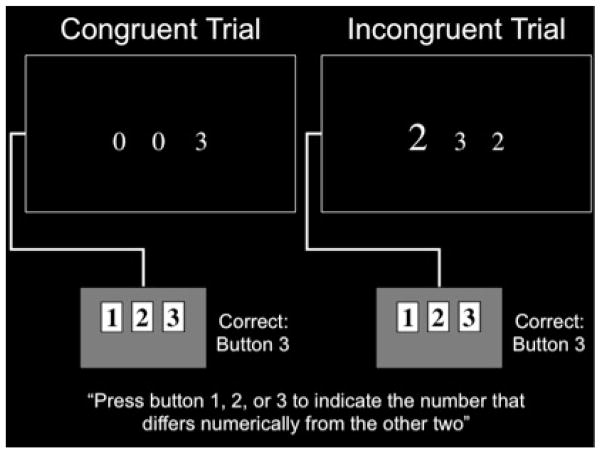
All experimental procedures occurred within a single scanning session. Participants performed the Multi-Source Interference Task (MSIT; Bush and Shin 2006), illustrated in Figure 1, during event-related fMRI. On each MSIT trial, participants pressed one of three buttons to indicate which of three concurrently presented digits differed numerically from the other two. There were two trial types, “Incongruent” and “Congruent.” Incongruent trials included distractor number choices that were distinctive due to size but not numerically different from each other and therefore not the correct choice. These numbers were salient because they were potentially valid choices (1, 2, or 3), and because their distinct sizes drew attention. Thus, Incongruent trials elicited attentional conflict, requiring the use of executive attention to overcome distractions in order to focus on the correct answer. Congruent trials did not involve attentional conflict. Specifically, all numbers other than the correct choice were the same size and were always 0s, designed to be minimally salient. However, all characteristics of the task appearance were the same for Incongruent and Congruent trials, and the instructions did not vary. Thus, the Incongruent > Congruent contrast provides an index of executive attention in MSIT with other task demands held constant. MSIT was selected because it is a well-validated measure of executive attention that demonstrates reliable recruitment across individuals (Bush & Shin, 2006), and has been successfully used to measure the effect of genotype on brain function (Green, Kraemer, Deyoung, Fossella, & Gray, 2012). We have reported portions of data from this MSIT dataset previously (Green, et al., 2012; Shehzad, DeYoung, Kang, Grigorenko, & Gray, 2012) for separate investigations, fully outside the context of any AD risk factor.
Figure 1.
Examples of MSIT Congruent and Incongruent trials participants performed during event-related fMRI. A between-condition contrast of Incongruent > Congruent trials provided a behavioral and brain-imaging index of executive attention with other task demands held constant.
2.4 MRI Imaging
Scanning was performed on a 3-T Trio System (Siemens, Erlangen, Germany) to collect whole-brain T2*-weighted blood oxygenation level-dependent (BOLD) functional images (asymmetric spin-echo echo-planar sequence; whole-brain repetition time, TR = 2,000 ms; echo time = 25ms; field of view = 256mm; flip angle = 80°; matrix = 64 × 64; axial slices 4 mm thick). The functional run comprised 183 sequential whole-brain volumes (32 contiguous slices). Ninety-six task trials were presented for 1750 ms each during the functional run, with jittered intertrial intervals (in which a centralized fixation cross was presented), across a range from 250 to 4,250 ms in steps of 2,000 ms (1 TR). The scanning run began with an unanalyzed fixation period equal to 3 TRs, which allowed the scanner to reach steady state. A whole-brain T1-weighted magnetization-prepared rapid gradient-echo (MPRAGE) image was acquired for each subject (FOV = 256 mm; 256 × 256 matrix; 1 × 1 mm in-plane resolution, 1.25 mm thick axial slices, 1 average).
2.5 Data Analysis
fMRI data processing was carried out using fMRI Expert Analysis Tool, Version 5.98, part of FMRIB’s Software Library (www.fmrib.ox.ac.uk/fsl). The following pre-statistics processing were applied: motion correction using MCFLIRT; spatial smoothing using a Gaussian kernel of 5mm FWHM; grand-mean intensity normalization of the entire 4D dataset by a single multiplicative factor; high-pass temporal filtering (Gaussian-weighted least-squares straight line fitting, with sigma = 50.0s). ICA-based exploratory data analysis was carried out using MELODIC (Beckmann & Smith, 2004), in order to investigate the possible presence of unexpected artifacts or activation. Registration to high resolution structural and, subsequently, standard space images was carried out using FLIRT. Higher-level analyses were carried out using FMRIB’s Local Analysis of Mixed Effects. Z (Gaussianized T/F) statistic images were thresholded using clusters determined by a minimum Z > 2.3 and a minimum significance threshold of p = .001.
For structural MPRAGE data, Voxel-Based Morphometry (VBM) was performed using the DARTEL toolbox for SPM (Ashburner, 2007; Ashburner & Friston, 2000). All default settings were used except were noted otherwise. Images were aligned into AC/PC orientation and then segmented into grey matter, white matter, and cerebrospinal fluid (CSF). All participants grey and white matter images were then simultaneously registered together to create a study specific template to reduce between-participant variability. The template was then used to normalize all images into the standard MNI space using the DARTEL Normalize to MNI Space program, utilizing the “preserve amount” option to retain the volumetric data of the original images, and the images were smoothed using a Gaussian kernel with 8 mm full-width half maximum (FWHM). Medial temporal lobe grey matter volume was extracted for a combined hippocampus and parahippocampus region via automated anatomical labeling in the Wake Forest University Pick Atlas (http://www.fmri.wfubmc.edu/download.htm).
A planned group-level Incongruent > Congruent within-subjects contrast was performed to identify brain regions recruited for executive attention in MSIT. The sample of participants was then divided based on combined APOE and CLU genotype and the results of the Incongruent > Congruent contrast were used for subsequent between-group contrast and parametric analyses.
An a priori region of interest (ROI) in MTL was defined as an anatomical probabilistic atlas-derived mask of bilateral parahippocampal gyrus and hippocampus from the Harvard-Oxford Cortical and Subcortical Atlases. This ROI was selected because MTL has been the region most consistently implicated by extant brain imaging studies of AD risk in healthy people (e.g., (S. Bokheimer & Burggren, 2009; S. Y. Bookheimer, et al., 2000; Dennis, et al., 2010; Filippini, et al., 2009; Mondadori, et al., 2007; Scarmeas, et al., 2005), and is also the region first affected by AD pathogenesis (Braak, Thal, Ghebremedhin, & Del Tredici, 2011; Gomez-Isla, et al., 1996; Hyman, Van Hoesen, Damasio, & Barnes, 1984). BOLD signal change averaged across voxels within this ROI was extracted in order to investigate genotype-related differences.
3. Results
3.1 Genotyping
We isolated DNA from saliva and genotyped two genetic risk factors for late-onset AD, APOE and CLU (the APOE-ε4 and CLU-C alleles are associated with an increased risk of AD) in a superset cohort of 160 individuals. The distribution of the six APOE genotypes was typical of numerous populations in the United States, with allele frequencies of 0.10 (ε2), 0.76 (ε3), and 0.14 (ε4) (Myers, et al., 1996). We also measured the distribution of the two common alleles of CLU. The allele frequencies were 0.58 (CLU-C) and 0.42 (CLU-T).
3.2 Behavioral
Genotype groups did not differ on MSIT performance. Response times for Congruent trials were, APOE-ε3ε3/CLU-NonC: M = 676.48 ms, SD = 96.74 ms; APOE-ε3ε3/CLU-C: M = 643.84 ms, SD = 79.25 ms; APOE-ε4ε3/CLU-NonC: M = 727.56 ms, SD = 172.86 ms; and APOE-ε4ε3/CLU-C: M = 613.82 ms, SD = 99.36 ms. Response times for Incongruent trials were, APOE-ε3ε3/CLU-NonC: M = 1041.65 ms, SD = 175.68 ms; APOE-ε3ε3/CLU-C: M = 998.43 ms, SD = 171.82 ms; APOE-ε4ε3/CLU-NonC: M = 1044.84 ms, SD = 223.42 ms; and APOE-ε4ε3/CLU-C: M = 927.21 ms, SD = 169.31 ms. The difference in response time for Incongruent > Congruent trials as a proportion of total response time represents the executive attentional component of MSIT and is the primary measure of MSIT performance (Bush & Shin, 2006). This measure did not show an overall effect of genotype, F(3,54) = .421, p = .74, nor did any group differ from any other group on this performance measure (all between-group p > .3). Accuracy for MSIT is typically near ceiling, as it was in this cohort (all groups > 98% accuracy), and did not differ between genotype groups, F(3,54) = .40, p = .76; all between-group p > .5. MSIT performance also did not differ between APOE-ε4-positive and APOE-ε4-negative individuals (Incongruent > Congruent response time as a proportion of total response time: t(56) = 1.02, p = .31; Accuracy: t(56) = .05, p = .67), or between CLU-C-positive and CLU-C-negative individuals (Incongruent > Congruent response time as a proportion of total response time: t(56) =.13, p = .90; Accuracy: t(56) = .89, p = .38). These data indicate that performance differences are not a confound for interpreting the brain-imaging data.
3.3 fMRI
Across all participants, the whole-brain analysis of Incongruent > Congruent MSIT trials demonstrated recruitment of brain regions previously linked to executive function and especially executive attention including anterior cingulate, inferior frontal gyrus, and superior parietal cortex. Results of this contrast (Supplementary Table 1) were consistent with our prediction based on prior reports (Bush & Shin, 2006), indicating that the task was successfully assaying executive attention in this cohort.
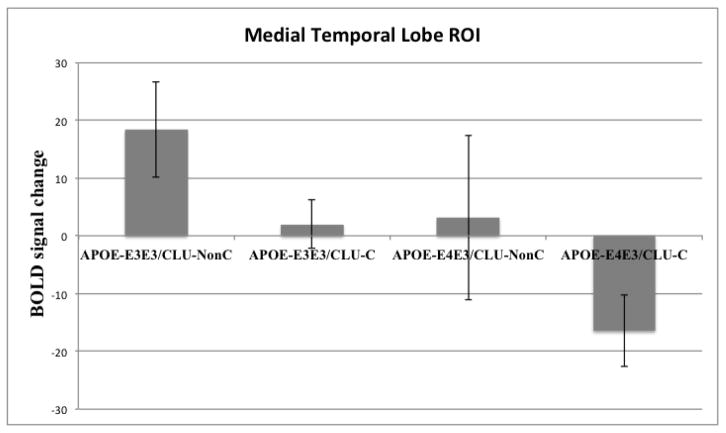
To directly examine the combined effect of CLU-C and APOE-ε4 risk alleles on executive attention-related brain function in the area where AD pathogenesis begins (Braak, et al., 2011; Gomez-Isla, et al., 1996; Hyman, et al., 1984), we extracted parameter estimates representing average activation for the Incongruent > Congruent contrast across voxels within an atlas-defined bilateral MTL region of interest for each genotype group. Results (Figure 3) indicate a cumulative effect of risk alleles on MTL activity such that the lowest genetic risk group showed the highest level of MTL activity, the intermediate risk groups showed an intermediate level of MTL activity, and the highest risk group showed the lowest level of MTL activity. One-way ANOVA revealed a significant effect of combined APOE and CLU risk status on MTL activity during executive attention, F(3, 54) = 5.12, p = .003. As an additional check, we found a similar effect of combined genotype on MTL activity when the full group of eighty-three APOE-ε3ε3/CLU-C individuals was used instead of the randomly selected group of sixteen of these individuals, F(3, 121) = 3.48, p = .018.
Figure 3.
Extracted activity across all voxels of a bilateral medial temporal lobe ROI, comprised of hippocampus and parahippocampal gyrus, in groups of subjects defined by combined APOE and CLU risk allele status. Error bars represent standard errors of the means. APOE-ε4 and CLU-C show an additive effect whereby increasing AD genetic risk is associated with decreasing bilateral medial temporal lobe activity during executive attention.
To characterize the nature of the combined effect of APOE and CLU risk genotype on MTL, we tested additive and multiplicative genetic “risk scores” as predictors of activity extracted from the MTL ROI. Risk scores were weighted by the AD-associated odds ratio for each risk allele (OR for APOE-ε4 = 4.3 (Bertram, et al., 2007); OR for CLU-C = 1.22 (Carrasquillo, et al., 2010)). Values for the additive and multiplicative risk scores were assigned to each participant as the sum or product, respectively, of the natural log of OR for each risk gene. The additive risk score was significantly negatively associated with MTL activity (β = −.43, t(57) = −2.39, p = .001). The multiplicative risk score did not predict MTL activity (β = −.15, t(57) = −1.11, p = .271). To determine whether the effect of the additive risk score was distinct from main effects of APOE-ε4 and CLU-C, MTL activity was regressed on main effect regressors for each risk allele along with the additive risk score. The additive risk score remained a significant predictor of MTL activity after accounting for the main effects of APOE-ε4 (β = −2.12, t(57) = −2.05, p = .045) and CLU-C (β = −.31, t(57) = −2.25, p = .029). Neither main effect remained significant when risk score was included as a regressor (both p > .1), however, the main effects of APOE-ε4 and CLU-C on MTL activity were each significant separately (APOE-ε4: β = −.40, t(57) = −3.28, p = .002; CLU-C: β = −.39, t(57) = −3.13, p = .003). The effect of additive risk score on MTL activity also remained significant when average MTL volume was included in the regression (β = −.45, t(57) = −3.49, p = .001) indicating that volume did not account for the observed effect. MTL volume did not differ between the four genotype groups F(3, 121) = .486, p = .694.
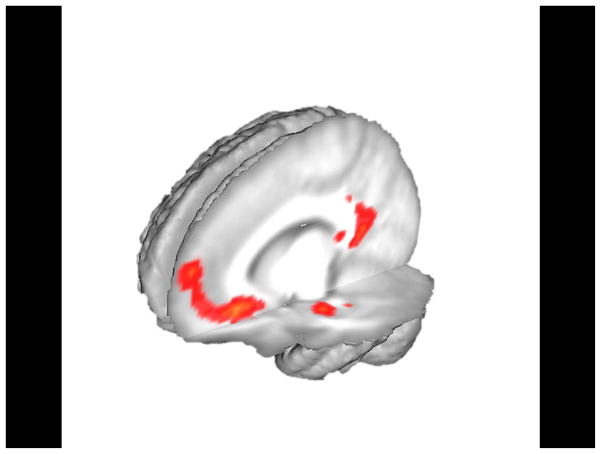
To test the combined effect of APOE-ε4 and CLU-C possession during executive attention across the whole brain, we included the additive risk score as a parametric regressor in our design matrix. This whole-brain analysis revealed activity negatively associated with risk score in left MTL, as well as posterior cingulate gyrus and medial prefrontal cortex (Figure 2; Table 2). These results provide evidence that risk alleles for APOE and CLU additively affect activity in the identified brain regions. Alteration of MTL activity in young people by AD genetic risk is particularly noteworthy given the early involvement of this region in AD pathogenesis (Braak, et al., 2011; Gomez-Isla, et al., 1996; Hyman, et al., 1984). A posteriori investigation of the observed posterior cingulate and medial prefrontal activity was conducted by defining regions of interest as 10mm-radius spheres surrounding the functional peaks identified in these two regions for the risk score parametric analysis and extracting executive attention-related activity for the Incongruent > Contrast (see Supplementary Materials). Activity in these regions demonstrated a trend that was similar (though less definitive) to the trend observed in MTL whereby higher combined APOE and CLU genetic risk was associated with decreased activity.
Figure 2.
Whole-brain parametric analysis of additive genetic “risk wcore” using combined APOE and CLU genotype (p < .001) to predict activation during MSIT executive attention. Risk score represented a de-meaned regressor after assignment of odds ratio-weighted scores to each participant. Risk score was strongly predictive of activity in left parahippocampal gyrus, posterior cingulate, and medial prefrontal cortex.
Table 2.
Whole-brain parametric analysis to identify activation in the Incongruent > Congruent contrast that was associated with the odds ratio-weighted additive risk score.
| Anatomical region | BA | z | Talairach Coordinates
|
||
|---|---|---|---|---|---|
| x | y | z | |||
| Left Medial Frontal Gyrus | 11 | 5.35 | −10 | 27 | −10 |
| Left Posterior Cingulate | 29 | 5.09 | −5 | −53 | 12 |
| Left Parahippocampal Gyrus | 35 | 4.30 | −23 | −14 | −22 |
| Left Caudate Tail | NA | 3.92 | −23 | −43 | 10 |
Results thresholded at p < .001
Convergent with the risk score parametric analysis, a whole-brain direct contrast between the highest-risk APOE-ε4ε3/CLU-C group and lowest-risk APOE-ε3ε3/CLU-NonC group revealed greater executive attention-related activity for the Incongruent > Congruent contrast in left MTL in the lowest risk group (Table 3). Additional whole-brain contrasts between 1) the highest risk and intermediate risk groups, and 2) between the intermediate risk groups and lowest risk group did not yield any activation meeting our conservative exploratory whole-brain threshold. This pattern of data again indicates that the effects of the APOE-ε4 and CLU-C alleles are additive (i.e., the difference between highest and lowest risk is greater than the difference between intermediate risk and either extreme).
Table 3.
Whole-brain between-group contrast of APOE-ε3ε3/CLU-NonC > APOE-ε4ε3/CLU-C for activation yielded by the Incongruent > Congruent contrast.
| Anatomical region | BA | z | Talairach Coordinates
|
||
|---|---|---|---|---|---|
| x | y | z | |||
| Left Parahippocampal Gyrus | 36 | 3.01 | −24 | −34 | −8 |
| Left Transverse Temporal Gyrus | 42 | 3.31 | −10 | 27 | −10 |
| Right Posterior Cingulate | 29 | 3.26 | 4 | −49 | 9 |
| Right Superior Temporal Gyrus | 22 | 3.26 | 60 | −8 | 8 |
Results thresholded at p < .001
A contrast of APOE-ε4-positive individuals vs. APOE-ε4-negative individuals, collapsed across CLU genotype, revealed greater activity for ε4-negative individuals in posterior cingulate gyrus (Talairach coordinates of peak voxel: x = −9, y = −39, z = 14). APOE-ε4-positive individuals did not show greater activity above the exploratory threshold for any region. A contrast of CLU-C-positive vs. CLU-C-negative identified significantly greater activity among CLU-C-negative individuals in right insula (x = 38, y = −13, z = 24) and superior parietal cortex (x = 31, y = −22, z = 36). CLU-C-positive individuals did not show greater activity for any region. It is notable that MTL did not show different activation above threshold for APOE-ε4 or CLU-C status independently within our sample, but did show the additive effect of the two risk alleles.
Because sex was not matched between the groups (4 females in the APOE-ε4ε3/CLU-C group and only 1 female in each of the other two groups), we sought to test whether sex was driving the observed effects through an additional set of analyses with female participants removed from the APOE-ε4ε3/CLU-C group. These analyses revealed very little change in regional brain activity in the Risk Score parametric analysis when the female subjects were removed. The effect of risk allele possession, calculated via one-way ANOVA on extracted activity in our a priori region of interest in MTL, remained significant after removing female participants F(3, 48) = 3.31, p = .028. These analyses enable us to conclude that sex is generally not driving the observed effects within our sample. Nonetheless, the paucity of female participants, and the relatively small sample size overall, are limitations for our study’s applicability to the general population.
4. Discussion
Two genetic risk factors for AD additively affected brain activity, including in the medial temporal lobe, during executive attention in a population of young adults. Our results demonstrate combined influence of APOE and CLU genotype on brain activity long before frank AD symptoms arise, and indicate executive attention as a neural marker of AD genetic risk in young people.
4.1 Effects of genetic risk of AD in young adults
The effects of APOE and CLU on risk of AD risk may be due to their effects on normal brain throughout life. In young people, APOE-ε4 status is associated with differences in neural activity (for review, see (Trachtenberg, et al., 2012)). Similarly, the CLU gene affects normal brain function and structure. In healthy young and middle-aged adults, the high-risk CLU-C variant was associated with increased brain activity during high memory demands (Lancaster, et al., 2011), reduced white matter integrity (Braskie, et al., 2011) and disrupted brain connectivity (Erk, et al., 2011). The effects of CLU on the progression from healthy brain function to MCI may be related to processes that affect regional cerebral blood flow (Thambisetty, et al., 2013) and brain atrophy (Thambisetty, et al., 2012). Grouping participants by combined APOE and CLU genotype, we found that individuals with higher combined AD genetic risk showed lower levels of activation in MTL, an area involved very early in AD as evidenced by accumulation of AD-related neuropathological lesions and atrophy (Braak, et al., 2011; Gomez-Isla, et al., 1996; Hyman, et al., 1984). Pathogenesis of AD begins at least twenty years before clinical symptoms (Bateman, et al., 2012), demonstrating that AD develops over long periods of time. The present finding that AD risk genotype is associated with reduced neural activity during executive attention is generally consistent with prior evidence that young APOE-ε4 carriers show reduced (putatively more efficient) neural activity as compared to APOE-ε4 noncarriers during memory function (Mondadori, et al., 2007; Scarmeas, et al., 2005), albeit without differences in performance. The antagonistic plieotropy account of APOE-ε4 (Han & Bondi, 2008) posits that APOE-ε4 confers an advantage in younger life (for reproductive selection) and then a disadvantage later in life. This account is supported by previously reported performance advantages for young APOE-ε4 carriers on executive tasks (Han & Bondi, 2008), and with evidence that, later in life, APOE-ε4 carriers show less efficient medial temporal lobe (MTL) activity (S. Y. Bookheimer, et al., 2000). However, there is some ambiguity surrounding this issue as several studies have reported increased MTL activity in young APOE-ε4 carriers during memory encoding (Borghesani, et al., 2008; Bunce, et al., 2011; Dennis, et al., 2010; Filippini, et al., 2009; Scarmeas, et al., 2005).
The addition of CLU to APOE in our analyses provides new insight into the independent and additive effects of the APOE gene, the strongest genetic risk factor for AD. APOE genotype had an independent effect on brain activity in our sample, including in posterior cingulate gyrus, as prior studies have indicated (Dennis, et al., 2010; Filbey, Slack, Sunderland, & Cohen, 2006; Filippini, et al., 2009). Analysis of combined CLU and APOE genotype showed an additive effect leading to alterations of both posterior cingulate and MTL function. This combined genotype analysis suggests that CLU-C may exacerbate the effects of APOE-ε4 in young people that ultimately lead to greater susceptibility to AD pathogenesis. CLU genotype has previously been shown to affect structural and functional connectivity in healthy young people (Erk, et al., 2011), which could plausibly interact with effects of APOE genotype on the integrity of white matter tracts (Nierenberg, et al., 2005). Further study in larger sample sizes will be required to meaningfully assess additional potential exacerbating factors (e.g., APOE-ε4ε4 homozygosity).
4.2 Neuroimaging of executive attention as a marker of AD risk
Executive attention assays are known to track AD progression in the early, pre-clinical stages (Belleville, Chertkow, & Gauthier, 2007; Neufang, et al., 2013; Saunders & Summers, 2011; Schroeter, et al., 2012; Wylie, et al., 2007). However, brain-imaging studies of AD risk in young people have not yet investigated executive attention, and the combined effects of APOE-ε4 and CLU-C risk alleles on executive attention-related brain activity has not been previously investigated in any population. Here, we found that combined CLU and APOE genotype was predictive of altered brain function in young people during executive attention, including in MTL, an early cortical site of AD pathogenesis. Thus, our data indicate that executive attention may be a sensitive early assay of risk-related cognitive and neural alterations long before the onset of AD pathogenesis (Braak, et al., 2011; Gomez-Isla, et al., 1996; Hyman, et al., 1984).
Parahippocampal activation has been observed in previous investigations of executive attention (Banich, et al., 2009; Casey, et al., 2002). In the present study, it is possible that the spatial processing component of the MSIT task (i.e., encoding the positions of the three numbers on the screen relative to each other and to the positions of the corresponding buttons on the keypad) accounts for the involvement of parahippocampal gyrus (Epstein, et al., 1999; Preston & Gabrieli, 2008; Ryan, et al., 2010). The genotype-dependent differences in brain function in the present study were overlapping with the parahippocampal “place area,” which contributes to processing of spatial layout information (Epstein, et al., 1999), as is required to process the spatial layout of numbers on each MSIT trial. Nonetheless, MTL is not among the regions most centrally involved in executive attention (Kane & Engle, 2003). The fact that a task that relies most heavily on regions outside MTL elicits genotype-related differences in MTL activity may indicate the strength of AD genetic risk effects on MTL cells in healthy young adults decades before the onset of AD. Likewise, in the context of prior AD risk research that has used memory assays to target the memory-critical MTL, it is noteworthy that a task (executive attention), which does not assay memory, nonetheless reveals altered medial temporal lobe function in individuals at genetic risk for AD. This finding provides a new and potentially important indication that the effect of AD genetic risk on MTL in young people are at least somewhat generalizable, rather than being specific to memory function, and again may point to an underlying effect on cellular physiology.
Regions identified in our analyses of APOE and CLU genotype are consistent with those identified in previous studies of APOE genotype in young people, including right superior temporal gyrus (Scarmeas, et al., 2005) and medial prefrontal cortex (Filippini, et al., 2009). The genotype-related differences we observed in the activity of posterior cingulate gyrus support a growing body of evidence that APOE-ε4 status affects the activity and connectivity of this region in young people (Dennis, et al., 2010; Filbey, et al., 2006; Filippini, et al., 2009). This effect may be related to decreased mitochondrial activity within this region in young APOE-ε4 carriers (Valla, et al., 2010). The present study extends the evidence concerning posterior cingulate and AD risk by revealing that decreased activity in this region is associated with combined APOE-ε4 and CLU-C carrier status. Posterior cingulate and medial prefrontal cortex are both prominent nodes in the so-called default mode network (Laird, et al., 2009), and MTL is also frequently considered part of the default mode network (Greicius, Srivastava, Reiss, & Menon, 2004; but see Laird, et al., 2009). That we observed effects in all three of these regions informs prior evidence indicating that default mode network activity is decreased in the early stages of AD (Greicius, Srivastava, Reiss, & Menon, 2004; Wang, et al., 2006).
As noted above, decreased medial temporal activity associated with AD risk genotype in the present study is consistent with a previous study of APOE genotype in young people (Mondadori et al., 2007). However several other studies, all of which have used memory paradigms, have indicated increased medial temporal activity associated with the APOE-ε4 allele (Borghesani, et al., 2008; Bunce, et al., 2011; Dennis, et al., 2010; Filippini, et al., 2009; Scarmeas, et al., 2005). Though more research will be necessary to resolve this, it is possible that this difference arises from the use of an executive attention paradigm in the present study, which may further indicate the usefulness of executive attention as a marker of additional variance in young people at risk for AD. The results of the combined APOE and CLU genotype analysis further develop MSIT executive attention as a marker of genetic risk by showing that it is sensitive to a combined risk profile stronger than APOE-ε4 carrier status alone. Further replication in larger samples and further study of the heritability of MSIT activation is needed before such activation can be recommended for extensive investigation as a neurocognitive marker of genetic risk.
4.3 Summary
We observed that inheritance of two AD risk alleles, APOE-ε4 and CLU-C additively decreased brain activity in the medial temporal lobe during a task of executive attention in young adults. Developing brain-imaging assays that are sensitive to combined AD risk genotype is an important direction for establishing early biomarkers of increased AD risk, and could open novel avenues for testing the efficacy of preventive treatments in healthy people before disease symptoms take hold.
Supplementary Material
APOE and CLU have a combined effect on MTL function
Executive attention may provide a viable neural marker of genetic AD risk in young people
Additivity of the genotype effects suggests a common pathway for APOE and CLU
MTL effect during non-memory task indicates domain general effect on MTL physiology
This is the first fMRI study of effects of CLU in young people
Acknowledgments
This work was supported by NIH R01 AG035379, P01 AG030128 (GWR), NSF DRL 0644131 (JRG) and NSF REC 0634025 (JRG), and a grant from Partners in Research (TRM and AEG). Samples from the National Cell Repository for Alzheimer’s Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study. We wish to thank M.H.E. Johnson and D.P. Kraemer for thoughtful comments on an earlier version of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38:95–113. doi: 10.1016/j.neuroimage.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Voxel-based morphometry--the methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Banich MT, Burgess GC, Depue BE, Ruzic L, Bidwell LC, Hitt-Laustsen S, Du YP, Willcutt EG. The neural basis of sustained and transient attentional control in young adults with ADHD. Neuropsychologia. 2009;47:3095–3104. doi: 10.1016/j.neuropsychologia.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman RJ, Xiong C, Benzinger TL, Fagan AM, Goate A, Fox NC, Marcus DS, Cairns NJ, Xie X, Blazey TM, Holtzman DM, Santacruz A, Buckles V, Oliver A, Moulder K, Aisen PS, Ghetti B, Klunk WE, McDade E, Martins RN, Masters CL, Mayeux R, Ringman JM, Rossor MN, Schofield PR, Sperling RA, Salloway S, Morris JC. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Belleville S, Chertkow H, Gauthier S. Working memory and control of attention in persons with Alzheimer’s disease and mild cognitive impairment. Neuropsychology. 2007;21:458–469. doi: 10.1037/0894-4105.21.4.458. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nature Genetics. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bookheimer S, Burggren A. APOE-4 genotype and neurophysiological vulnerability to Alzheimer’s and cognitive aging. Annu Rev Clin Psychol. 2009;5:343–362. doi: 10.1146/annurev.clinpsy.032408.153625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC, Small GW. Patterns of brain activation in people at risk for Alzheimer’s disease. N Engl J Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesani PR, Johnson LC, Shelton AL, Peskind ER, Aylward EH, Schellenberg GD, Cherrier MM. Altered medial temporal lobe responses during visuospatial encoding in healthy APOE*4 carriers. Neurobiol Aging. 2008;29:981–991. doi: 10.1016/j.neurobiolaging.2007.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Thal DR, Ghebremedhin E, Del Tredici K. Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J Neuropathol Exp Neurol. 2011;70:960–969. doi: 10.1097/NEN.0b013e318232a379. [DOI] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW, Thompson PM. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. J Neurosci. 2011;31:6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce D, Anstey KJ, Burns R, Christensen H, Easteal S. Does possession of apolipoprotein E varepsilon4 benefit cognitive function in healthy young adults? Neuropsychologia. 2011;49:1693–1697. doi: 10.1016/j.neuropsychologia.2011.02.042. [DOI] [PubMed] [Google Scholar]
- Bush G, Shin LM. The Multi-Source Interference Task: an fMRI task that reliably activates the cingulo-frontal-parietal cognitive/attention network. Nat Protoc. 2006;1:308–313. doi: 10.1038/nprot.2006.48. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff-Radford NR, Petersen RC, Morgan K, Younkin SG. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Archives of Neurology. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Thomas KM, Davidson MC, Kunz K, Franzen PL. Dissociating striatal and hippocampal function developmentally with a stimulus-response compatibility task. J Neurosci. 2002;22:8647–8652. doi: 10.1523/JNEUROSCI.22-19-08647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Seeley WW, Zhou J, Shirer WR, Coppola G, Karydas A, Rosen HJ, Miller BL, Kramer JH, Greicius MD. Gender modulates the APOE epsilon4 effect in healthy older adults: convergent evidence from functional brain connectivity and spinal fluid tau levels. J Neurosci. 2012;32:8254–8262. doi: 10.1523/JNEUROSCI.0305-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41:193–202. doi: 10.1016/s0896-6273(03)00850-x. [DOI] [PubMed] [Google Scholar]
- Dennis NA, Browndyke JN, Stokes J, Need A, Burke JR, Welsh-Bohmer KA, Cabeza R. Temporal lobe functional activity and connectivity in young adult APOE varepsilon4 carriers. Alzheimers Dement. 2010;6:303–311. doi: 10.1016/j.jalz.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanis SB, DiBattista AM, Miessau M, Moussa CE, Rebeck GW. APOE genotype affects the pre-synaptic compartment of glutamatergic nerve terminals. J Neurochem. 2013;124:4–14. doi: 10.1111/j.1471-4159.2012.07908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumanis SB, Tesoriero JA, Babus LW, Nguyen MT, Trotter JH, Ladu MJ, Weeber EJ, Turner RS, Xu B, Rebeck GW, Hoe HS. ApoE4 decreases spine density and dendritic complexity in cortical neurons in vivo. J Neurosci. 2009;29:15317–15322. doi: 10.1523/JNEUROSCI.4026-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Weickert CS, Garner B. Apolipoproteins in the brain: implications for neurological and psychiatric disorders. Clin Lipidol. 2010;51:555–573. doi: 10.2217/CLP.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein R, Harris A, Stanley D, Kanwisher N. The parahippocampal place area: recognition, navigation, or encoding? Neuron. 1999;23:115–125. doi: 10.1016/s0896-6273(00)80758-8. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Opitz von Boberfeld C, Esslinger C, Schnell K, Kirsch P, Mattheisen M, Muhleisen TW, Cichon S, Witt SH, Rietschel M, Nothen MM, Walter H. Hippocampal function in healthy carriers of the CLU Alzheimer’s disease risk variant. J Neurosci. 2011;31:18180–18184. doi: 10.1523/JNEUROSCI.4960-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. Jama. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Fei M, Jianhua W. Apolipoprotein epsilon4-Allele as a Significant Risk Factor for Conversion from Mild Cognitive Impairment to Alzheimer’s disease: a Meta-analysis of Prospective Studies. J Mol Neurosci. 2012 doi: 10.1007/s12031-012-9934-y. [DOI] [PubMed] [Google Scholar]
- Filbey FM, Slack KJ, Sunderland TP, Cohen RM. Functional magnetic resonance imaging and magnetoencephalography differences associated with APOEepsilon4 in young healthy adults. Neuroreport. 2006;17:1585–1590. doi: 10.1097/01.wnr.0000234745.27571.d1. [DOI] [PubMed] [Google Scholar]
- Filippini N, MacIntosh BJ, Hough MG, Goodwin GM, Frisoni GB, Smith SM, Matthews PM, Beckmann CF, Mackay CE. Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc Natl Acad Sci U S A. 2009;106:7209–7214. doi: 10.1073/pnas.0811879106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghebremedhin E, Schultz C, Thal DR, Rub U, Ohm TG, Braak E, Braak H. Gender and age modify the association between APOE and AD-related neuropathology. Neurology. 2001;56:1696–1701. doi: 10.1212/wnl.56.12.1696. [DOI] [PubMed] [Google Scholar]
- Goldberg TE, Weinberger DR. Genes and the parsing of cognitive processes. Trends Cogn Sci. 2004;8:325–335. doi: 10.1016/j.tics.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Gomez-Isla T, Price JL, McKeel DW, Jr, Morris JC, Growdon JH, Hyman BT. Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J Neurosci. 1996;16:4491–4500. doi: 10.1523/JNEUROSCI.16-14-04491.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AE, Fugelsang JA, Kraemer DJ, Dunbar KN. The Micro-Category account of analogy. Cognition. 2008;106:1004–1016. doi: 10.1016/j.cognition.2007.03.015. [DOI] [PubMed] [Google Scholar]
- Green AE, Kraemer DJ, Deyoung CG, Fossella JA, Gray JR. A Gene-Brain-Cognition Pathway: Prefrontal Activity Mediates the Effect of COMT on Cognitive Control and IQ. Cereb Cortex. 2012 doi: 10.1093/cercor/bhs035. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SD, Bondi MW. Revision of the apolipoprotein E compensatory mechanism recruitment hypothesis. Alzheimers Dement. 2008;4:251–254. doi: 10.1016/j.jalz.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nature Genetics. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JM, Cirrito JR, Restivo JL, Kinley RD, Sullivan PM, Holtzman DM, Koger D, Delong C, Lin S, Zhao L, Liu F, Bales K, Paul SM. Emergence of a seizure phenotype in aged apolipoprotein epsilon 4 targeted replacement mice. Brain Res. 2012;1467:120–132. doi: 10.1016/j.brainres.2012.05.048. [DOI] [PubMed] [Google Scholar]
- Hyman BT, Van Hoesen GW, Damasio AR, Barnes CL. Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science. 1984;225:1168–1170. doi: 10.1126/science.6474172. [DOI] [PubMed] [Google Scholar]
- Kane MJ, Engle RW. Working-memory capacity and the control of attention: the contributions of goal neglect, response competition, and task set to Stroop interference. J Exp Psychol Gen. 2003;132:47–70. doi: 10.1037/0096-3445.132.1.47. [DOI] [PubMed] [Google Scholar]
- Kang SW, Shin YJ, Shim YJ, Jeong SY, Park IS, Min BH. Clusterin interacts with SCLIP (SCG10-like protein) and promotes neurite outgrowth of PC12 cells. Exp Cell Res. 2005;309:305–315. doi: 10.1016/j.yexcr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer’s disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Donarski N, Goetze K, Kreckel M, Stuerenburg HJ, Buhmann C, Beisiegel U. Characterization of four lipoprotein classes in human cerebrospinal fluid. J Lipid Res. 2001;42:1143–1151. [PubMed] [Google Scholar]
- Kounnas MZ, Loukinova EB, Stefansson S, Harmony JA, Brewer BH, Strickland DK, Argraves WS. Identification of glycoprotein 330 as an endocytic receptor for apolipoprotein J/clusterin. J Biol Chem. 1995;270:13070–13075. doi: 10.1074/jbc.270.22.13070. [DOI] [PubMed] [Google Scholar]
- Laird AR, Eickhoff SB, Li K, Robin DA, Glahn DC, Fox PT. Investigating the functional heterogeneity of the default mode network using coordinate-based meta-analytic modeling. Journal of Neuroscience. 2009;29:14496–14505. doi: 10.1523/JNEUROSCI.4004-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lancaster TM, Baird A, Wolf C, Jackson MC, Johnston SJ, Donev R, Thome J, Linden DE. Neural hyperactivation in carriers of the Alzheimer’s risk variant on the clusterin gene. Eur Neuropsychopharmacol. 2011;21:880–884. doi: 10.1016/j.euroneuro.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Lehmann DJ, Refsum H, Nurk E, Warden DR, Tell GS, Vollset SE, Engedal K, Nygaard HA, Smith AD. Apolipoprotein E epsilon4 and impaired episodic memory in community-dwelling elderly people: a marked sex difference. The Hordaland Health Study. J Neurol Neurosurg Psychiatry. 2006;77:902–908. doi: 10.1136/jnnp.2005.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci. 2006;7:818–827. doi: 10.1038/nrn1993. [DOI] [PubMed] [Google Scholar]
- Mondadori CR, de Quervain DJ, Buchmann A, Mustovic H, Wollmer MA, Schmidt CF, Boesiger P, Hock C, Nitsch RM, Papassotiropoulos A, Henke K. Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers. Cereb Cortex. 2007;17:1934–1947. doi: 10.1093/cercor/bhl103. [DOI] [PubMed] [Google Scholar]
- Myers RH, Schaefer EJ, Wilson PW, D’Agostino R, Ordovas JM, Espino A, Au R, White RF, Knoefel JE, Cobb JL, McNulty KA, Beiser A, Wolf PA. Apolipoprotein E epsilon4 association with dementia in a population-based study: The Framingham study. Neurology. 1996;46:673–677. doi: 10.1212/wnl.46.3.673. [DOI] [PubMed] [Google Scholar]
- Nathan BP, Bellosta S, Sanan DA, Weisgraber KH, Mahley RW, Pitas RE. Differential effects of apolipoproteins E3 and E4 on neuronal growth in vitro. Science. 1994;264:850–852. doi: 10.1126/science.8171342. [DOI] [PubMed] [Google Scholar]
- Neufang S, Akhrif A, Riedl V, Forstl H, Kurz A, Zimmer C, Sorg C, Wohlschlager AM. Predicting effective connectivity from resting-state networks in healthy elderly and patients with prodromal Alzheimer’s disease. Hum Brain Mapp. 2013 doi: 10.1002/hbm.22226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierenberg J, Pomara N, Hoptman MJ, Sidtis JJ, Ardekani BA, Lim KO. Abnormal white matter integrity in healthy apolipoprotein E epsilon4 carriers. Neuroreport. 2005;16:1369–1372. doi: 10.1097/01.wnr.0000174058.49521.16. [DOI] [PubMed] [Google Scholar]
- Pena-Gomez C, Sole-Padulles C, Clemente IC, Junque C, Bargallo N, Bosch B, Molinuevo JL, Valls-Sole J, Pascual-Leone A, Bartres-Faz D. APOE status modulates the changes in network connectivity induced by brain stimulation in non-demented elders. PLoS One. 2012;7:e51833. doi: 10.1371/journal.pone.0051833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston AR, Gabrieli JD. Dissociation between explicit memory and configural memory in the human medial temporal lobe. Cereb Cortex. 2008;18:2192–2207. doi: 10.1093/cercor/bhm245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan L, Lin CY, Ketcham K, Nadel L. The role of medial temporal lobe in retrieving spatial and nonspatial relations from episodic and semantic memory. Hippocampus. 2010;20:11–18. doi: 10.1002/hipo.20607. [DOI] [PubMed] [Google Scholar]
- Saunders NL, Summers MJ. Longitudinal deficits to attention, executive, and working memory in subtypes of mild cognitive impairment. Neuropsychology. 2011;25:237–248. doi: 10.1037/a0021134. [DOI] [PubMed] [Google Scholar]
- Scarmeas N, Habeck CG, Hilton J, Anderson KE, Flynn J, Park A, Stern Y. APOE related alterations in cerebral activation even at college age. J Neurol Neurosurg Psychiatry. 2005;76:1440–1444. doi: 10.1136/jnnp.2004.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeter ML, Vogt B, Frisch S, Becker G, Barthel H, Mueller K, Villringer A, Sabri O. Executive deficits are related to the inferior frontal junction in early dementia. Brain. 2012;135:201–215. doi: 10.1093/brain/awr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Z, DeYoung CG, Kang Y, Grigorenko EL, Gray JR. Interaction of COMT val158met and externalizing behavior: relation to prefrontal brain activity and behavioral performance. Neuroimage. 2012;60:2158–2168. doi: 10.1016/j.neuroimage.2012.01.097. [DOI] [PubMed] [Google Scholar]
- Silveri MC, Reali G, Jenner C, Puopolo M. Attention and memory in the preclinical stage of dementia. J Geriatr Psychiatry Neurol. 2007;20:67–75. doi: 10.1177/0891988706297469. [DOI] [PubMed] [Google Scholar]
- Tan HY, Callicott JH, Weinberger DR. Intermediate phenotypes in schizophrenia genetics redux: is it a no brainer? Mol Psychiatry. 2008;13:233–238. doi: 10.1038/sj.mp.4002145. [DOI] [PubMed] [Google Scholar]
- Thambisetty M, An Y, Kinsey A, Koka D, Saleem M, Guntert A, Kraut M, Ferrucci L, Davatzikos C, Lovestone S, Resnick SM. Plasma clusterin concentration is associated with longitudinal brain atrophy in mild cognitive impairment. Neuroimage. 2012;59:212–217. doi: 10.1016/j.neuroimage.2011.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thambisetty M, Beason-Held LL, An Y, Kraut M, Nalls M, Hernandez DG, Singleton AB, Zonderman AB, Ferrucci L, Lovestone S, Resnick SM. Alzheimer Risk Variant CLU and Brain Function During Aging. Biol Psychiatry. 2013;73:399–405. doi: 10.1016/j.biopsych.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg AJ, Filippini N, Ebmeier KP, Smith SM, Karpe F, Mackay CE. The effects of APOE on the functional architecture of the resting brain. Neuroimage. 2012;59:565–572. doi: 10.1016/j.neuroimage.2011.07.059. [DOI] [PubMed] [Google Scholar]
- Valla J, Yaari R, Wolf AB, Kusne Y, Beach TG, Roher AE, Corneveaux JJ, Huentelman MJ, Caselli RJ, Reiman EM. Reduced posterior cingulate mitochondrial activity in expired young adult carriers of the APOE epsilon4 allele, the major late-onset Alzheimer’s susceptibility gene. J Alzheimers Dis. 2010;22:307–313. doi: 10.3233/JAD-2010-100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zang Y, He Y, Liang M, Zhang X, Tian L, Wu T, Jiang T, Li K. Changes in hippocampal connectivity in the early stages of Alzheimer’s disease: evidence from resting state fMRI. Neuroimage. 2006;31:496–504. doi: 10.1016/j.neuroimage.2005.12.033. [DOI] [PubMed] [Google Scholar]
- Wu ZC, Yu JT, Li Y, Tan L. Clusterin in Alzheimer’s disease. Adv Clin Chem. 2012;56:155–173. doi: 10.1016/b978-0-12-394317-0.00011-x. [DOI] [PubMed] [Google Scholar]
- Wylie SA, Ridderinkhof KR, Eckerle MK, Manning CA. Inefficient response inhibition in individuals with mild cognitive impairment. Neuropsychologia. 2007;45:1408–1419. doi: 10.1016/j.neuropsychologia.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Zahs KR, Ashe KH. ‘Too much good news’ - are Alzheimer mouse models trying to tell us how to prevent, not cure, Alzheimer’s disease? Trends Neurosci. 2010;33:381–389. doi: 10.1016/j.tins.2010.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.