Abstract
RhoGTPases play important roles in the regulation of protein transport and membrane recycling. Little is known, however, about how RhoGTPases affect HIV-1 virion production, which is dependent on the endosomal sorting pathway. We report that ectopic expression of citron kinase (citron-K), a RhoA effector, preferentially enhances HIV-1 virion production. Depletion of endogenous citron-K inhibits HIV-1 virion production. Citron-N, which lacks the kinase domain, also enhances HIV-1 virion production. The leucine zipper, Rho-binding and zinc finger domains of citron-N are necessary for the enhancement activity. Citron-K also enhances murine leukemia virion production and the HIV-1 late domain is not required for the citron-K-mediated enhancement. Ectopic expression of citron-K leads to the formation of cytoplasmic structures containing citron-K and HIV-1 Gag proteins. HIV-1 and citron-K cooperatively enhance acidic endosome and lysosome compartments. Finally, citron-K promotes exocytosis of microvesicles or exosomes that co-purify with HIV-1 virions. We conclude that citron-K enhances HIV-1 virion production by stimulating the endosomal compartments and exocytosis.
Keywords: citron kinase, exocytosis, exosome, HIV-1 egress, RhoA
HIV-1 replication is modulated by a number of cellular signaling pathways regulated by both host and viral factors (1). HIV-1 assembly and release occur in a series of essential steps mediated by the viral Gag precursor protein, Pr55Gag (2,3). HIV-1 Gag is organized into four distinct regions: matrix, capsid, nucleocapsid and late (L) domain. The L domain catalyzes the pinching off and detachment of virus particles from the cell surface and each other (2). The L domain is a highly conserved Pro-rich motif found in enveloped viruses. There are three classes of tetrapeptide motifs: PT/SAP (4,5), PPXY (6–8) and YPXL (9). In the case of HIV-1, the L domain is encoded by a small peptide motif, Pro-Thr/Ser-Ala-Pro (PTAP) in the C-terminal, p6 domain of Pr55Gag. The L domains of retroviruses, despite differences in amino acid sequence and location within their respective viral structural proteins, are functionally interchangeable, suggesting commonality of function, perhaps as docking sites for host proteins (10–12). Mutations in the PTAP motif of p6 or deletion of the p6 domain produced a striking defect in the production of virus particles where viral particles remain tethered to each other and the plasma membrane (4,5), thereby identifying the p6 domain as playing a critical role in HIV-1 budding.
Viruses like HIV-1 do not encode their own machinery for viral budding and must usurp existing cellular pathways to facilitate viral release. The PTAP motif of HIV-1 Gagp6 was found to bind to the ubiquitin enzyme 2 variant domain of Tsg101 (13–16). Tsg101 is a component of the endosomal sorting complex required for transport (ESCRT-I), a 350 kDa cellular complex essential in the vacuolar protein-sorting (VPS) pathway. Point mutations in HIV-1 PTAP motif block virus release at late stages (4,5) and disrupt binding to Tsg101 (14). Small inhibitory RNA-mediated Tsg101 depletion potently blocks HIV-1 release resulting in the virus forming stalks of tethered virus at the plasma membrane (14). Moreover, overexpression of the dominant-negative form of VPS4, an ESCRT recycling factor, inhibits particle release of HIV-1 and other enveloped viruses, such as murine leukemia virus (MLV) (14). Moreover, a second region of HIV-1 Gagp6 has been defined to contribute to viral release and interacts with AIP1, a host protein (17,18). AIP1 interacts with Tsg101 and CHMP proteins of ESCRT-III complex, coupling HIV-1 Gagp6 to the early and late-acting endosomal sorting complexes and binds directly to HIV-1 Gagp6 Leu-Arg-Ser-Leu (LRSL) motif (17,18). Taken together, these data suggest that the VPS machinery and perhaps other host factors are involved in facilitating the budding of retroviruses.
The endosomal sorting pathway controls a variety of cellular processes and plays a role in the sorting of ubiquitinated cargo proteins into the lumen of the multi-vesicular bodies (MVB) (19,20). Ubiquitinated proteins are recognized on the limiting endosomal membrane and sorted resulting in either MVB fusion with the lysosome to degrade contents or release of material into the extra-cellular environment via exosomal vesicles (21–23). ESCRT-I, composed of Tsg101, Vps28 and Vps37, recognize the ubiquitinated protein cargo and recruit two more class E protein cargos (ESCRT-II/III) that participate in protein sorting and vesicle formation (24–26). HIV-1 may bind Tsg101 and AIP1 to gain access to downstream machinery involved in catalyzing MVB vesicle budding, a mechanism topologically similar to viral budding from the plasma membrane.
RhoGTPases play a pivotal role in the dynamic regulation of actin cytoskeleton (27–29) and through this, control cell morphology (30,31), motility, adhesion and activation of transcription factors such as NF-κB (32) and serum response factor (33). Although RhoGTPases have been implicated in various steps of T-cell activation (34–36), little is known about how RhoGTPases affect HIV-1 replication. We have previously shown that the cytoplasmic tail of the HIV-1 transmembrane glycoprotein interacts with the carboxy-terminus of p115RhoGEF (37), a guanine nucleotide exchange factor and activator of RhoA. Activation of RhoA by p115RhoGEF or Gα13 leads to inhibition of HIV-1 gene expression in a RhoA-dependent manner (38). The RhoA effectors involved in modulating HIV-1 gene expression are not defined.
To investigate how RhoA-signaling pathways modulate HIV-1 replication, we tested various RhoA effectors in 293T and human T cells. We determined that citron kinase (citron-K), a Ser/Thr kinase, enhances HIV-1 virion production with no significant effect on HIV-1 gene expression. Knockdown of citron-K by short-hairpin RNA (shRNA) reduced HIV-1 virion production but not HIV-1 gene expression. Citron-K also enhanced MLV production, as well as virion production from a HIV-1 GagΔp6 construct, suggesting that citron-K mediates virion production independently of the HIV-1 L domain. Citron-K induced intra-cellular compartments and colocalized with Gag in these compartments. HIV-1 and citron-K cooperatively enhanced acidic (late) endosome and lysosome compartments. Furthermore, citron-K enhanced exocytosis of microvesicles or exosomes that co-purify with HIV-1 virions.
Results
Citron-K preferentially enhances HIV-1 viral replication
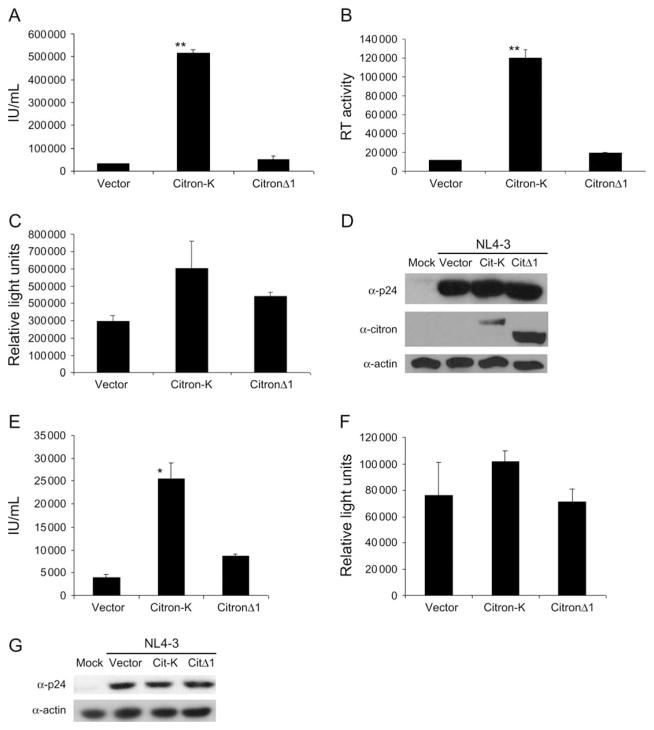
To investigate how RhoA-signaling pathways modulate viral replication, we tested various RhoA effectors for their effect on HIV-1 viral replication in 293T and human T cells. Ectopic expression of citron-K, a RhoA effector involved in cytokinesis (39) and membrane vesicle transport (40–42), was shown to preferentially enhance HIV-1 virion production without significantly affecting HIV-1 gene expression in 293T (Figure 1A–D), Jurkat T (Figure 1E–G) and HeLa cells (data not shown). Both infectious virions (Figure 1A, E) and total virions (virion-associated RT activity or p24; Figure 1B and data not shown) demonstrated a similar enhancement (7- to 15-fold), while expression of HIV-1 reporter gene (Figure 1C, F) and cell-associated viral proteins (Figure 1D, G) showed minimal enhancement (<2.5-fold in 293T cells and no change in Jurkat T cells). The C-terminal truncation mutant of citron-K, citronΔ1, did not enhance HIV-1 virion production. We conclude that citron-K preferentially enhances HIV-1 virion production with minimal effect on HIV-1 gene expression and that activity depends on the C-terminal domains of citron-K.
Figure 1. Citron-K preferentially enhances HIV-1 virion production.
(A and B) 293T cells were co-transfected with proviral DNA NL4-3 and vector, citron-K or citronΔ1. Supernatant was harvested at 48 h post-transfection to determine infectious units/mL (A) or virion-associated RT activity (B). (C and D) 293T cells were co-transfected with pNL4.Luc and vector, citron-K or citronΔ1. Cells were harvested at 48 h post-transfection to determine luciferase activity (C). (D) Cell-associated viral Gag proteins, Myc-tagged citron-K and actin in 293T cells were analyzed using anti-p24, anti-Myc and anti-actin antibodies, respectively. (E and F) Jurkat T cells were co-transfected with pNL4-3 (E) or pNL4.Luc (F) and vector, citron-K or citronΔ1. Supernatants and cells were harvested at 48 h post-transfection to determine infectious units/mL (E) or luciferase activity (F). (G) Cell-associated viral Gag proteins (p55) and actin in Jurkat T cells were analyzed using anti-p24 and anti-actin antibodies. Error bars are standard deviation of duplicate samples and at least three independent experiments were performed. *P < 0.05, **P < 0.005. enhancement (Figure 3). All citron truncation mutant proteins were efficiently expressed (Figure S2A, B).
To evaluate a role for endogenous citron-K in HIV-1 virion production, we depleted endogenous citron-K from 293T and Jurkat T cells using shRNA constructs that targeted various regions of citron-K (Figure S1). The 293T cells were transfected twice, first with control or citron-targeted shRNA constructs only and 24 h later, cells were co-transfected with citron-targeted shRNA constructs and pNL4GFP. Supernatant and cells were harvested 48 h after the second transfection and analyzed. We saw efficient knockdown of citron-K protein (65–90%; Figure 2A) and a 75–85% reduction in virion production by all three constructs as determined by p24 enzyme-linked immunosorbent assay (Figure 2B). HIV-1 gene expression, as measured by cell-associated Gag, was not affected by depletion of citron-K (Figure 2A). Jurkat T cells were also co-transfected with pNL4GFP and the citron-specific shRNA or control constructs. Supernatants and cells were collected 48 h post-transfection. Virion production was inhibited by 60% in citron-K-depleted cells compared with the control (Figure 2C). The inhibition of viral particle release correlated with reduction in expression of endogenous citron-K (Figure 2D). Therefore, citron-K is required for efficient HIV-1 virion production.
Figure 2. Depletion of endogenous citron-K inhibits HIV-1 virion production.
(A and B) Knockdown of endogenous citron-K using citron-targeted shRNA constructs in 293T cells inhibited HIV-1 virion production. 293T cells were transfected with citron-targeted shRNA or control plasmid constructs, 24 h later, cells were co-transfected with pNL4GFP and citron-targeted shRNA or control plasmid constructs. Supernatant and cells were harvested 48 h after the second transfection and analyzed for cell-associated gene expression of citron-K, p24 and actin (A) and relative HIV-1 virion production in the supernatant (p24 pg/mL) (B). (C and D) Knockdown of endogenous citron-K in T cells inhibited HIV-1 virion production. Jurkat T cells were transfected with pNL4GFP and citron-targeted shRNA or control plasmid constructs. Supernatant and cells were harvested at 48 h post-transfection. The effect of citron-K knockdown on HIV-1 virion production in the supernatant was measured by p24 enzyme-linked immunosorbent assay (C). *P < 0.05. Cells were lysed and analyze with anti-citron and anti-actin antibodies (D).
The leucine zipper, Rho-binding and zinc finger domains, but not kinase activity, of citron-K are necessary for enhancing HIV-1 virion production
Deletion mutants were generated to map the domain(s) of citron-K involved in the enhancement of HIV-1 virion production. Citron-N, a naturally occurring splice variant of citron-K lacking the N-terminal kinase domain, showed similar enhancement activity as citron-K, indicating that the kinase domain was dispensable (Figure 3A). Next, we made a series of C-terminal truncation mutants in the citron-N gene. The PH, SH3 and PDZ domains were not required for the activity. The zinc finger motif of citron was necessary for the enhancement of virion production because deletion of the zinc finger domain from citron-N (CNΔZnF) abolished its ability to enhance virion production (Figure 3A). The zinc finger domain alone was unable to enhance virion production (Figure 3A). Further deletion of the N-terminal α-helix demonstrated that two copies of the leucine zipper motif and the Rho-binding domain (RBD), in addition to the zinc finger, were sufficient for citron-mediated viral
Figure 3. Leucine zipper, RBD and zinc finger motifs, but not the kinase domain, are necessary for citron-K to enhance HIV-1 viral release.
(A) 293T cells were co-transfected with pNL4-3 and citron truncation constructs. Supernatants were harvested at 48 h for infectious unit assays. Graph depicts fold enhancement of infectious units by various citron truncation mutants compared with vector control. *P < 0.05. (B) Schematic diagram of the citron-K constructs and of results shown in (A). +, enhancement of viral release.
Citron-K enhances release of MLV and of HIV-1 Gag lacking the L domain
The L domain is important for retroviral release (4,5). HIV-1 viral release is dependent on the unique HIV-1 L domain to interact with the endosomal sorting protein Tsg101 and subsequently the endosomal sorting pathway (14). Release of MLV, which contains a different L domain tetrapeptide sequence (PPPY) than HIV-1 (PTAP), is unaffected by depletion of Tsg101. To determine if citron-K enhancement of virion production is HIV-1 specific, we evaluated whether citron-K was able to enhance MLV production. Citron-K enhanced MLV virion production to comparable levels as HIV-1 (Figure 4A), suggesting that citron-K has a general enhancement activity on retroviral production.
Figure 4. Citron-K enhanced release of MLV and HIV-1 GagΔp6.
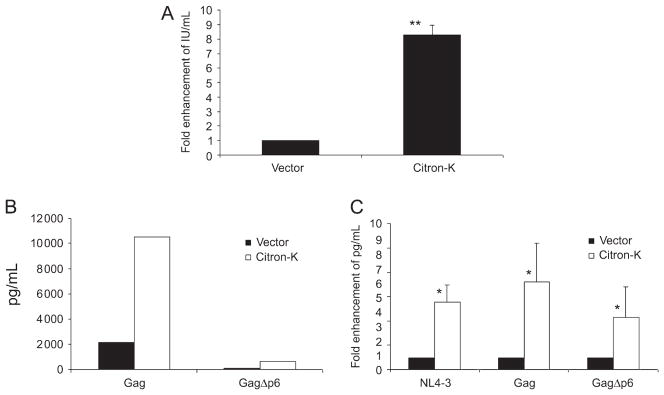
(A) MLV virus-like particle production was similarly enhanced by citron-K. 293T cells were co-transfected with MLV Gag-Pol, VSV-G-env and HSCG (an MLV-based retroviral vector expressing GFP) with citron-K or control vector. Supernatant was collected at 48 h post-transfection and titered for GFP expression on 3T3 cells to determine infectious units/mL. (B and C) Citron-K enhancement of virion production is L domain independent. 293T cells were co-transfected with Gag or GagΔp6 and vector or citron-K. Supernatant and cells were harvested 48 h post-transfection for analysis. (B) Citron-K enhances virion production of both HIV-1 Gag and GagΔp6. Virion production of GagΔp6 alone is significantly reduced compared with Gag but is enhanced by citron-K. (C) Citron-K similarly enhances virion production of HIV-1 Gag, GagΔp6 and NL4-3. Shown is relative enhancement of virion production of Gag, GagΔp6 and NL4-3 from two independent experiments. *P < 0.05, **P < 0.005.
To further determine the role of HIV-1 L domain in citron-K-mediated virion production, we transfected 293T cells with Gag or GagΔp6 constructs (43). The GagΔp6 construct, which lacks the entire L domain, showed significantly reduced virion production compared to full-length Gag (Figure 4B). Citron-K enhanced virus-like particle (VLP) production similarly from HIV-1 Gag proteins with or without the L domain (5–7 fold; Figure 4B, C). These results demonstrate that citron-K enhances HIV-1 virion production independent of the viral L domain.
Citron-K induces formation of intracellular compartments and co-localizes with HIV-1 Gag
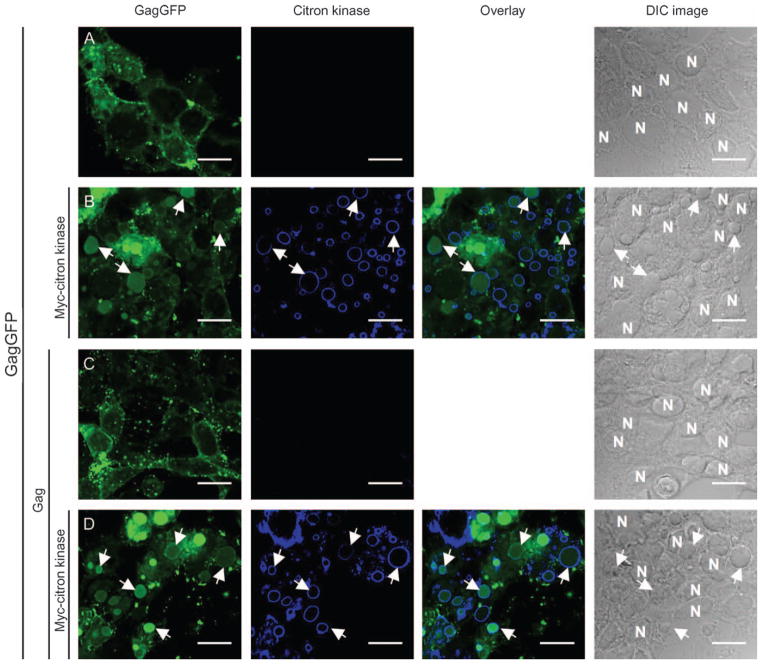
To define a possible mechanism by which citron-K enhances HIV-1 virion production, we evaluated the subcellular localization of citron-K and HIV-1 Gag in 293T cells by confocal microscopy. Citron-K localized to unique compartments that accumulated in the cytoplasm [data not shown, previously described in (44)]. When GagGFP was expressed alone or GagGFP and Gag were co-expressed in 293T cells (Figure 5A, C), Gag exhibited cytoplasmic and plasma membrane localization, the primary site for HIV-1 release. However, when GagGFP or GagGFP/Gag were co-expressed with citron-K, Gag localized to the same dense protein compartments that expressed citron-K (Figure 5B, D). Citron-K mediated relocalization of Gag to these citron-K-induced intracellular structures may mediate enhancement of virion production.
Figure 5. Citron-K and Gag colocalize to citron-K-induced cytoplasmic compartments.
(A and C) 293T cells were transfected with GagGFP or GagGFP/Gag in a 10:1 ratio and protein localization was determined by confocal microscopy. GagGFP localizes primarily to the plasma membrane in the absence of exogenous citron-K. (B and D) 293T cells were co-transfected with GagGFP or GagGFP/Gag combination and citron-K. The localization of GagGFP is altered in the presence of citron-K. Instead of localizing to the plasma membrane, GagGFP localized intracellularly with citron-K into ‘citron-induced cytoplasmic compartments’ (indicated by arrows). N; nucleus. Bars represent 17.5 μm.
Citron-K enhances HIV-1 virion production by modulating late endosomal compartments and exocytosis
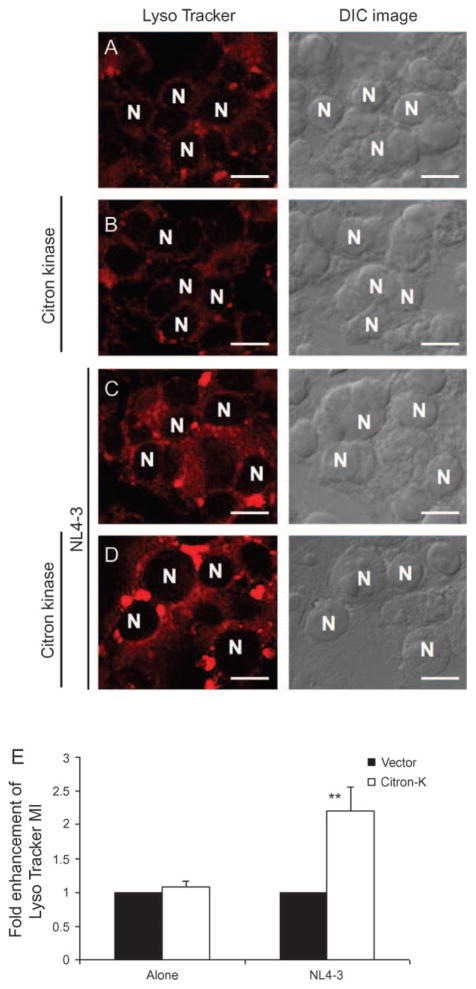
Next, we determined the effect of citron-K on the late endosomal and lysosomal compartments during HIV-1 replication because all retroviruses are dependent on the late endosomal sorting pathway for virion release. Cells were co-transfected with citron-K and/or HIV-1 proviral DNA (pNL4-3) and labeled with LysoTracker, a dye that marks acidic endosomal/lysomal compartments (i.e. low internal pH). Citron-K or HIV-1 alone showed no significant effect on these compartments (Figure 6A–C). Citron-K and HIV-1, however, cooperatively enhanced the acidic late endosomal and lysosomal compartments (Figure 6D). The results from confocal microscopy were supported by fluorescence-activated cell sorter (FACS) analysis of the LysoTracker signal (Figure 6E). These findings suggest that citron-K interacts with HIV-1 to enhance activity of the late endosomal sorting pathway to promote HIV-1 virion production.
Figure 6. Citron-K and HIV-1 cooperatively enhance acidic (late) endosomal and lysosomal compartments.
293T cells were transfected with pNL4-3 and vector or citron-K. Cells were incubated with 60 μM LysoTracker for 90 min before fixation at ~48 h post-transfection. (A–D) Images of LysoTracker signals were collected via confocal microscopy. N, nucleus. Bars represent 7 μm. (E) Relative LysoTracker signals of 293T cells were measured by FACS analysis (average of four independent experiments). **P < 0.005.
In the endosomal sorting pathway, the MVB either fuses with the lysosome to degrade its contents or is directed to the plasma membrane where its contents are released via an exocytic pathway. Interestingly, HIV-1 virions have been shown to resemble exosomes (or exocytosed microvesicles) in both size and components (45–48). We hypothesized that citron-K modulates the exocytic pathway to promote HIV-1 virion production as well as MVB exocytosis. To test this, we purified microvesicles or exosomes secreted from cells transfected with HIV-1 proviral DNA in the presence and absence of citron-K. Our results indicate that citron-K enhanced the secretion of microvesicles with exosomal markers (i.e. hsc70, CD82 and LAMP-1), as well as HIV-1 virions, in HeLa (Figure 7A) and 293T cells (Figure 7C). The intracellular exosomal markers in the presence of citron-K and/or HIV-1 were not significantly altered (Figure 7B, D). We conclude that citron-K enhances HIV-1 virion production by promoting exocytosis of microvesicles/exosomes, including HIV-1 virions.
Figure 7. Citron-K enhances exocytosis.
(A and B) HeLa cells or (C and D) 293T cells were co-transfected with pNL4-3 and vector or citron-K. Supernatant and cells were collected 48 h post-transfection for analysis. The microvesicles and virion pellets in the supernatant were purified and analyzed for late endosomal/exosomal markers and HIV-1 Gag p24 (A and C). Exosomal markers analyzed include hsc70, LAMP-1 and CD82. Cell-associated proteins were analyzed for late endosomal/exosomal markers (B and D).
Discussion
We investigated the role of citron-K, a RhoA effector, in modulating HIV-1 replication. Ectopic expression of citron-K and the kinase-deficient splice variant citron-N preferentially enhanced HIV-1 virion production but not HIV-1 gene expression. Knockdown of endogenous citron-K by shRNA inhibited HIV-1 virion production, demonstrating an important role for endogenous citron-K in viral budding. Citron-K also enhanced MLV production and the HIV-1 L domain was not required for the enhancement. Citron-K forms cytoplasmic structures (44) and, when both HIV-1 Gag and citron-K are co-expressed, Gag is colocalized to these citron-K-induced cytoplasmic structures. Further analysis of the endosomal sorting pathway indicated that citron-K, together with HIV-1, enhanced the compartments of acidic late endosomes and lysosomes. We demonstrated that production of microvesicles or exosomes and HIV-1 virions were both enhanced by citron-K. These results suggest that citron-K is involved in modulating exocytosis and HIV-1 virion production.
RhoGTPases play a central role in the dynamic regulation of the actin cytoskeleton and through this, control cell morphology, motility and adhesion (49,50). RhoGTPases act locally to control individual trafficking events, but also act globally to control the spatial organization of membrane traffic in response to cues from the extracellular environment. In addition to interacting with the endocytic pathway to modulate clathrin-independent internalization from the cell surface (51,52), RhoGTPases are involved in mediating trafficking/sorting decisions at a number of distinct endocytic subcompartments. For example, RhoD is localized to both the plasma membrane and early endocytic vesicles, resulting in activation of an isoform of Diaphanous and tyrosine kinase c-Src (53). RhoB, which is highly homologous to RhoA, is localized to the plasma membrane and the bounding membrane of MVBs (54,55) and is activated by internalized epidermal growth factor receptors, as they enter the late endosomal compartment (56). These findings suggest that RhoGTPases interface the actin cytoskeleton with endocytic trafficking events. Activated RhoA mediates a number of effectors that modulate distinct cellular processes. For example, an undefined RhoA effector inhibits HIV-1 gene expression (38), making it difficult to evaluate the effect of activating citron-K specifically via RhoA on HIV-1 virion budding or production.
There are two endogenous forms of citron, the kinase form (citron-K) and the non-kinase form (citron-N). Citron-K is ubiquitously expressed in most cell types with cell cycle-dependent cellular localization (57). RhoA co-localizes with citron-K in the cell cortex during anaphase and both are enriched in the cleavage furrow and midbody during telophase (39). Citron-K modulates the contractile motion required for separation of the two daughter cells during cytokinesis through phosphorylation of the regulatory light chain of myosin II at both Ser-19 and Thr-18 (58). It is feasible that citron-K enhances virion budding from host cells in a similar process as cytokinesis. However, our observation that the kinase domain is not required to enhance HIV-1 virion production indicates that a distinct mechanism is involved.
Citron-N is a splice variant of citron-K lacking the kinase domain (59). Actin polymerization and cytoskeleton are regulated by RhoGTPases and are essential for the organization and dynamics of membrane organelles such as endosomes and the Golgi complex (40–42). Citron-N is enriched and associated with the Golgi apparatus of hippocampal neurons in culture (60). Suppression of citron-N or expression of a mutant lacking Rho-binding activity leads to dispersion of the Golgi apparatus (60), suggesting that citron-N functions as a scaffolding molecule on Golgi membranes, organizing Rho-mediated actin polymerization locally by assembling the actin polymerizing complexes together (ROCK II and profilin-IIa) (60). Citron-N also interacts with the postsynaptic scaffold protein, PSD-95/SAP-90, a member of the membrane-associated guanylate kinase protein family (MAGUK) (61,62). This interaction is important for neural plasticity and its localization at postsynaptic sites in all hippocampal neurons (61,62). Therefore, citron-N may play a role in organization of the endosomal sorting pathway and regulation of the actin cytoskeleton to facilitate exocytosis and viral release. However, neither monomeric nor polymerized actin appears to be altered by citron-K and/or HIV-1 in transfected 293T or HeLa cells (R. Loomis and L. Su, UNC-CH, Chapel Hill, NC, unpublished data).
We determined that the region of citron-K necessary for enhancement of virion production contained the leucine zipper, the RBD and the zinc finger domains. It has been speculated that the leucine zipper domain may be a scaffold for multimeric structures functioning through conformational changes and interactions with additional partners (59). The zinc finger (C6H2) in citron-K is believed to bind lipid second messengers or to recruit additional proteins (59). Taken together, citron-K may enhance virion production by binding or recruiting other cellular (perhaps endosomal sorting proteins) or viral components to facilitate viral release.
Recent evidence has shown that Tsg101, a component of ESCRT-I, binds specifically to the L domain of HIV-1 Gag to promote viral release (13–15). Small inhibitory RNA-mediated depletion of Tsg101 potently blocked HIV-1 release but not MLV release (14). We showed that citron-K enhanced virion production of MLV. Citron-K also enhanced VLP production of HIV-1 GagΔp6, which lacks the L domain. Therefore, this citron activity probably affects a step of HIV-1 viral release that is distinct from the Tsg101-dependent step. Dominant-negative mutants of Vps4 inhibited viral release of both HIV-1 and MLV, indicating the essential role of the late endosomal pathway in mediating retroviral release (14). Co-expression of dominant-negative Vps4 mutants also blocked HIV-1 virion production in the absence or presence of citron-K (R. Loomis and L. Su, UNC-CH, Chapel Hill, NC, unpublished data). Therefore, citron-K enhanced virion production is still dependent on the late endosomal sorting pathway.
HIV-1 can bud from both the plasma membrane and internal endosomal membranes (14,18,43,63–67). Citron-K may either recruit late endosomal factors to the plasma membrane or promote utilization of the endosomal pathway for virion production. Cells transfected with both citron-K and HIV-1 lead to increased late endosome and lysosome compartments compared to cells transfected with HIV-1 or citron-K alone. Based on the localization of citron-K and colocalization of Gag, it is likely that citron-K promotes a step late in the endosomal sorting pathway to enhance virion production. HIV-1 virions share similar characteristics with exocytosed membrane microvesicles or exosomes (66,68). They are similar in size and contain exosome-specific markers, as well as common cellular membrane proteins. Although exosome production occurs more prominently in macrophages, B cells and dendritic cells, recent evidence indicates that the exosomal pathway operates in most blood cell types, including T cells (69). Our data suggest that citron-K is involved in modulating the exocytosis process to enhance HIV-1 virion production. In addition, HIV-1 may interact functionally with citron-K to modulate endosomal sorting and HIV-1 virion release.
Remarkably, citron-K−/− mice are viable, although they grow at slower rates, are severely ataxic and die before adulthood as a consequence of fatal seizures (70). Since knockdown of endogenous citron-K expression in 293T and T cells inhibited HIV-1 virion production, citron-K may provide a new host target for the development of anti-HIV-1 therapeutics. In addition to affecting HIV-1 viral production, the altered release of exosomes/microvesicles may also have significant immunological consequences, as exosomes have been shown to function as antigen-presenting cells to interact with T cells (66,68,69,71).
Materials and Methods
Reagents, plasmids and cell lines
The pNL4-3 plasmid encodes the entire HIV-1 genome DNA in pUC18 (24). The pNL4.Luc.R-E plasmid was obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program (72). The pNL4GFP plasmid encodes Gag, Pol, tat and rev and was kindly provided by Dr Dan Littman, NYU, NY, NY. pCAG (vector), pCAG/citron-K, pCAG/citronΔ1 plasmids were previously reported (57). All citron truncation mutants were generated by polymerase chain reaction of the citron-N template with an N-terminal Myc-tag added and inserted into a retroviral vector, pHSCG (73). The control (pSUPER.Retro.Puro; OligoEngine, Seattle, WA, USA) and citron short-hairpin constructs (cit A, cit B and cit C) were a kind gift from Dr Alan Hall, University College London, London, UK. Gag and GagΔp6 constructs were a kind gift from Dr Paul Bieniasz, Aaron Diamond AIDS Research Center, NY, NY (43) and GagGFP was a kind gift from Dr Marilyn Resh, Sloan-Kettering Institute, NY, NY.
The 293T, HeLa and HeLa-MAGI cells [NIH AIDS Research and Reference Reagent Program; (73)] were maintained in DMEM supplemented with 10% FBS, penicillin/streptomycin and L-glutamine. Jurkat T cells were maintained in RPMI-1640 supplemented with 10% FBS, penicillin/streptomycin and L-glutamine. The NIH-3T3 cells were maintained in DMEM supplemented with 10% FBS, penicillin/streptomycin and L-glutamine.
HIV-1 production and replication in transfected human cells
Transient production of HIV-1 was performed by transfecting NL4-3 (0.1 μg) with pCAG, pCAG/citron-K, pCAG/citronΔ1 or citron truncation plasmids (0.2 μg) in 293T cells in 48-well plates at 2.5 × 104 cells/well using Effectene (Qiagen, Santa Clarita, CA, USA) or NL4-3 (0.3 μg) with pCAG, pCAG/citron-K, pCAG/citronΔ1 or citron truncation plasmids (0.6 μg) in Jurkat T cells in 48-well plates at 4 × 105 cells/well using GenePorter (Gene Therapy Systems, San Diego, CA, USA). At 48 h post-transfection, HIV-1 virions in cell supernatant were measured by p24 or RT assays (74) and infectious units were determined by titering the supernatant on HeLa-CD4-LTR-lacZ cells (MAGI) as described previously (73).
To analyze HIV-1 gene expression in transfected cells, pNL4.Luc (0.1 μg) was co-transfected into 2.5 × 104 293T cells/well together with pCAG, pCAG/citron-K, pCAG/citronΔ1 or citron truncation plasmids (0.2 μg) or pNL4.Luc (0.3 μg) was co-transfected into 4 × 105 Jurkat T cells/well with pCAG, pCAG/citron-K, pCAG/citronΔ1 or citron truncation plasmids (0.6 μg). Cell extracts were analyzed at 48 h after transfection for luciferase activity (EG&G berthold, Oak Ridge, TN and FluoStar Optima; BMG LabTech, Durham, NC). In some experiments, we used pGag, pGagΔp6 or pNL4GFP, in place of NL4-3 or pNL4.Luc.
Western blotting
Transfected 293T or Jurkat T cells were lysed and resolved (50–100 μg of total protein) on a 10% SDS–PAGE. Gels were transferred to polyvinylidene fluoride (PVDF) membranes (Amersham Biosciences, Piscataway, NJ) and blocked with 5% non-fat dry milk. The membrane was probed with mouse anti-p24 (NIH AIDS Research and Reference Reagent Program), mouse anti-Myc (AbCam, Cambridge, MA, USA), rabbit anti-citron [a kind gift from Dr Lola Reid, UNC-CH, as described in (75)], mouse anti-actin (Sigma-Aldrich, St Louis, MO), mouse anti-tubulin, mouse anti-hsc70 (BD Biosciences, San Jose, CA), mouse anti-LAMP1 (Santa Cruz, Santa Cruz, CA) or mouse anti-CD82 (BD Biosciences, San Jose, CA) specific antibodies and visualized using an enhanced chemiluminescence Kit (Amersham Biosciences, Piscataway, NJ).
shRNA knockdown of citron-K
The citron-K shRNA and control constructs are in the pSUPER.Retro.Puro vector from OligoEngine. The citron-K shRNA constructs target different regions of citron-K. Cit A RNA targets the C-terminus of citron-K beginning at base pair 6085, Cit B and Cit C RNA target the N-terminus of citron-K beginning at base pair 359 and 244, respectively, (Figure S1). For experiments in 293T cells, 293T cells were plated in a 24-well plate at 5 × 104 cells/well and transfected with 0.4 μg control or citron-targeted shRNA constructs using Effectene. Twenty-four hours later, another transfection was performed with 0.1 μg pNL4GFP and 0.2 μg control or citron-targeted shRNA constructs. Two days after the second transfection, supernatants and cells were harvested for further analysis. For experiments in Jurkat T cells, 0.3 μg of pNL4GFP and 0.6 μg of control or citron-targeted shRNA constructs were transfected via GenePorter into 4 × 105 cells/well. Supernatants and cells were harvested at 48 h post-transfection for further analysis.
MLV production in transfected human cells
The 293T cells were plated in a 6-well plate at 4 × 105 cells/well and transfected with 2 μg HSCG (a retroviral vector with MSCV LTR, 34), 1.5 μg vesicular stomatitis virus-glycoprotein (VSV-G), 1.5 μg murine leukemia virus (MLV) Gag-Pol and 2 μg vector or citron-K DNA using the calcium phosphate transfection method as described previously (76,77). Retrovirus was harvested 48 h after transfection. Virus was titered by seeding NIH-3T3 cells at 5 × 104 cells/well in a 24-well plate, adding serial dilution of virus and counting GFP-positive cells 48 h after infection on the Guava EasyCyte (Guava Technologies, Hayward, CA).
Confocal microscopy
The 293T cells were plated on coverslips at 1 × 105 cells/well in a 24-well plate and transfected as described previously (Effectene, Qiagen). For the GagGFP confocal experiments, we used 0.2 μg of GagGFP, 0.02 μg vector, 0.4 μg vector or citron-K and for the samples with wild-type Gag – 0.2 μg of GagGFP, 0.02 μg of Gag and 0.4 μg vector or citron-K were used in the transfections. At 48 h post-transfection, cells were fixed with 4% paraformaldehyde for 15 min at 4°C, washed once with cold 1×PBS, permeabilized with ice-cold methanol for 30 seconds, washed once with cold 1×PBS, washed three times with Quench Buffer (1% milk, 150 mM NaOAc pH 7, 1×PBS), washed three times with wash buffer (1% milk, 1×PBS), incubated with primary antibody for 1 h at room temperature, washed three times with wash buffer, incubated with secondary antibody for 1 h at room temperature, washed five times with wash buffer and mounted on slide with polyvinyl alcohol. The primary antibody used was chicken anti-Myc (Molecular Probes, Carlsbad, CA) at 1:1000 dilution. The secondary antibody used was anti-chicken-Cy5 (Molecular Probes, Carlsbad, CA) at 1:200 dilution.
For LysoTracker experiment, cells were incubated with 60 μM LysoTracker (Molecular Probes, Carlsbad, CA) for 90 min then fixed with 4% paraformaldehyde 48 h post-transfection. Images were collected using Olympus confocal microscope and software.
FACS
Each sample (~5 × 105 cells) that had been incubated with 60 μM LysoTracker (Molecular Probes, Carlsbad, CA) for 90 min were washed in PBS with 2% FBS and stained with 7AAD (1 μL/mL) for 15 min on ice, then rinsed and resuspended in PBS with 2% FBS. Cells were analyzed using FACScan (Becton Dickinson, San Jose, CA).
Purification of exosomal/microvesicles/HIV-1 virions
Transfections were performed as described previously (Effectene, Qiagen), except in 100-mm dishes using 1 × 106 cells/plate of 293T cells or 5 × 105 cells/plate of HeLa cells. The FBS used to culture the cells was depleted of exosomes or microvesicles by spinning at 100 000 × g overnight at 4°C. Supernatant (10 mL) were collected at 48 h post-transfection. The exosome purification protocol is similar to those described previously (66,71). At the time of collection, supernatants were spun at 800 × g for 10 min to remove cells and large cell debris. Supernatants were then added to Beckman Ultra Clear Centrifuges (11 × 89 mm) with 2 mL 25% sucrose. Tubes were spun in the Beckman L7 Ultracentrifuge at 100 000 × g for 2 h at 4°C. Supernatant was discarded and exosomal pellet was resuspended in 1% Triton-X-100 and analyzed by SDS–PAGE.
Supplementary Material
All citron-K shRNA constructs were made in the pSUPER.Retro.Puro vector. The start site base pairs are from the beginning of the coding sequence. Italics, target sequence, Underline, hairpin.
(A and B) Expression of citron constructs was verified by analyzing cell-associated proteins with anti-Myc, anti-p24 and anti-actin antibodies. All citron constructs are Myc-tagged at the N-terminus.
Acknowledgments
We thank Dr Lola Reid (UNC-CH) for the citron antibody, Dr Robert Bagnell and Vicki Madsen (UNC-CH) for help with confocal microscopy, Robin Hunt, Robert Hales and Dedeke Brouwer for technical support; Drs JoAnn Trejo, Ron Swanstrom and Blossom Damania (UNC-CH) for helpful discussions. We also thank Drs Paul Bieniasz (Aaron Diamond AIDS Research Center), Alan Hall (University College London), Chris Aiken (Vanderbilt University), Wesley Sundquist (University of Utah) and Marilyn Resh (Sloan-Kettering Institute) for reagents and discussions. This work was supported by NIH grants to L. S. (AI/GM 48407 and CA92240).
Footnotes
Supplemental materials are available as part of the online article at http://www.blackwell-synergy.com
References
- 1.Fauci AS. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 2.Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 3.Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 4.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiang Y, Cameron CE, Wills JW, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76Gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan B, Li X, Goff SP. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puffer BA, Watkins SC, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, Wilson CB, Puffer BA, Montelaro RC, Wills JW. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Strack B, Calistri A, Accola MA, Palu G, Gottlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci U S A. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc Natl Acad Sci U S A. 2001;98:7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 16.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002;99:955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strack B, Calistri A, Craig S, Popova E, Gottliner HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 18.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung H-Y, Morita E, Wang HE, Cimbora DM, Scott A, Krausslich H-G, Kaplan J, Morham SG, Sundquist WI. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 19.Bishop N, Horman A, Woodman P. Mammalian class E vps proteins recognize ubiquitin and act in the removal of endosomal protein-ubiquitin conjugates. J Cell Biol. 2002;57:91–101. doi: 10.1083/jcb.200112080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 21.Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop N, Woodman P. TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem. 2001;276:11735–11742. doi: 10.1074/jbc.M009863200. [DOI] [PubMed] [Google Scholar]
- 23.Lemmon SK, Traub LM. Sorting in the endosomal system in yeast and animal cells. Curr Opin Cell Biol. 2000;12:457–466. doi: 10.1016/s0955-0674(00)00117-4. [DOI] [PubMed] [Google Scholar]
- 24.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- 26.Babst M, Katzmann DJ, Snyder WB, Wendland B. Emr SD Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 27.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 28.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 29.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 30.Khosravi-Far R, Solski PA, Clark GJ, Kinch MS, Der CJ. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson MF, Ashworth A, Hall A. An essential role for Rho, Rac, and Cdc42 GTPases in cell cycle progression through G1. Science. 1995;269:1270–1272. doi: 10.1126/science.7652575. [DOI] [PubMed] [Google Scholar]
- 32.Perona R, Montaner S, Saniger L, Sanchez-Perez I, Bravo R, Lacal JC. Activation of the nuclear factor-kappaB by Rho, CDC42, and Rac-1 proteins. Genes Dev. 1997;11:463–475. doi: 10.1101/gad.11.4.463. [DOI] [PubMed] [Google Scholar]
- 33.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 34.Coffield VM, Helms WS, Jiang Q, Su L. Galpha13 mediates a signal that is essential for proliferation and survival of thymocyte progenitors. J Exp Med. 2004;200:1315–1324. doi: 10.1084/jem.20040944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galandrini R, Henning SW, Cantrell DA. Different functions of the GTPase Rho in prothymocytes and late pre-T cells. Immunity. 1997;7:163–174. doi: 10.1016/s1074-7613(00)80519-1. [DOI] [PubMed] [Google Scholar]
- 36.Henning SW, Galandrini R, Hall A, Cantrell DA. The GTPase Rho has a critical regulatory role in thymus development. EMBO J. 1997;16:2397–2407. doi: 10.1093/emboj/16.9.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, Wang L, Kao S, Whitehead IP, Hart MJ, Liu B, Duus K, Burridge K, Der CJ, Su L. Functional interaction between the cytoplasmic leucine-zipper domain of HIV-1 gp41 and p115-RhoGEF. Curr Biol. 1999;9:1271–1274. doi: 10.1016/s0960-9822(99)80511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, Zhang H, Solski PA, Hart MJ, Der CJ, Su L. Modulation of HIV-1 Replication by a novel RhoA effector activity. J Immunol. 2000;164:5369–5374. doi: 10.4049/jimmunol.164.10.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eda M, Yonemura S, Kato T, Watanabe N, Ishizaki T, Maduale P, Narumiya S. Rho-dependent transfer of Citron-kinase to the cleavage furrow of dividing cells. J Cell Sci. 2001;114:3273–3284. doi: 10.1242/jcs.114.18.3273. [DOI] [PubMed] [Google Scholar]
- 40.Allan VJ, Schroer TA. Membrane motors. Curr Opin Cell Biol. 1999;11:476–482. doi: 10.1016/s0955-0674(99)80068-4. [DOI] [PubMed] [Google Scholar]
- 41.Allan VJ, Thompson HM, McNiven MA. Motoring around the Golgi. Nat Cell Biol. 2002;4:E236–242. doi: 10.1038/ncb1002-e236. [DOI] [PubMed] [Google Scholar]
- 42.di Campli A. Morphological changes in the Golgi complex correlate with actin cytoskeleton rearrangements. Cell Motil Cytoskeleton. 1999;43:334–348. doi: 10.1002/(SICI)1097-0169(1999)43:4<334::AID-CM6>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 43.Martin-Serrano J, Zang T, Bieniasz PD. Role of ESCRT-1 in retroviral budding. J Virol. 2003;77:4794–4804. doi: 10.1128/JVI.77.8.4794-4804.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Di Cunto F, Calautti E, Hsiao J, Ong L, Topley G, Turco E, Dotto GP. Citron Rho-interacting kinase, a novel tissue-specific Ser/Thr kinase encompassing the Rho-Rac-binding protein citron. J Biol Chem. 1998;273:29706–29711. doi: 10.1074/jbc.273.45.29706. [DOI] [PubMed] [Google Scholar]
- 45.Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Couzin J. Cell biology: The ins and outs of exosomes. Science. 2005;308:1862–1863. doi: 10.1126/science.308.5730.1862. [DOI] [PubMed] [Google Scholar]
- 47.Kramer B, Pelchen-Matthews A, Deneka M, Garcia E, Piguet V, Marsh M. HIV interaction with endosomes in macrophages and dendritic cells. Blood Cells Mol Dis. 2005;35:136–142. doi: 10.1016/j.bcmd.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen DG, Booth A, Gould SJ, Hildreth JE. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- 49.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 50.Ridley AJ. Rho GTPases and cell migration. J Cell Sci. 2001;114:2713–2722. doi: 10.1242/jcs.114.15.2713. [DOI] [PubMed] [Google Scholar]
- 51.Lamaze C, Dujeancourt A, Baba T, Lo CG, Benmerah A, Dautry-Varsat A. Interleukin 2 receptors and detergent-resistant membrane domains define a clathrin-independent endocytic pathway. Mol Cell. 2001;7:661–671. doi: 10.1016/s1097-2765(01)00212-x. [DOI] [PubMed] [Google Scholar]
- 52.Sabharanjak S, Sharma P, Parton RG, Mayor S. GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev Cell. 2002;2:411–423. doi: 10.1016/s1534-5807(02)00145-4. [DOI] [PubMed] [Google Scholar]
- 53.Murphy C, Saffrich R, Grummt M, Gournier H, Rybin V, Rubino M, Auvinen P, Lutcke A, Parton RG, Zerial M. Endosome dynamics regulated by a Rho protein. Nature. 1996;384:427–432. doi: 10.1038/384427a0. [DOI] [PubMed] [Google Scholar]
- 54.Adamson P, Paterson HF, Hall A. Intracellular localization of the P21rho proteins. J Cell Biol. 1992;119:617–627. doi: 10.1083/jcb.119.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robertson D, Paterson HF, Adamson P, Hall A, Monaghan P. Ultra-structural localization of ras-related proteins using epitope-tagged plasmids. J Histochem Cytochem. 1995;43:471–480. doi: 10.1177/43.5.7537292. [DOI] [PubMed] [Google Scholar]
- 56.Gampel A, Mellor H. Small interfering RNAs as a tool to assign Rho GTPase exchange-factor function in vivo. Biochem J. 2002;366:393–398. doi: 10.1042/BJ20020844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maduale P, Eda M, Watanabe N, Fujisawa K, Matsuoka T, Bito H, Ishizaki T, Narumiya S. Role of citron kinase as a target of the small GTPase Rho in cytokinesis. Nature. 1998;394:491–494. doi: 10.1038/28873. [DOI] [PubMed] [Google Scholar]
- 58.Yamashiro S, Totsukawa G, Yamakita Y, Sasaki Y, Maduale P, Ishizaki T, Narumiya S, Matsumura F. Citron kinase, a Rho-dependent kinase, induces di-phosphorylation of regulatory light chain of Myosin II. Mol Biol Cell. 2003;14:1745–1756. doi: 10.1091/mbc.E02-07-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maduale P, Furuyashiki T, Reid T, Ishizaki T, Watanabe G, Morii N, Narumiya S. A novel partner for the GTP-bound forms of rho and rac. FEBS. 1995;377:243–248. doi: 10.1016/0014-5793(95)01351-2. [DOI] [PubMed] [Google Scholar]
- 60.Camera P, Da Silva JS, Griffiths G, Giuffrida MMG, Ferrara L, Schubert V, Imarisio S, Silengo L, Dotti CG, Di Cunto F. Citron-N is a neuronal Rho-associated protein involved in Golgi organization through actin cytoskeleton regulation. Nat Cell Biol. 2003;5:1071–1078. doi: 10.1038/ncb1064. [DOI] [PubMed] [Google Scholar]
- 61.Furuyashiki T, Fujisawa K, Fujita A, Maduale P, Uchino S, Mishina M, Bito H, Narumiya S. Citron, a Rho-Target, interacts with PSD-95/SAP-90 at glutamatergic synapses in the thalamus. J Neurosci. 1999;19:109–118. doi: 10.1523/JNEUROSCI.19-01-00109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang W, Vazquesz L, Apperson M, Kennedy MB. Citron binds to PSD-95 at glutamatergic synapses on inhibitory neurons in the hippocampus. J Neurosci. 1999;19:96–108. doi: 10.1523/JNEUROSCI.19-01-00096.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Conibear E. An ESCRT into the endosome. Mol Cell. 2002;10:215–216. doi: 10.1016/s1097-2765(02)00601-9. [DOI] [PubMed] [Google Scholar]
- 64.Freed E. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 2003;11:56–59. doi: 10.1016/s0966-842x(02)00013-6. [DOI] [PubMed] [Google Scholar]
- 65.Freed EO. Viral late domains. J Virol. 2002;76:4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen DG, Booth A, Gould SJ, Hildreth JEK. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J Biol Chem. 2003;278:52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- 67.Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 2002;3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 68.Pelchen-Matthews A, Kramer B, Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol. 2003;162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thery C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 70.Di Cunto F, Imarisio S, Hirsch E, Broccoli B, Bulfone A, Migheli A, Atzori C, Turco E, Triolo R, Dotto GP, Silengo L, Altruda F. Defective neurogenesis in citron kinase knockout mice by altered cytokinesis and massive apoptosis. Neuron. 2000;28:115–127. doi: 10.1016/s0896-6273(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 71.Wubbolts R, Leckie RS, Veenhuizen PTM, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot J-W, Geuze HJ, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. J Biol Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 72.He J, Landau NR. Use of a novel human immunodeficiency virus type 1 reporter virus expressing human placental alkaline phosphatase to detect an alternative viral receptor. J Virol. 1995;69:4587–4592. doi: 10.1128/jvi.69.7.4587-4592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su L, Kaneshima H, Bonyhadi M, Salimi S, Kraft D, Rabin L, McCune JM. HIV-1-induced thymocyte depletion is associated with indirect cytopathogenicity and infection of progenitor cells in vivo. Immunity. 1995;2:25–36. doi: 10.1016/1074-7613(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 75.Liu H, Di Cunto F, Imarisio S, Reid LM. Citron kinase is a cell cycle-dependent, nuclear protein required for G2/M transition of hepatocytes. J Biol Chem. 2003;278:2541–2548. doi: 10.1074/jbc.M210391200. [DOI] [PubMed] [Google Scholar]
- 76.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Coffield VM, Jiang Q, Su L. A genetic approach to inactivating chemokine receptors using a modified viral protein. Nat Biotechnol. 2003;21:1321–1327. doi: 10.1038/nbt889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All citron-K shRNA constructs were made in the pSUPER.Retro.Puro vector. The start site base pairs are from the beginning of the coding sequence. Italics, target sequence, Underline, hairpin.
(A and B) Expression of citron constructs was verified by analyzing cell-associated proteins with anti-Myc, anti-p24 and anti-actin antibodies. All citron constructs are Myc-tagged at the N-terminus.