Nucleophosmin 1 gene (NPM1) mutations are found in approximately 50% of cytogenetically normal acute myeloid leukemia (CN-AML).1 NPM1-mutated CN-AML is one provisional entity of the revised World Health Organization (WHO) 2008 classification. Numerous studies have consistently established that NPM1 mutations are associated with a better outcome. Internal tandem duplications (ITDs) of the FLT3 gene are also frequent in CN-AML, but associated with a poor prognosis. Due to their high frequencies and consistent impacts, both NPM1 mutations and FLT3-ITDs contribute to the European LeukemiaNet (ELN) classification of CN-AML.2 Nonetheless, these observations have been made before the description of other mutations also frequently found in CN-AML and potentially capable to alter patient outcome. This includes epigenetic IDH,3,4 TET2,5 and DNMT3A gene mutations,6–8 all found in 20%–30% of CN-AML cases. With the aim of assessing the role of these additional mutations, we performed a multivariable analysis in a cohort of 393 adult patients of all ages with NPM1-mutated CN-AML intensively treated in consecutive trials from the Acute Leukemia French Association (ALFA).
Cytogenetic results were centrally reviewed. Normal karyotype was defined as 20 or more normal metaphases without abnormal metaphases. Detection of exon 12 NPM1 mutation was centrally performed on genomic DNA by polymerase chain reaction (PCR) and fragment analysis, as described. In addition, the following gene mutations were centrally screened using standard-PCR methods and Sanger sequencing: FLT3-ITD, FLT3-TKD (D835/I836), IDH1-R132, IDH2-R172, IDH2-R140, TET2 and DNMT3A (exons 8–9, 11–23). We analyzed recurrent R882 mutations and all other DNMT3A mutations apart, as it was suggested their biology and clinical impacts may differ.7,8 Patients were treated between 1999 and 2012 in the ALFA-9801 (clinicaltrials.gov identifier 00931138),9 ALFA-9802 (clinicaltrials.gov identifier 00880243),10 ALFA-9803 (clinicaltrials.gov identifier 00363025),11 ALFA-0701 (clinicaltrials.gov identifier 00927498) trials,12 and the unpublished ALFA-0702 trial (clinicaltrials.gov identifier 00932412). Trials were conducted in accordance with the Declaration of Helsinki. All patients gave informed consent for treatment and mutation screening. Approval was obtained from the ethical review boards of the participating institutions.
Primary end point was cumulative incidence of relapse (CIR). Other end points were complete remission (CR) achievement, relapse-free (RFS) and overall survival (OS). Failure time data were analyzed without censoring patients who received allogeneic stem cell transplant (SCT). A sensitivity analysis was also performed after censoring patients transplanted in first CR at date of SCT. RFS and OS were estimated by the Kaplan-Meier method and compared by the log rank test. CIR was estimated taking into account death in first CR as a competing risk. Stepwise logistic regression and Cox models were used to analyze associations between base-line characteristics and outcome. Cause-specific hazard ratios (HRs) were given as measures of association between each variable and CIR. The following mutations entered the models: FLT3-ITD, FLT3-TKD, IDH1-R132 mutations, IDH2-R140 mutation, DNMT3A-R882 mutation, other DNMT3A mutations, and TET2 mutations in the subset of 232 patients tested for TET2. Analysis was performed in the whole population, then repeated in patients not presenting FLT3-ITD. We estimated missing data for mutations by using 50 multiple imputations in chain equations incorporating predictive mean matching. In addition to the molecular markers, age (using a 60-year cutoff) and the logarithm of white blood cell count (WBC) were added to all models, which were also adjusted on treatment protocol. Hazard ratios and cause-specific HRs were given with 95% confidence interval (CI). Statistical analysis was performed on the STATA/IC 12.1 software package (StataCorp, College Station, TX, USA).
The patient population was heterogeneous with regard to age and treatment. Median age was 52.8 years (range 17–79). Median WBC was 23.5×109/L (range 0.8–377), with 171 males and 222 females. Distribution across ALFA protocols was: 23%, 23%, 5%, 21% and 28% in the ALFA 9801, 9802, 9803, 0701 and 0702, respectively. Median follow up was 2.2 years. Most patients were tested for FLT3, DNMT3A, and IDH gene mutations, while only 232 (57%) were tested for TET2 gene mutations. The lack of available material was the main reason for not testing all patients for all mutations. Incidences of gene mutations are indicated in Table 1. Notably, no patient had IDH2-R172 mutation. Coexistence of gene mutations is illustrated in Figure 1. A higher WBC was associated with FLT3-ITD, TET2 and IDH2-R140 mutations (P<0.001, P=0.002, and P<0.001, respectively). Non-R882 DNMT3A mutations were more frequently observed in older patients (P =0.004), found for instance in 24% of patients aged 60 years or over versus 12.5% in younger patients.
Table 1.
Associated gene mutations (n=393 patients).
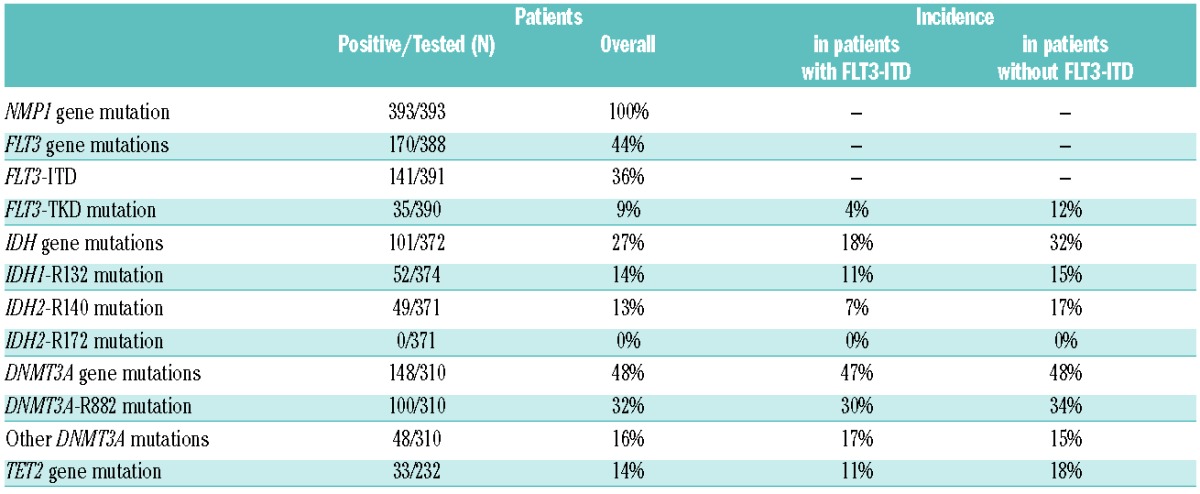
Figure 1.
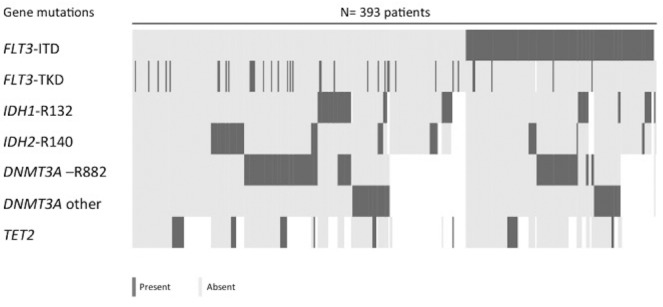
Co-existence of associated gene mutations (n=393 patients). In patients with FLT3-ITD, FLT3-TKD and IDH2-R140 mutations were less frequently observed (P=0.005 and 0.004, respectively). Patients with IDH2-R140 mutation presented less frequently IDH1-R132 and DNMT3A-R882 mutations (P=0.001 and 0.011, respectively). No patients had both DNMT3A-R882 and non-R882 DNMT3A mutations (P<0.001). No patients had both IDH1-R132 and TET2 mutations (P=0.030), while 5 patients had both IDH2-R140 and TET2 mutations.
Overall CR rate was 91% (359 of 393 patients). Among CR patients, 146 relapsed and 111 died, including 19 deaths in first CR. A total of 69 patients (18%) received allogeneic SCT in first CR. At three years, CIR was estimated at 53% (95%CI: 47–60) in the entire patient cohort, while RFS and OS were respectively estimated at 47% (95%CI: 41–53) and 56% (95%CI: 49–61). Advanced age was the only factor associated with a lower CR rate in multivariable analysis [HR 4.63 (95%CI: 2.13–10.07) for patients aged 60 years or over; P<0.001], while associated gene mutations did not influence CR achievement. CR rate was 81% in the 108 patients aged 60 years or over versus 95% in the 285 younger patients.
In a first analysis not considering TET2 status, advanced age, FLT3-ITD and non-R882 DNMT3A mutations were independently associated with a higher hazard of relapse. At three years, CIR was estimated at 66% (95%CI: 54–78) in patients aged 60 years or over versus 49% (95%CI: 42–57) in younger patients (specific HR 2.04 [95%CI: 1.44–2.90]; P<0.001). Specific HRs for FLT3-ITD and non-R882 DNMT3A mutations were 1.73 (95%CI: 1.24–2.43; P=0.001) and 2.06 (95%CI: 1.28–3.32; P=0.003), respectively. WBC did not influence CIR in this multivariable setting. In a second analysis performed in the subset of patients tested for TET2 mutations, age, FLT3-ITD and non-R882 DNMT3A mutation, but not TET2 mutation, were still identified as independent factors. These three variables were also identified as the variables that influenced RFS in multivariable analysis. Again, TET2 mutation did not impact RFS. Results were roughly unchanged when the 69 patients transplanted in first CR were censored at SCT time.
Repeating these analyses in the subset of patients without FLT3-ITD confirmed the bad-prognosis value of non- R882 DNMT3A mutations in these patients, for CIR [specific HR 1.95 (95%CI: 1.02–3.70); P=0.042] as well as for RFS. This analysis also evidenced IDH1-R132 mutation, but not IDH2-R140 mutation, as poor prognostic factor in these patients, for CIR [specific HR, 1.74 (95%CI: 1.02–2.97); P=0.041] as well as for RFS. Again, TET2 mutation was not shown to significantly impact the outcome of these patients and results were similar when the 69 patients transplanted in first CR were censored at time of SCT.
Overall, these results indicated that, among patients with NPM1-mutated CN-AML, not only FLT3-ITD, but also non-R882 DNMT3A and IDH1-R132 mutation could represent additional high-risk genetic alterations. This is important as these patients are currently considered to be favorable-risk patients in the current ELN classification if no FLT3-ITD (Figure 2). As shown in Figure 2A, if FLT3-ITD mutations were indeed associated with an increased incidence of early relapses, the two other types of mutations were associated with an increased incidence of later relapses. This translated into a shorter RFS in the latter subsets as well (Figure 2B). However, due to a relatively good salvage rate and post-relapse survival (data not shown), this did not translate into a significantly shorter OS for patients with non-R882 DNMT3A or IDH1-R132 mutations as it did for patients with FLT3-ITD (Figure 2C).
Figure 2.
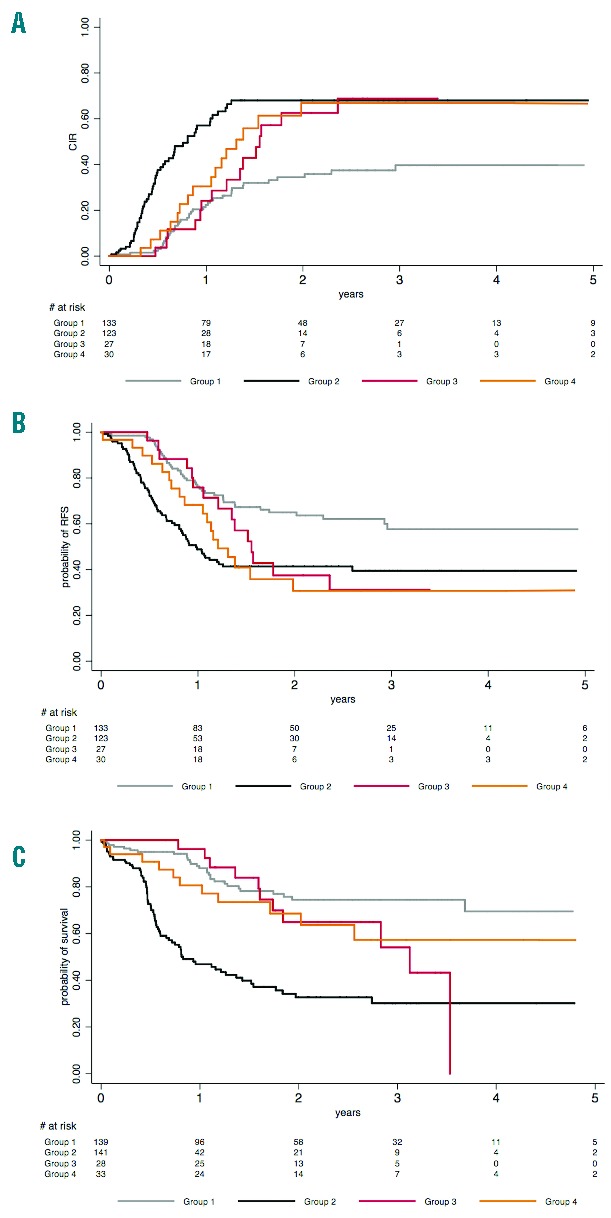
Effects of FLT3-ITD, non-R882 DNMT3A mutation and IDH1-R132 mutation in patients with de novo NPM1-mutated cytogenetically normal acute myeloid leukemia. The patient cohort was divided here into four distinct groups: 1) patients without FLT3-ITD, non-R882 DNMT3A mutation, nor IDH1-R132 mutation (group 1, 41% of the patients); 2) patients with FLT3-ITD (group 2, 41% of the patients); 3) patients without FLT3-ITD but non-R882 DNMT3A mutation (group 3, 8% of the patients); and 4) patients without FLT3-ITD but with IDH1-R132 mutation (group 4, 10% of the patients). Note that 3 patients with both non-R882 DNMT3A and IDH1-R132 mutations were classified within group 3. (A) 2-year cumulative incidence of relapse (CIR) was 34% (95%CI: 26–45) in group 1 versus 68% (95%CI: 57–78), 63% (95%CI: 43–82) and 67% (95%CI: 47–85) in groups 2, 3 and 4, respectively. As compared to group 1 patients, specific HR for group 2, 3 and 4 patients were 3.18 (95%CI: 2.11–4.77; P<0.001), 1.35 (95%CI: 1.00–1.81; P=0.05) and 1.27 (95%CI: 1.04–1.54; P=0.017), respectively. (B) 2-year relapse-free survival (RFS) was 65% (95%CI: 55–73) in group 1 versus 41% (95%CI: 32–50), 37% (95%CI: 18–57) and 31% (95%CI: 14–49) in groups 2, 3 and 4, respectively. As compared to group 1 patients, HR for group 2, 3 and 4 patients were 2.25 (95%CI: 1.54–3.29; P<0.001), 1.33 (95%CI: 0.99–1.78; P =0.059) and 1.28 (95%CI: 1.07–1.54; P=0.008), respectively. (C) 2-year overall survival (OS) was 74% (95%CI: 65–82) in group 1 versus 33% (95%CI: 23–42), 65% (95%CI: 42–81) and 69% (95%CI: 48–83) in groups 2, 3 and 4, respectively. As compared to group 1 patients, HR for group 2, 3 and 4 patients were 4.28 (95%CI: 2.77–6.60; P<0.001), 1.32 (95%CI: 0.93–1.87; P=0.12) and 1.17 (95%CI: 0.93–1.48; P=0.18), respectively.
Reviewing the literature, the Cancer and Leukemia Group B (CALGB) and the German AML Study Group have already reported an unfavorable impact of IDH1-R132 or IDH1/2 mutations in general in similar favorable-ELN patients.3,4 In contrast, the 16-gene analysis from the Eastern Cooperative Oncology Group (ECOG), identification of an IDH1/2 mutation was associated with a better prognosis in these patients.13 Nonetheless, in a recent meta-analysis, IDH1-mutated patients had an inferior event-free survival, especially in the subset with NPM1 mutation but no FLT3-ITD.14 It has also been reported that DNMT3A mutations may negatively influence the survival of patients with CN-AML,7,8 but up till now not specifically in those with the favorable NPM1-mutated/no FLT3-ITD genotype. As in our study, different prognostic values of R882 versus non-R882 mutations have been observed in two studies.7,8 In one of these, only non-R882 mutations were associated with an increased risk of relapse in younger patients with CN-AML.7
To conclude, there is accumulating evidence, including the observations made in the present study, to suggest that taking into account epigenetic non-R882 DNMT3A and IDH1-R132 gene mutations might help refine the current ELN classification, even if some discrepancies are observed from one study to another. The most striking observation of our study was the occurrence of delayed relapses in NPM1-mutated CN-AML patients without FLT3-ITD but with non-R882 DNMT3A or IDH1-R132 mutations, as compared to earlier relapses in NPM1-mutated CN-AML patients with FLT3-ITD. This observation fits nicely with the recent reports showing that IDH, TET2, or DNMT3A mutations may precede the appearance of NPM1 mutations in CN-AML, be present in pre-leukemic stem cells, and even to persist in hematologic remission in numerous cases.15
Acknowledgments
We thank all participating investigators from the ALFA-9801, ALFA-9802, ALFA-9803, ALFA-0701 and ALFA-0702 protocols.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Falini B, Mecucci C, Tiacci E, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. [DOI] [PubMed] [Google Scholar]
- 2.Dohner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. [DOI] [PubMed] [Google Scholar]
- 3.Marcucci G, Maharry K, Wu YZ, et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(14):2348–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. [DOI] [PubMed] [Google Scholar]
- 5.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–2301. [DOI] [PubMed] [Google Scholar]
- 6.Renneville A, Boissel N, Nibourel O, et al. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: a study by the Acute Leukemia French Association. Leukemia. 2012;26(6):1247–1254. [DOI] [PubMed] [Google Scholar]
- 7.Marcucci G, Metzeler KH, Schwind S, et al. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. J Clin Oncol. 2012;30(7):742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaidzik VI, Schlenk RF, Paschka P, et al. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG). Blood. 2013; 121(23):4769–4777. [DOI] [PubMed] [Google Scholar]
- 9.Pautas C, Merabet F, Thomas X, et al. Randomized study of intensified anthracycline doses for induction and recombinant interleukin-2 for maintenance in patients with acute myeloid leukemia age 50 to 70 years: results of the ALFA-9801 study. J Clin Oncol. 2010;28(5):808–14. [DOI] [PubMed] [Google Scholar]
- 10.Thomas X, Elhamri M, Raffoux E, et al. Comparison of high-dose cytarabine and timed-sequential chemotherapy as consolidation for younger adults with AML in first remission: the ALFA-9802 study. Blood. 2011;118(7):1754–1762. [DOI] [PubMed] [Google Scholar]
- 11.Gardin C, Turlure P, Fagot T, et al. Postremission treatment of elderly patients with acute myeloid leukemia in first complete remission after intensive induction chemotherapy: results of the multicenter randomized Acute Leukemia French Association (ALFA) 9803 trial. Blood. 2007;109(12):5129–3515. [DOI] [PubMed] [Google Scholar]
- 12.Castaigne S, Pautas C, Terre C, et al. Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): a randomised, open-label, phase 3 study. Lancet. 2012;379(9825):1508–1516. [DOI] [PubMed] [Google Scholar]
- 13.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou KG, Jiang LJ, Shang Z, Wang J, Huang L, Zhou JF. Potential application of IDH1 and IDH2 mutations as prognostic indicators in non-promyelocytic acute myeloid leukemia: a meta-analysis. Leuk Lymphoma. 2012;53(12):2423–2429. [DOI] [PubMed] [Google Scholar]
- 15.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci USA. 2014;111(7):2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]


