Abstract
In α-complementation, inactive N-terminal (α-domain) and C-terminal (ω-domain) fragments of β-galactosidase associate to reconstitute the active protein. To date, the effect of α-domain size on α-complementation activity has not been systematically investigated. In this study, we compared the complementation activities of α-domains of various sizes using an in vitro system. We found that the complementation activities are similar for α-domains comprising between 45 and 229 N-terminal residues but are significantly decreased for those containing less than 37 residues. However, these smaller α-domains (15 and 25 residues) exhibited sufficient α-complementation activity for application as reporters.
Keywords: α-complementation, β-galactosidase, reporter gene, cell-free translation system, reporter peptide
Introduction
β-Galactosidase is widely used as a reporter protein both in vivo1–3 and in vitro,4–6 and its structure has been investigated in detail.7–9 This enzyme can be separated into two fragments: an N-terminal fragment of 45–200 residues known as the α-domain, and a C-terminal fragment that lacks residues 11–41, known as the ω-domain. These two domains are inactive when separated but can combine to regenerate active β-galactosidase in a process known as α-complementation.10–14 Through α-complementation, a small gene encoding the α-domain can be used as a reporter for gene expression if the ω-domain protein is supplemented in advance.
Owing to its small size, the α-domain has been utilized for various applications. For example, the α-domain is included in various plasmid vectors for blue-white screening15 because the insertion of this small domain negligibly affects the plasmid size, which affects the growth rate of host bacteria.16,17 The α-domain is also a useful reporter for gene expression in certain in vitro systems: we have previously demonstrated that using the α-domain rather than full-length β-galactosidase significantly improves the efficiency of translation-coupled RNA replication18 because replication efficiency is heavily dependent on RNA size.
Previous studies have used N-terminal sequences of various lengths as the α-domain.18–22 However, the relationship between α-domain size and α-complementation activity has not been widely explored, and thus, the minimal α-domain that retains the ability to function as a reporter remains unknown. In this study, we compared the α-complementation activities of α-domains of different sizes in vitro and developed a smaller reporter peptide than previously reported, which may be useful for various applications.
To assay the α-complementation activity of α-domains of various sizes, we utilized a reconstituted transcription/translation system in Escherichia coli (the PURE SYSTEM23). As the original system exhibits high β-galactosidase activity because of contamination, we used a customized system consisting of highly purified components that exhibit negligible β-galactosidase activity.24 We amplified DNA fragments encoding α-domains comprising between 27 and 229 N-terminal residues from β-galactosidase using PCR and incubated these fragments with the transcription/translation system containing the purified ω-domain protein and a fluorescent substrate, 5-chloromethylfluorescein di-β-D-galactopyranoside (CM-FDG). The fluorescence produced by β-galactosidase activity was measured every minute. A representative example of the raw data is shown in Figure 1(A). As an index of α-complementation activity, we determined the maximum rate of the increase in the fluorescence emission for each α-domain [Fig. 1(B)]. The α-domains comprising between 45 and 229 residues (45–229AA) exhibited similar levels of activity, whereas α-domains containing less than 45 residues (27AA and 37AA) exhibited drastically reduced activities. These reductions may be reasonably expected because these smaller α-domains do not perfectly complement the sequence that is absent (residues 11–41) in the ω-domain [Fig. 1(C)]. Unexpectedly, 27AA and 37AA exhibited significantly higher α-complementation activities than the control without DNA (blank). This result suggests that α-complementation by much smaller α-domains may still be possible.
Figure 1.
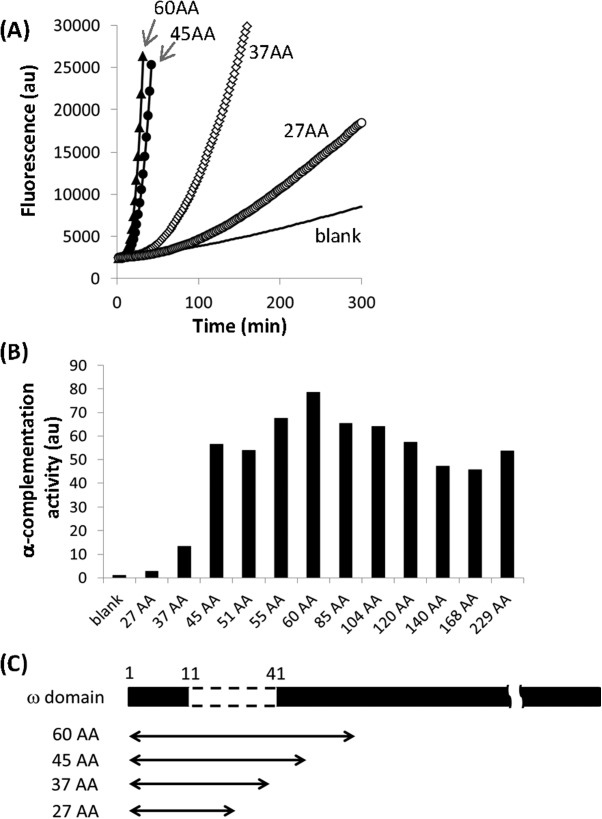
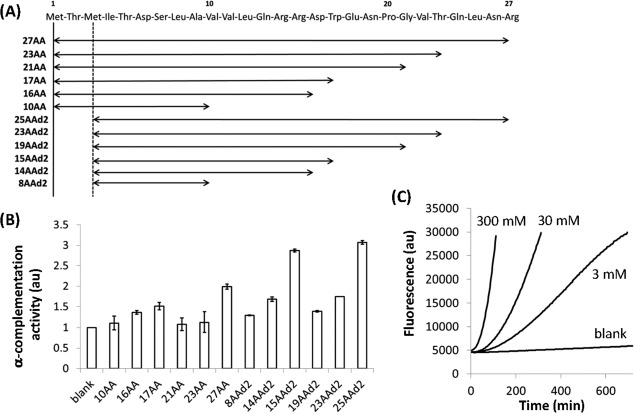
α-Complementation activities of various α-domains. (A) Representative fluorescence kinetics of α-complementation by DNAs encoding different α-domains. The background fluorescence level in the absence of DNA is also shown (blank). (B) α-Complementation activities of DNAs encoding α-domains of various sizes. As an index of the α-complementation activity, the maximum slopes of the fluorescence increases were normalized to the blank and plotted. The numbers before “AA” indicate the number of N-terminal β-galactosidase residues. (C) Relationship between the absent region of the ω-domain and the coding region of each α-domain.
To examine whether the α-domain can be further reduced in length, we amplified DNA fragments encoding shorter N-terminal regions of β-galactosidase [10, 16, 17, 21, 23, and 27AA; Fig. 2(A)] using PCR and determined their α-complementation activities as described above [Fig. 2(B)]. The observed α-complementation activities did not correlate with size, although certain α-domains (27AA, 17AA, and 16AA) exhibited significantly higher activities than the blank. We further deleted the first 2 N-terminal residues (Met and Thr), which are reportedly dispensable for α-complementation,25 to obtain 25AAd2, 21AAd2, 19AAd2, 15AAd2, 14AAd2, and 8AAd2 [Fig. 2(A)], all of which exhibited increased α-complementation activity relative to the blank [Fig. 2(B)].
Figure 2.
α-Complementation activities of small α-domains. (A) Amino acid sequences of each α-domain. (B) α-Complementation activity of each α-domain normalized to the fluorescence of the blank. The activities were determined as described in Figure 1. (C) Fluorescence kinetics for α-complementation by the chemically synthesized 25AAd2 peptide.
These experiments were all performed using DNA fragments encoding α-domains. To confirm that the translated proteins possess α-complementation activity, we chemically synthesized the 25AAd2, 15AAd2, and 8AAd2 peptides and determined their α-complementation activities using the same method as described above, with the exception that the peptides were added to the reaction mixture instead of DNA. The 25AAd2 peptide amplified fluorescence in a concentration-dependent manner [Fig. 2(C)], confirming that the 25AAd2 peptide possesses α-complementation activity. Addition of the 8AAd2 peptide did not result in increased fluorescence, suggesting that the α-complementation activity of 8AAd2 was too weak to be detected using this method. The 15AAd2 peptide was insoluble in water and thus could not be assayed.
Next, we evaluated the ability of the 25AAd2 and 15AAd2 α-domain fragments to serve as reporter genes by performing gene expression experiments in lipid bilayer membranes (liposomes), which are widely used as microreactors. We encapsulated DNA fragments (5 nM) encoding 15AAd2 or 25AAd2 into liposomes together with the transcription/translation system, the ω-domain protein, and the fluorescent substrate. We measured the fluorescence intensity of each liposome using flow cytometry following a 20-h incubation at 37°C (Fig. 3). For both DNA fragments, most of the liposomes exhibited higher fluorescence intensities than the blank, irrespective of the liposome volume. This result demonstrates that 15AAd2 and 25AAd2 are sufficiently active for use as reporter genes in liposomes.
Figure 3.
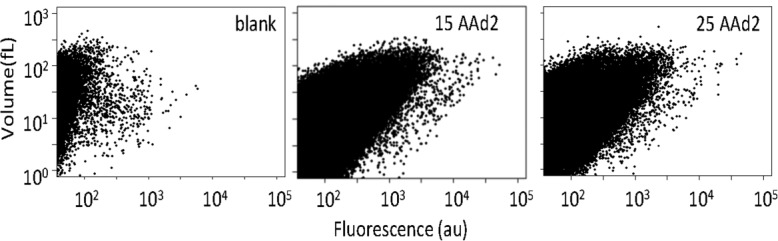
Expression of the small α-domains in liposomes. DNA fragments encoding 15AAd2 and 25AAd2 were encapsulated into liposomes together with the α-complementation assay mix. The fluorescence of each liposome was measured using flow cytometry.
In this study, we investigated the α-complementation activities of variously sized α-domains. We found that α-domains composed of 45–229 residues exhibited similar α-complementation activities but that smaller α-domains, i.e., 37AA and 27AA, exhibited 4-fold and 20-fold decreased activity, respectively, compared with that of 45AA. This result is largely consistent with previous observations based on the protein structure of the α-domain; residues 13–23 directly contribute to the subunit-subunit interface, whereas residues 29–33 serve as “anchors” to connect the interface to each subunit.7 In this study, the α-domains that were smaller than 37AA lacked these anchor regions, and thus, the observed decreases in α-complementation activity could be expected. However, two results in this study are inconsistent with those from previous studies. The first result is that the activity of 37AA was 4-fold lower than that of 45AA, although 37AA contains the entire “anchor” sequence. This result suggests that residues 38–45 may also serve important roles in anchoring the α-domain. The second result is that the α-domains that lack the entire ‘anchor’ sequence (i.e., smaller than 37AA) still exhibited modest but significant α-complementation activity, which may be explained by their structures (Fig. 4). The 25AAd2 α-domain (green) primarily consists of the subunit–subunit interface and does not contain the anchor region (cyan), although it still associates with the ω-domain (gray) via the other side of the interface. This weak association with the ω-domain likely accounts for the weaker but persistent α-complementation activity of the small α-domains.
Figure 4.
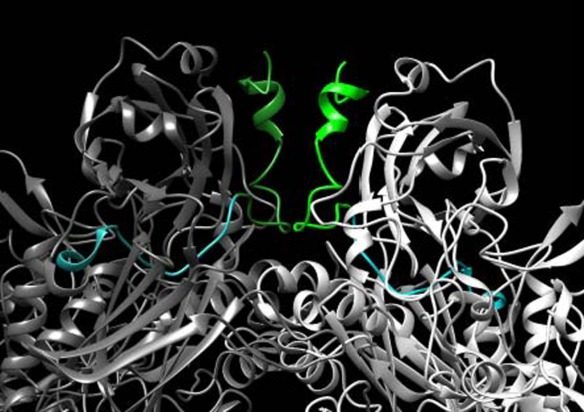
Mapping the region complemented by 25AAd2 in the β-galactosidase tetramer. A portion of the interface structure of the β-galactosidase tetramer9 is shown. The ω-domain protein lacks the colored regions (green and cyan). The region complemented by 25AAd2 (residues 3–25, green) and the absent region (residues 26–41, cyan) are marked. The lighter and darker colors represent the same domain in different monomers. This image was rendered using UCSF Chimera.30
Unexpectedly, we also found that small α-domains (25AAd2 and 15AAd2) retained a certain level of α-complementation activity and could be used as reporter genes. These small reporters can potentially be used for novel applications. For example, protein size is a critical parameter for in vitro translation using nonbiological amino acids26,27 or tRNAs,28 as well as prebiological ribozymes,29 which only permit the translation of small peptides. These smaller reporter peptides may be useful for monitoring translation activity that cannot be detected using conventional reporters.
Materials and Methods
The ω-protein was purchased from Clontech (Mountain View, CA, USA), as was the EA reagent of the ProLabel Detection Kit II. The DNA fragments encoding various α-domains were PCR amplified from the pET-lacZ plasmid using the primers shown in Supporting Information Table S1. The pET-lacZ plasmid was constructed as follows, and the partial sequence around lacZ is shown in the Supporting Information: A DNA fragment containing the lacZ gene was amplified from the plasmid encoding Rep(+)Gal(-) RNA6 and inserted into pET-21a (Novagen). The T7 tag and histidine tag fused to the lacZ gene were then removed. To prepare the α-domains lacking the two N-terminal residues, the plasmid pET-lacZd2 was constructed by PCR using the pET-lacZ template and the primers GAAGGAGATATACATATGATTACGGATTCACTGGCCG and ATGTATATCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGAGGGG, followed by self-ligation using the InFusion cloning kit (Takara, Japan). The chemically synthesized α-domains were purchased from Gene Design (Japan).
The α-complementation assay was performed as described previously.18 The α-complementation reaction contained the customized transcription/translation system, 1 nM α-domain DNA, 50 µM CM-FDG (Life Technologies), and 2 µL of EA reagent (ω-domain protein) in a 20-µL reaction volume. Reactions were incubated at 37°C for 3 h, and fluorescence was measured every minute. For α-complementation assays using the chemically synthesized peptides, the peptides were added instead of DNA. For the liposome experiments, the reactions additionally contained a red fluorescent compound (1.9 µM transferrin Alexa 647) as a volume marker, and the DNA concentration was increased to 5 nM. The mixture was incubated at 37°C for 20 h and subjected to flow cytometry. The composition and preparation of the customized transcription/translation system was described previously.24
Acknowledgments
We thank R. Otsuki and T. Sakamoto for technical assistance. We also thank Dr. Y. Shimizu (RIKEN) for helpful comments on this study.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
References
- Lis JT, Simon JA, Sutton CA. New heat shock puffs and beta-galactosidase activity resulting from transformation of Drosophila with an hsp70-lacZ hybrid gene. Cell. 1983;35:403–410. doi: 10.1016/0092-8674(83)90173-3. [DOI] [PubMed] [Google Scholar]
- Bronstein I, Kricka LJ. Clinical applications of luminescent assays for enzymes and enzyme labels. J Clin Lab Anal. 1989;3:316–322. doi: 10.1002/jcla.1860030511. [DOI] [PubMed] [Google Scholar]
- Tung CH. In vivo imaging of beta-galactosidase activity using far red fluorescent switch. Cancer Res. 2004;64:1579–1583. doi: 10.1158/0008-5472.can-03-3226. , Zeng Q, Shah K, Kim DE, Schellingerhout D, Weissleder R ( [DOI] [PubMed] [Google Scholar]
- Schulz VP, Reznikoff WS. In vitro secondary structure analysis of mRNA from lacZ translation initiation mutants. J Mol Biol. 1990;211:427–445. doi: 10.1016/0022-2836(90)90363-Q. [DOI] [PubMed] [Google Scholar]
- Seeber F, Boothroyd JC. Escherichia coli beta-galactosidase as an in vitro and in vivo reporter enzyme and stable transfection marker in the intracellular protozoan parasite Toxoplasma gondii. Gene. 1996;169:39–45. doi: 10.1016/0378-1119(95)00786-5. [DOI] [PubMed] [Google Scholar]
- Kita H. Replication of genetic information with self-encoded replicase in liposomes. Chembiochem. 2008;9:2403–2410. doi: 10.1002/cbic.200800360. , Matsuura T, Sunami T, Hosoda K, Ichihashi N, Tsukada K, Urabe I, Yomo T ( [DOI] [PubMed] [Google Scholar]
- Matthews BW. The structure of E. coli beta-galactosidase. C R Biol. 2005;328:549–556. doi: 10.1016/j.crvi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Juers DH. High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation. Protein Sci. 2000;9:1685–1699. doi: 10.1110/ps.9.9.1685. , Jacobson RH, Wigley D, Zhang XJ, Huber RE, Tronrud DE, Matthews BW ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S. Structure of beta-galactosidase at 3.2-A resolution obtained by cryo-electron microscopy. Proc Natl Acad Sci USA. 2014;111:11709–11714. doi: 10.1073/pnas.1402809111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann A, Perrin D, Jacob F, Monod J. Identification, by in vitro complementation and purification, of a peptide fraction of Escherichia coli beta-galactosidase. J Mol Biol. 1965;12:918–923. doi: 10.1016/s0022-2836(65)80338-2. [DOI] [PubMed] [Google Scholar]
- Lin S, Villarejo M, Zabin I. Beta-galactosidase: Alpha-complementation of a deletion mutant with cyanogen bromide peptides. Biochem Biophys Res Commun. 1970;40:249–254. doi: 10.1016/0006-291x(70)91002-8. [DOI] [PubMed] [Google Scholar]
- Langley KE, Zabin I. beta-Galactosidase alpha complementation: properties of the complemented enzyme and mechanism of the complementation reaction. Biochemistry. 1976;15:4866–4875. doi: 10.1021/bi00667a018. [DOI] [PubMed] [Google Scholar]
- Gallagher CN, Huber RE. Studies of the M15 beta-galactosidase complementation process. J Prot Chem. 1998;17:131–141. doi: 10.1023/a:1022579416300. [DOI] [PubMed] [Google Scholar]
- Gallagher CN, Huber RE. Stabilities of uncomplemented and complemented M15 beta-galactosidase (Escherichia coli) and the relationship to alpha-complementation. Biochem Cell Biol. 1999;77:109–118. doi: 10.1139/o99-025. [DOI] [PubMed] [Google Scholar]
- Ruther U. Construction and properties of a new cloning vehicle, allowing direct screening for recombinant plasmids. Mol Gen Genet. 1980;178:475–477. doi: 10.1007/BF00270503. [DOI] [PubMed] [Google Scholar]
- Warnes A, Stephenson JR. The insertion of large pieces of foreign genetic material reduces the stability of bacterial plasmids. Plasmid. 1986;16:116–123. doi: 10.1016/0147-619x(86)90070-3. [DOI] [PubMed] [Google Scholar]
- Ryan W, Parulekar SJ, Stark BC. Expression of beta-lactamase by recombinant Escherichia coli strains containing plasmids of different sizes—effects of pH, phosphate, and dissolved oxygen. Biotechnol Bioeng. 1989;34:309–319. doi: 10.1002/bit.260340306. [DOI] [PubMed] [Google Scholar]
- Nishiyama K, Ichihashi N, Matsuura T, Kazuta Y, Yomo T. Alpha-complementation in an artificial genome replication system in liposomes. Chembiochem. 2012;13:2701–2706. doi: 10.1002/cbic.201200586. [DOI] [PubMed] [Google Scholar]
- Langley KE, Villarejo MR, Fowler AV, Zamenhof PJ, Zabin I. Molecular basis of beta-galactosidase alpha-complementation. Proc Natl Acad Sci USA. 1975;72:1254–1257. doi: 10.1073/pnas.72.4.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard C. Dissecting virus entry: Replication-independent analysis of virus binding, internalization, and penetration using minimal complementation of beta-galactosidase. PLoS One. 2014;9:e101762. doi: 10.1371/journal.pone.0101762. , Bloyet LM, Wicht O, van Kuppeveld FJ, Rottier PJ, de Haan CA, Bosch BJ ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkaar F, Blankesteijn WM, Smits JF, Zaman GJ. Beta-Galactosidase enzyme fragment complementation for the measurement of Wnt/beta-catenin signaling. FASEB J. 2010;24:1205–1217. doi: 10.1096/fj.09-141671. [DOI] [PubMed] [Google Scholar]
- Gallagher CN, Roth NJ, Huber RE. A rapid method for the purification of large amounts of an alpha-complementing peptide derived from beta-galactosidase (Ecoli). Prep Biochem. 1994;24:297–304. doi: 10.1080/10826069408010101. [DOI] [PubMed] [Google Scholar]
- Shimizu Y. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. , Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T ( [DOI] [PubMed] [Google Scholar]
- Kazuta Y, Matsuura T, Ichihashi N, Yomo T. Synthesis of milligram quantities of proteins using a reconstituted in vitro protein synthesis system. J Biosci Bioeng. 2014;118:554–557. doi: 10.1016/j.jbiosc.2014.04.019. [DOI] [PubMed] [Google Scholar]
- Langley KE, Fowler AV, Zabin I. Amino acid sequence of beta-galactosidase. IV. Sequence of an alpha-complementing cyanogen bromide peptide, residues 3 to 92. J Biol Chem. 1975;250:2587–2592. [PubMed] [Google Scholar]
- Ohta A, Yamagishi Y, Suga H. Synthesis of biopolymers using genetic code reprogramming. Curr Opin Chem Biol. 2008;12:159–167. doi: 10.1016/j.cbpa.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Hipolito CJ, Suga H. Ribosomal production and in vitro selection of natural product-like peptidomimetics: the FIT and RaPID systems. Curr Opin Chem Biol. 2012;16:196–203. doi: 10.1016/j.cbpa.2012.02.014. [DOI] [PubMed] [Google Scholar]
- Terasaka N, Hayashi G, Katoh T, Suga H. An orthogonal ribosome-tRNA pair via engineering of the peptidyl transferase center. Nat Chem Biol. 2014;10:555–557. doi: 10.1038/nchembio.1549. [DOI] [PubMed] [Google Scholar]
- Zhang B, Cech TR. Peptide bond formation by in vitro selected ribozymes. Nature. 1997;390:96–100. doi: 10.1038/36375. [DOI] [PubMed] [Google Scholar]
- Pettersen EF. UCSF chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. , Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE ( [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information