Abstract
Invading pathogens manipulate cellular process of the host cell to establish a safe replicative niche. To this end they secrete a spectrum of proteins called effectors that modify cellular environment through a variety of mechanisms. One of the most important mechanisms is the manipulation of cellular signaling through modifications of the cellular phosphoproteome. Phosphorylation/dephosphorylation plays a pivotal role in eukaryotic cell signaling, with ∼500 different kinases and ∼130 phosphatases in the human genome. Pathogens affect the phosphoproteome either directly through the action of bacterial effectors, and/or indirectly through downstream effects of host proteins modified by the effectors. Here we review the current knowledge of the structure, catalytic mechanism and function of bacterial effectors that modify directly the phosphorylation state of host proteins. These effectors belong to four enzyme classes: kinases, phosphatases, phospholyases and serine/threonine acetylases.
Keywords: bacterial effector kinases, bacterial phosphatases, bacterial phospholyases, bacterial acetylases, structure-function relationship, noneukaryotic-like kinases
Introduction
Pathogenic bacteria have evolved diverse mechanisms to invade and establish a replicative niche within eukaryotic cells, and in higher eukaryotes in particular. These mechanisms include, but are not limited to, quorum sensing, biofilm formation, cell adhesion, secretion of toxins and other virulence proteins to the host cell. Most Gram-negative pathogens utilize either Type 3 or Type 4 secretion systems (T3SS and T4SS),1,2 which act as macromolecular syringes allowing bacteria to inject a set of effector proteins inside the host cell. These effectors modify normal cellular processes to sustain bacterial proliferation. T3SS is evolutionary related to the bacterial flagellum, and T4SS to the conjugation apparatus. Depending on the bacteria replication cycle, the number of secretion systems varies from one [T3SS—Shigella, enteropathogenic Escherichia coli (EPEC); T4SS—Legionella], two [T3SS—Yersinia, Salmonella, enterohaemorrhagic E. coli (EHEC)], and up to three (T3SS—Burkholderia pseudomallei). The multiple secretion systems are activated at different stages of infection. For example, Salmonella Pathogenicity Island 1 (SPI1) T3SS is turned on early, during bacterial internalization. The SPI2 T3SS is activated later during infection and delivers effectors necessary for the formation of the Salmonella-containing vacuole (SCV), where the bacterium survives and replicates, and Salmonella-induced filaments (SIFs).3
Although the effectors are tailored precisely to the bacterial life cycle and the host cellular environment, nevertheless common themes occur among various pathogens.4 Some of the effectors are enzymes, for example, proteases, phosphatases, glycosylases, acetylases, or lipases, others mimic functions of host proteins, yet others are transcription factors or protein–protein interaction partners.5 The effectors interfere in various host cellular processes, such as cytoskeleton rearrangement, signaling, cellular adherence, transcription, vesicular trafficking, membrane biogenesis, apoptosis and metabolism. Some of the characterized effectors display new folds and catalyze previously undiscovered reactions. The number of effectors secreted by pathogens varies greatly from ∼20 in EPEC,6 to ∼60 in EHEC7 and as many as ∼300 in Legionella.8 The unusual abundance of effectors in Legionella is thought to be related to the diversity of Legionella primary hosts, that is amoebas, in which the bacterium replicates.9
Phosphorylation/dephosphorylation plays a pivotal role in eukaryotic cell signaling, with ∼500 different kinases in the human genome, representing around 1.7% of all genes.10 There are ∼30 catalytic domains of dual-specificity phosphatases, which are paired with multiple specificity domains, and ∼100 tyrosine phosphatases.11 Although the ability of pathogenic bacteria to influence host phosphorylation was known for a long time, it is only recently that extensive phosphoproteome studies shed light on substantial changes in the host phosphoproteome caused by the pathogens. Tracking of phosphorylation changes in HeLa cells during 2 h after Shigella flexneri infection using a label-free quantitative proteomics approach showed that the changes in the phosphorylation profile are substantial and dynamic, changing with time.12 Gene ontology mapping of the proteins differentially phosphorylated during infection classified them by function into signal transduction (50 proteins), actin cytoskeleton (40), exocytosis, endocytosis and intracellular transport (24), RNA processing (24), and cell cycle (23). The consensus phosphorylation motifs suggested that the changes were most likely associated with ACG kinases AKT, PKA/PHC/PKG, RSK, CamKII, as well as MAPK, CDC2, CDK, CK2 and ATM/ATR/DNAPK. The mTOR pathway was overrepresented in the phosphoproteome and important for activation of both S6 and AKT kinases.12 Only few Shigella effectors are known to interfere with host protein phosphorylation (OspF (PDB: 3I0U), OspG; see below). Comparison of the phosphoproteome of HeLa cells infected with wild-type or ΔospF Shigella strains identified ∼140 proteins that showed differences in their phosphorylation,12 providing evidence for the global role of this effector.
The phosphoproteome of Salmonella-infected HeLa cells was investigated 20 minutes post-infection13 and in RAW264.7 microphages 8 h postinfection14 using SILAC quantitative mass spectrometry. At 20 minutes post-infection ∼500 proteins showed changes in their phosphorylation, with the nuclear and membrane fractions enriched in proteins with decreased phosphorylation. Gene Ontology mapping showed that peptides with increased phosphorylation participate in apoptosis, transmembrane transport, nuclear organization and cell proliferation, while peptides with decreased phosphorylation are involved in cytoskeleton organization, protein complex assembly and cell polarity. The phosphorylation consensus sequences indicated increased activity of PKC/PKG, AKT, PIM family kinases, S6 kinase, mTOR, Src, ERK, and others. Comparing the effects of Salmonella and Shigella at early post-infection times identified 57 common phosphorylated proteins, indicating overlap of the molecular mechanisms of these two pathogens in epithelial cell invasion.14
Similarly to ΔospF Shigella, the ΔsopB (inositol phosphate phosphatase) Salmonella strain showed a 35% decrease in the number of proteins with an altered phosphorylation pattern,13 even though the mechanism of these two proteins are different. Enrichment of peptides with a RxRxxS motif suggested significant involvement of AKT, Rsk and p70S6K kinases. An observed increase in BAD phosphorylation at Ser99, which inactivates its proapoptotic functions, and an increase in Ser126 and Ser129 phosphorylation of syntaxin 7, which promotes membrane fusion in the endosomal pathway, may be linked to the action of these kinases during Salmonella infection.
The Salmonella SPI2 T3SS is activated at a longer post-infection time. This T3SS is essential for the pathogen survival in macrophages but less crucial for survival in epithelial HeLa cells. Out of 442 proteins in RAW264.7 macrophages and 606 in Hela cells with changed phosphorylation profile there were 355 common proteins.14 There were however substantial differences in overall functions of the affected proteins. As deducted from the GO terms, proteins affected in macrophages are involved in protein transport, actin regulation and immune signaling, processes that were frequently associated with SPI2 effectors. In epithelial cells the most impacted proteins were involved in apoptosis and regulation of gene expression, in line with previous reports that in this type of cell apoptosis is attenuated. A thorough analysis of phosphorylation patterns in macrophages revealed that the ERK1/2, CDK. PKA, PKC, and MAPK kinases are the key regulators in macrophages in the late stages of infection affected by the SPI2 effectors.
In light of such dramatic rearrangement of host cell phosphoproteome, the question “What are the mechanisms by which pathogens modify host cell phosphorylation?” becomes paramount. These mechanisms may be classified as direct, caused by the bacterial effectors or indirect, caused by the downstream effects of host proteins modified by the effectors. Four direct mechanisms of bacterial effectors that influence host cell phosphoproteome have been discovered:
Phosphorylation. Kinases reversibly phosphorylate Ser and Thr residues of the host cell proteins.
Phosphate hydrolysis. Phosphatases dephosphorylate phosphoSer and phosphoThr residues.
Phosphate elimination. PhosphoSer- and phosphoThr-lyases eliminate the phosphate and convert Ser and Thr to dehydroalanine and methyldehydroalanine, respectively.
Acetylation. Acetylases irreversibly acetylate specific Ser or Thr residues in kinases.
The role of these bacterial effectors in host-pathogen interactions was discussed in a recent review.15–17 Here we review the structural information available for the above four classes of bacterial effectors and their catalytic mechanisms.
Bacterial Effector Kinase Family
Analysis of gene and protein sequences showed presence of eukaryotic-like proteins among bacterial effectors, indicating that they were acquired through lateral transfer from their hosts late during evolution.18,19 Several bacterial effector kinases belong to this category including Salmonella Typhimurium SteC,20 Yersinia pestis YpkA,21 and Legionella pneumophila LegK1, LegK3, LegK4.22 In addition, bacteria evolved their own effector kinases. These kinases belong to the kinase superfamily but are more similar to nonregulatory kinases. They include NleH1 (PDB: 4LRJ) and NleH2 (PDB: 4LRK) from pathogenic E. coli,23 OspG from Shigella,24 SboH from Salmonella bongori25 and YspK from Yersinia.26 Bioinformatics analysis shows that although the kinase domain is much smaller than the average human kinase, it contains all the essential elements characteristic of kinases [Fig. 1(a)]. Of the eleven motifs characteristic of regulatory kinases,27 the bacterial effector kinase domain spans motifs I–VIII but lacks the activation loop. Recent structural studies of NleH1, NleH2, and OspG revealed the structural organization of these kinases28–31 and allowed building a structure-based phylogenetic tree, which indicated that the bacterial effector kinases form a new family within the kinase superfamily28 [Fig. 1(b)]. Although Hervet et al.32 argued that Legionella LegK2 lacks the activation loop [Fig. 1(a)], the classification of LegK2 kinase in one of these two classes is presently uncertain. So far three-dimensional structures are known only for the noneukaryotic-like kinases.
Figure 1.
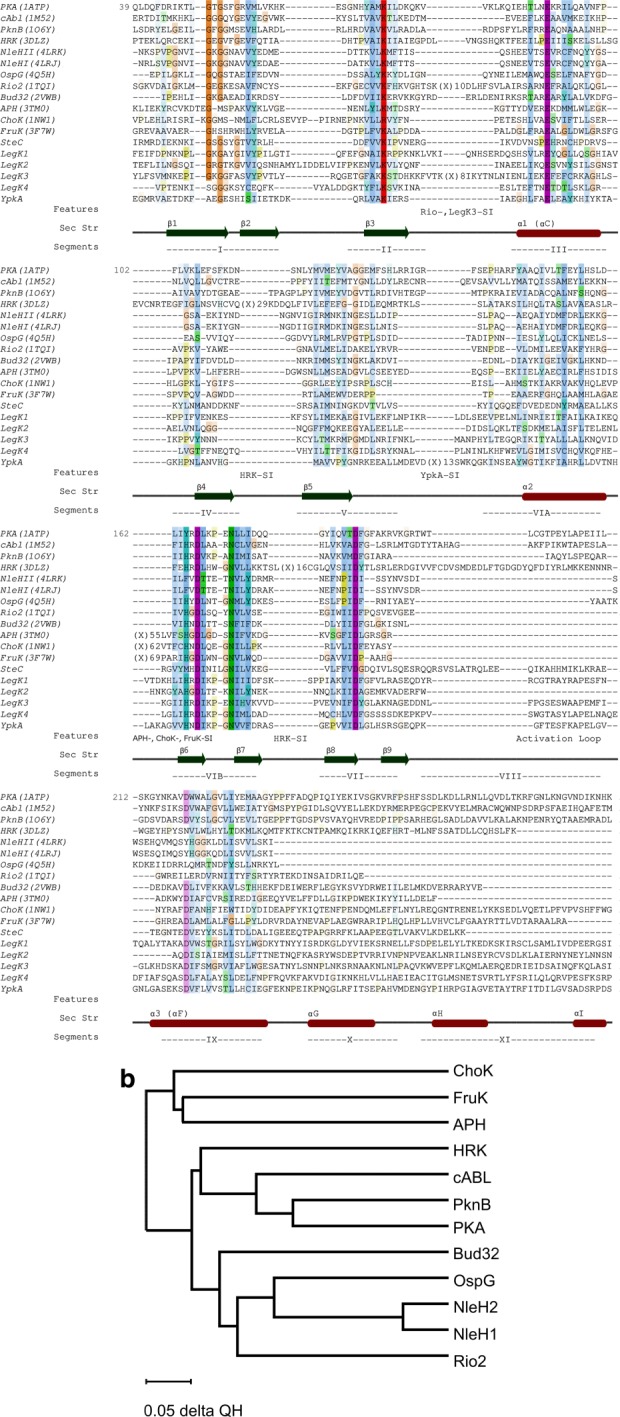
(A) Structure-based sequence alignment of several mammalian kinases and effector kinases with known structures. Highly conserved residues are color coded: hydrophobic residues are blue, His and Tyr—cyan, polar residues—green, Arg and Lys—red, Asp and Glu—magenta, Gly—orange, and Pro—olive. Higher color saturation reflects higher level of conservation. Other effector kinases were aligned based on their sequences. PKA—human protein kinase A, cABL—mouse cABL kinase, PknB—Mycobacterium tuberculosis kinase PknB, HRK—atypical human kinase haspin, NleHI and NleHII—E. coli O157:H7 kinases, OspG—Shigella flexneri kinase, Rio2—Archaeoglobus fulgidus Rio2 kinase, Bud32—Methanocaldococcus jannaschii Bud32 kinase, APH—Enterococcus faecalis 3′,5"-Aminoglycoside Phosphotransferase, CHOK—Caenorhabditis elegans choline kinase, FruK—Thermobifida fusca fructosamine-3-kinase, SteC—Salmonella Typhimurium kinase, LegK1, LegK2, LegK3, LegK4—Legionella pneumophila kinases, YpkA—Yersinia pestis kinase. The secondary structure elements are marked below the sequences together with the motifs I-XI 27; (B) evolutionary tree built based on structure conservation with program Multiseq.154
Noneukaryotic-like kinases
Functional aspects
Functional aspects of bacterial effector kinases were reviewed recently,17 therefore our discussion of functional aspects of effector kinases in this review is brief. NleH1 and NleH2 are homologous effector serine/threonine protein kinases (STPK) (84% sequence identity) from pathogenic E. coli strain.37 Related Citrobacter rodentium possesses only one copy of NleH.33 These proteins are composed of a C-terminal kinase module (∼160 amino acids) and an N-terminal segment of ∼140 amino acids with the “telltale” signature of an intrinsically disordered domain. The first 25 amino acids are believed to contain the T3SS secretion signal.34
The clearest function of NleHs is the inhibition of the NF-κB pathway.35 NleH1 was shown to bind the ribosomal protein 3 (RPS3) and prevent its phosphorylation by IKKβ.36 As a consequence, the RPS3 is not translocated to the nucleus to serve as a subunit for NF-κB transcriptional complexes. The kinase activity of NleH1 is also essential for this effect, since the active site mutant of NleH1 is not able to inhibit the NF-κB pathway.35 However, the phosphorylation target of NleH1 is not yet known. NleH2, despite being homologous to NleH1, has apparently an opposite effect and mildly activates the NF-κB pathway.37 In addition, NleHs were shown to bind PDZ2 domain of the Na+/H+ Exchanger Regulatory Factor 2 (NHERF2) via four C-terminal residues38 and deletion of this motif blocked the NF-κB inhibition activity of NleH1.37 Finally, NleHs were able to inhibit apoptosis by binding to the N-terminal segment of Bax Inhibitor 1.23
OspG is a Shigella effector kinase composed of only the kinase module (170 amino acids) and an N-terminal secretion signal (25 amino acids). Intriguingly, OspG also suppresses the NF-κB pathway but does it by stabilizing IκB.24 As with NleHs, the kinase activity was shown to be important for this biological function, however the phosphorylation target is also unknown. OspG binds ubiquitin39 and a series of E2∼Ub conjugates,24 however, the relevance of this interaction for the NF-κB inhibition is unclear.
The effector kinase domain
The ∼160 residues long effector kinase module of this protein family is significantly smaller than the eukaryotic regulatory kinases that are over 300 amino acids long. The module is composed of a mostly β-stranded N-terminal domain and a C-terminal mixed α/β domain, joined together by a short connector28 [Fig. 2(a)]. The N-terminal domain contains motifs I to IV,27 covering the glycine loop and the catalytic Lys, which is properly oriented via a salt bridge with a neighboring Glu. The C-terminal domain includes motifs V-IX that include the catalytic loop and the DFG-motif. Motif VIII, which comprises the activation loop, is very short in NleHs28 and slightly longer in OspG.29,31 The catalytic site is situated in a deep crevice between the two subdomains, next to the ATP and Mg2+ binding site. Site-directed mutagenesis showed the importance for catalysis of Lys169 (motif II), Glu183 (motif III), Asp249 (motif VIB), and Asp268 (motif VII)28 in NleH2 and of Lys53 (motif II) and Asp 138 (motif VIB) in OspG.39 Similarly to other kinases, NleHs and OspG possess two arrangements of hydrophobic residues called the catalytic and regulatory spines.40,41 The catalytic spine is formed by eight resides from both domains of the kinase that upon ATP binding create a contiguous stack with the adenine ring in the middle. The regulatory spine contains four hydrophobic residues that interact with each other only in the active conformation of regulatory kinases.
Figure 2.
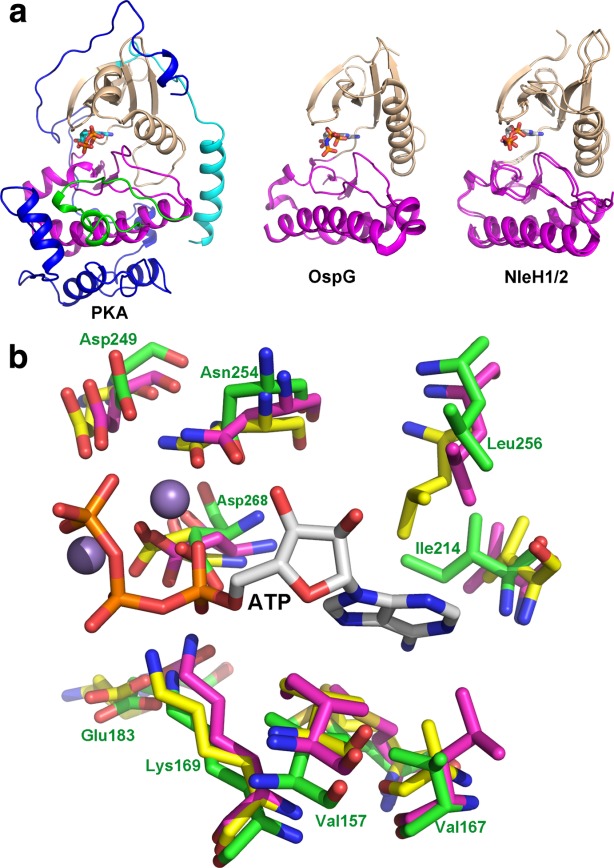
(A) Cartoon representation of kinase domains of OspG and NleH1/2 showing their overall similarity. The human protein kinase A is shown for comparison. The N-terminal lobe is painted wheat, the C-terminal is magenta and the activation loop in green. The N-terminal extension of PKA is in cyan and the C-terminal extension that embraces the N-terminal lobe is in dark blue; (B) superposition of the residues forming the ATP binding site. The carbons in PKA are painted yellow, in OspG they are magenta, in NleH2 they are green. The ATP (white carbons) and two Mn2+ ions (spheres) were taken from the PKA structure (PDB code 1ATP). The residue numbers are provided based on NleH2 structure. This and the following figures were prepared using PyMol (www.pymol.org). An interactive view is available in the electronic version of the article.
The shorter activation loop is devoid of the regulatory phosphorylation site, which poses the question if these kinases are constitutively active or are activated differently than the eukaryotic regulatory kinases. The crystal structures showed that the NleH1 and NleH2 kinase domains were already in the active conformation both in the apo- and AMPPNP-bound structures.28,30 Not only does the catalytic machinery align well with that of protein kinase A in the active conformation [Fig. 2(b)] but also the two hydrophobic spines are fully assembled. The adenine ring complements the catalytic spine upon ATP binding. The nucleotide binding proceeds with minimal structural rearrangements. The biochemical data support the notion that NleHs do not require a special activation mechanism. Even when purified from E. coli, NleHs are phosphorylated with an average of 2–3 phosphate groups per molecule, which was attributed to autophosphorylation.28 The activity assays with radioactive [γ-32P]ATP showed that both full-length NleHs could autophosphorylate and that both the NleHs full-length and kinase domains could phosphorylate the generic kinase substrate myelin basic protein (MBP). Mapping the phosphorylation sites has shown that most of them were located in the N-terminal unstructured segment of NleH.28
OspG was shown to bind Ub and the UbcH7∼Ub conjugate.24,39 By itself OspG showed low intrinsic activity against another generic kinase substrate—histones. However, the activity increased 8- and 20-fold upon binding to Ub and the UbcH7∼Ub conjugate, respectively,29,31,39 indicating a novel activation mechanism. Moreover, the affinity toward nonhydrolysable ATP-γS was higher when OspG was complexed with the UbcH5∼Ub (PDB: 4BVU) conjugate.31 The structures of the complexes of OspG with UbcH7∼Ub and UbcH5∼Ub have shown that UbcH5/7∼Ub conjugate binds to two sites on OspG, leaving the active site open for the interaction with the kinase substrate29,31 [Fig. 3(a)]. These observations suggest that the interactions of OspG with Ub and the ubiquitin conjugating enzyme shift the conformation of OspG toward a fully active conformation.31 Another possible explanation is that Ub binding affects the catalytic machinery of OspG to adopt productive conformation, as some Ub residues are less than 8 Å away from the catalytic loop.29 Another indication of conformational flexibility of OspG is the difficulty of obtaining crystals of this protein by itself or in the presence of ATP. These structures are essential to understand the details of the activation mechanism.
Figure 3.
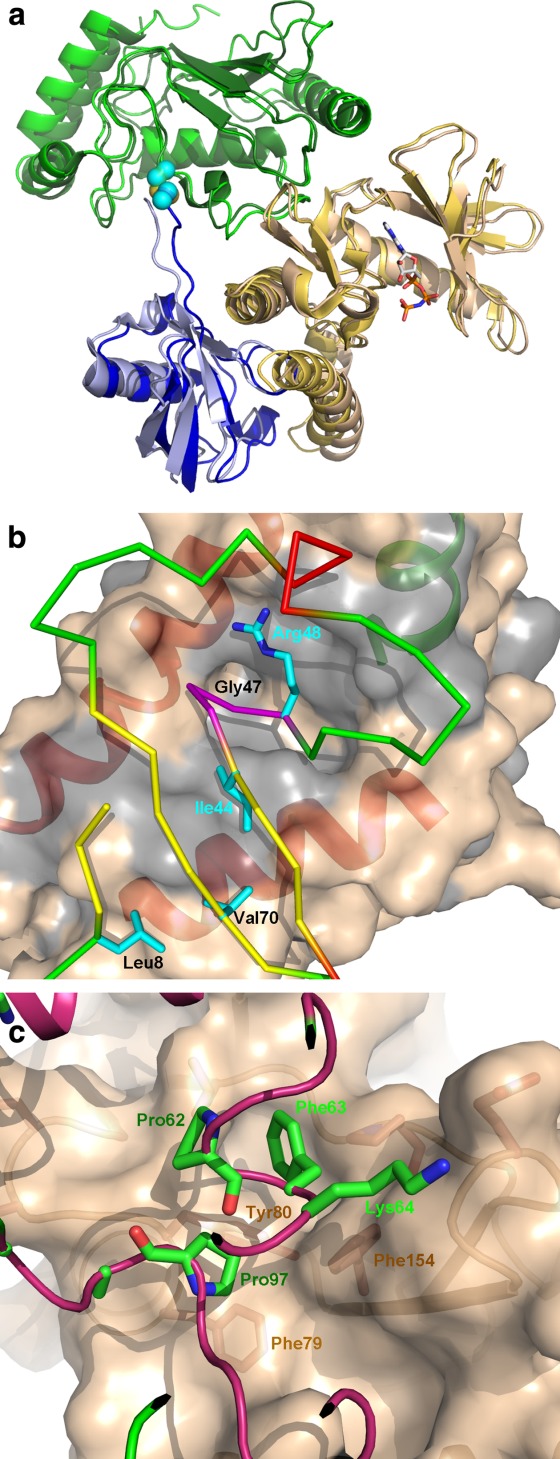
(A) Cartoon representation of OspG-E2∼Ub complexes. The UbcH5 and UbcH7 complexes are superimposed and are nearly identical. OspG is painted wheat, UbcH5/7 is green and Ub is blue. The darker shades show the UbcH7 complex; (B) the OspG-Ub interface. The OspG is shown as molecular surface colored wheat with gray marking hydrophobic residues. The Ub faces OspG with the Ile44 side and inserts a loop culminating with GlyXX into a deep depression in OspG surface; (C) the UbcH7 residue contacting OspG surface. Phe63 plays a key role in these interactions. An interactive view is available in the electronic version of the article.
Currently, the phosphorylation targets of effector kinases remain unknown. However, recently the v-crk Sarcoma Virus CT10 Oncogene-like Protein (CRKL) was proposed as the first NleH1 target.42
Structural aspects of protein–protein interactions
Notwithstanding a relatively small size, the effector kinases were shown to participate in several protein–protein interactions.23,36,38 No structural information on NleH-host protein complexes is yet available, and complex formation with previously identified host target proteins has proved to be challenging (Grishin AM, unpublished results). However, the structures of OspG with ubiquitin-conjugating enzyme conjugated with ubiquitin (E2∼Ub) have recently been determined.29,31 Several ubiquitinated E2s were initially identified by a yeast two-hybrid screen as the host proteins binding to OspG.24 Subsequently, it was shown that OspG also interacts with ubiquitin and polyubiquitin chains.39 The binding constant of OspG to UbcH7-Ub (580 nM) was 15 times higher than to Ub (9 µM) and 150 times higher than to UbcH7 (∼90 µM). Importantly, the UbcH5/7∼Ub increased the OspG kinase activity more than Ub alone. Taken together, it was concluded that the E2∼Ub conjugate is the OspG cellular binding target.29
UbcH5/7∼Ub associates with OspG in an open conformation, essentially the same as observed when bound to a HECT-type E3s,43 and with the same surfaces on E2s and Ub involved in both these interactions. Ub binds OspG through the well-known Ile44 hydrophobic patch (Leu8, Ile44, and Val70) and UbcHs through α-helix 1, loop4 and loop 729,31 [Fig. 3(b,c)]. These two surfaces have been previously characterized as involved in the recognition and functionality of the ubiquitination machinery.44–49 The OspG Ub-binding surface is a large concave hydrophobic surface formed between the C-terminal and the penultimate α-helices and the shortened “activation” loop [Fig. 3(b)]. In addition to hydrophobic contacts a salt bridge between Lys48Ub and Asp177OspG is formed. Comparison with NleHs and PKA showed that the OspG C-terminal helix rotates away from the penultimate helix, with one end moving by ∼7 Å, to create this surface.29,31 Whether this conformation of the C-terminal α-helix is induced by ubiquitin binding or is a feature of OspG will be answered by structure determination of OspG alone. UbcH5/7 binds through key residues Pro62, Phe63 and Pro97 to a hydrophobic patch on OspG involving residues from both lobes (Phe79, Tyr80, Leu99, Pro102, and Phe154) on the opposite side to the active site [Fig. 3(c)]. Residues located around this central hydrophobic zone formed additional polar interactions.29,31
Effects of protein–protein interactions
OspG binding to the same surface of E2∼Ub conjugate as do the E3 ligases suggested that OspG might interfere with the host E3 functions. Indeed, the addition of OspG to a parkin autoubiquitination assay completely inhibited the autoubiquitination in vitro at 1:1 OspG:UbcH7 (PDB: 4Q5H) ratio.29 Whether OspG affects the host ubiquitination machinery in vivo is unknown. However, several mutations disrupting OspG:E2 or OspG:Ub interfaces significantly reduced the inhibition of the NFκB pathway. For example, the OspG Leu190Asp/Leu191Asp double mutation, which targeted the interactions of the C-terminal α-helix with the rest of the molecule, rendered OspG incapable of binding to Ub and blocking IκB degradation.39 Similarly, the OspG Cys127Arg mutation on the Ub-binding interface not only blocks the activation of the kinase activity by UbcH5∼Ub conjugate but also results in the mouse infection phenotype, similar to the deletion of OspG altogether.31 Moreover, OspG Phe154Arg and Leu99Arg/Pro102Glu mutations within the E2 binding interface also affected OspG function. The Leu99Arg/Pro102Glu mutant was no longer activated by E2∼Ub while the Phe154Arg mutant was only marginally activated. The mutations also affected stability of OspG in vivo; the mutants, but not the wild-type OspG, were degraded quickly in a proteasome-dependent manner.29
Legionella kinase LegK2
LegK2 is a 62-kDa translocated effector of T4SS.50 Kinase domain of LegK2 resides in the N-terminal region (aa 77–232) and has all the consensus sequences in Hanks subdomains I-VII, required for catalytic activity.32 LegK2 undergoes autophosphorylation and phosphorylates the general eukaryotic protein kinase substrate MBP in vitro in the presence of Mn2+ ions. LegK2 is the only Legionella kinase that proved to be indispensable for virulence toward amoeba. Kinase activity of LegK2 is required for efficient recruitment of ER to Legionella-containing vacuoles, necessary for intracellular replication of bacteria.32 Substrates of LegK2 have not been identified yet, and functions of the C-terminal nonkinase domain of this effector remain to be elucidated.
Eukaryotic-like Ser/Thr protein kinases
Structural aspects of Legionella kinases
L. pneumophila strains encode three translocated eukaryotic-like serine/threonine protein kinases LegK1, LegK3 and LegK4.32,51 Multiple sequence alignments of kinase domains from L. pneumophila Paris with other prokaryotic and eukaryotic STPKs [Fig. 1(a)] revealed that LegK1 and LegK3 cluster in the group of eukaryotic protein kinases, close to STPKs from pathogenic anaerobic amoeba Entamoeba histolytica.18 This observation, together with the lower GC content (33%) of the LegK3 gene (lpp2626) compared to the average of 38% for protein-coding genes of L. pneumophila Paris, suggested it was acquired by a horizontal gene transfer (Garcia-Vallve et al. 2003).52 However, there is no direct evidence for a lateral transfer of LegK1 genes from eukaryotic organisms. The same phylogenetic analysis demonstrated that the STPK domain of LegK4 is closely related to PknG (from M. tuberculosis) and to the Y. pseudotuberculosis translocated STPK YpkA.18
Alignments with several prokaryotic and eukaryotic protein kinases revealed that all three kinases harbor highly conserved residues in subdomains I-IX [Fig. 1(a)].27,53 These include the glycine rich loop in subdomain I, the invariant Lys in subdomain II, which anchor and orients ATP in the kinase active site, and the conserved Glu (Lys in case of LegK3) in subdomain III that stabilizes this interaction. The HRDxKxxN motif in the catalytic loop and the conserved DF/YG triplet in subdomain VII participate in chelating Mg2+ ions. All the three kinases have an activation loop (subdomain VIII) with the conserved APE motif, which plays a significant role in substrate recognition.
Based on in vitro studies, LegK1 is expected to be constitutively active or use an activation loop-independent activation mechanism, since mutations of both Ser252 and Tyr256, the only potential phosphorylation sites in the activation loop, did not affect the enzyme activity.51 LegK3 and LegK4 undergo autophosphorylation and phosphorylate the general eukaryotic protein kinase substrate MBP, as demonstrated by in vitro kinase assay. LegK1 phosphorylates specifically its known substrate IkB, but not MBP. All three kinases require Mg2+ ions for kinase activity while LegK4 is also able to use Mn2+ with the same efficiency.32
Physiological role of Legionella eukaryotic-like kinases
Separate inactivation of LegK1, LegK3 or LegK4 kinase genes did not influence bacterial growth in liquid medium or cellular morphology, nor did it affect Legionella virulence toward amoeba.32 However, overexpression of one of the kinases, LegK1, in mammalian HEK293T cells significantly upregulated the NF-κB pathway, as demonstrated by NF-κB luciferase reporter studies.51,54 The kinase activity of LegK1 was responsible for this effect, since the mutation of a conserved catalytic Asp223 from the catalytic loop to an alanine completely abolished NF-kB reporter activation.54 Recombinant LegK1 triggered phosphorylation in cell-free extracts and also phosphorylated IκB on Ser32 and Ser36 in vitro. LegK1 efficiently phosphorylated four other IκB family members (IκBβ, p100, p105, and IκBε), which harbor the characteristic DSGXXS/T phospho-motif.51 Further truncation studies demonstrated that the N-terminal pre-kinase region of LegK1 was indispensable for IκBα phosphorylation, whereas the C-terminal domain was not strictly required for enzyme activity; full-length LegK1 and a truncated LegK1(1–386) phosphorylated IκBα with a comparable efficiency in radioactive 32P incorporation assay, however deletion of the first 45 residues abolished in vitro phosphorylation of IκBα.51
The functions of LegK3 and LegK4 remain largely unknown. A L. pneumophila mutant deficient in legK1, legK2 and legK3 genes still induced p38 and SAPK/JNK MAPK activation.50 In support of this result, expression of LegK1 and LegK3 in mammalian cells did not affect MAPKs activation. According to the same study, overexpression of these proteins did not activate the IFNβ promoter in two different luciferase reporter systems.51
Salmonella kinase SteC
SteC is the only secreted protein kinase effector of Salmonella enterica serovar Typhimurium and is delivered by SPI2 T3SS20 across the Salmonella-containing vesicle (SCV) vacuolar membrane. SCVs are surrounded by an actin cytoskeleton meshwork, presumably involved in maintaining the integrity of vacuolar membranes.55,56 Formation of this network is dependent on SteC and its kinase activity.20
The SteC C-terminal kinase domain (aa 201–457) contains the characteristic kinase subdomains I to IX with a ∼35 residue long activation loop and is most similar to the human RAF proto-oncogene STPK Raf-1.20,57 SteC and its kinase domain undergo autophosphorylation and phosphorylate the general kinase substrate MBP in vitro.20 This kinase may play a role in colonization of the chick intestine as demonstrated by gene inactivation studies.58 Moreover, SteC-null and kinase-deficient mutant strains displayed increased replication in infected cells and in a mouse model of systemic infection, suggesting that SteC could serve to restrain bacterial growth and thus regulate virulence.59 The mechanism of action of SteC involves actin rearrangement through activation of a signaling pathway containing the MAP kinases MEK and ERK, myosin light chain kinase (MLCK) and Myosin IIB.59 SteC phosphorylates MEK on Ser200, which leads to MEK autoactivation through autophosphorylation of Ser218 and Ser222.59
A recent phosphoproteomics analysis of macrophage RAW264.7 and HeLa cells infected with Salmonella identified chaperone HSP27 as another potential substrate of SteC. This chaperone is phosphorylated in vivo on Ser15 and on at least six other sites in vitro, indicating that it might be a direct substrate of SteC and a part of a parallel route to SteC-induced host actin manipulation.14 Moreover, the described interaction between the N-terminal non-kinase domain of SteC and the yeast guanine nucleotide exchange factor CDC24, the activator of a small GTPase CDC42, may suggest that both domains of SteC complement each other in actin modulation.60
Yersinia protein kinase A (YpkA)
YpkA from Y. pseudotuberculosis (YopO in Y. enterocolitica) is a 729 amino acids multidomain protein with a Ser/Thr protein kinase activity required for full virulence in mouse models of infection.61,62 The eukaryotic-like STPK domain encompasses residues 150–400. Upon translocation to a host cell YpkA is targeted to the inner side of the plasma membrane by a membrane localization domain (MLD, amino acids 20–90).63,64 This hydrophobic region is shielded inside the bacterium by the chaperon SycO to prevent aggregation.64 Recently, a phosphoinositide-binding domain (PBD) with conserved membrane localization GKxYxnF motif was mapped in the same region (aa 32–89).65
A Ser/Thr kinase domain of YpkA contains subdomains I-XI61 (Fig 1 1). YpkA is expressed in Yersinia in a catalytically inactive form and requires G-actin binding and autophosphorylation for activation.66,67 Multiple autophosphorylation sites have been identified in the N-terminal and kinase domains of YpkA,67,68 however the detailed activation mechanism and the role of phosphorylation of the activation loop is not yet known.
Heterotrimeric G protein was identified as a direct substrate of YpkA kinase. Phosphorylation of Ser47 in the consensus sequence, GXXXXGK(S/T) of the diphosphate binding loop disrupted GTP binding and prevented G protein activation.69 Residues 40–49 within the YpkA membrane-localization domain were shown to be critical for Gαq binding and phosphorylation.68 Recently, vasodilator-stimulated phosphoprotein (VASP) was shown to be phosphorylated by YpkA in vitro and in vivo, predominantly at S157. The STPK and actin-binding domains of YpkA were involved in its interaction with VASP, while GDI domain was dispensable.70
Phosphatases
Two bacterial effector phosphatases, YopH (PDB: 1YTW) and SptP (PDB: 1G4W), have been structurally characterized and are described below.
Y. pestis protein tyrosine phosphatase YopH
Functional aspects of YopH
YopH is indispensable for Yersinia virulence,71,72 it subdues host defenses by inhibiting phagocytosis and disrupting focal adhesion in macrophages.73,74 It also interferes with T and B lymphocyte activation and thus blocks adaptive immune response.75 In vivo studies in a mouse infection model demonstrated that YopH inactivated PRAM-1/SKAP-HOM and the SLP-76/Vav/PLCγ2 signal transduction axes, which might be critical for bacterial survival.76 Several direct substrates of YopH were identified in different cell types: p130Cas (p130Crk-associated substrate), paxillin and focal adhesion kinase FAK in HeLa cells,74,77 Fyn-binding protein and murine SKAP-HOM in macrophages,78,79 Lck, LAT, and SLP-76 in T-cells,80,81 p85 (regulatory subunit of the PI3K) in HEK293.82
Structure of YopH
YopH consists of two domains, the smaller N-terminal domain (aa 1–130) is connected by a an intrinsically disordered region to the larger C-terminal PTPase domain (aa 163–468). The PTPase domain bears a remarkable structural similarity to human PTP1B (PDB: 1PTY)83 despite only ∼20% sequence identity.84 It contains all the invariant residues present in eukaryotic PTPases.85 The catalytic domain forms an eight-stranded mixed β-sheet, flanked by five α-helices on one side and two α-helices on the other side86 (Fig. 4). The P-loop (phosphate binding), located between β-strand and α-helix, adheres to the signature PX(I/V)(I/V)HCSAGXGR(T/S)G PTPase motif. This motif includes the catalytically essential His402, nucleophile Cys40387 and the GXGXXG sequence, similar to the nucleotide-binding motif of dehydrogenases and kinases.
Figure 4.
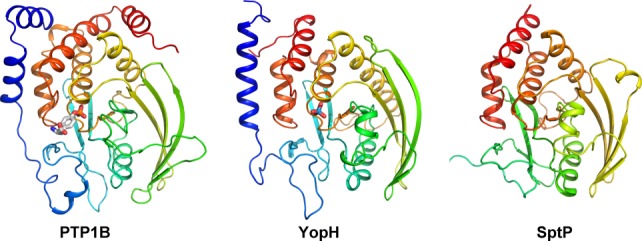
The structures of bacterial phosphatases YopH and SptP are compared to the human protein phosphatase 1B. An interactive view is available in the electronic version of the article.
The X-ray structures of the apo YopH PTPase domain and its complexes with various ligands mimicking intermediate and transition states88 together with kinetic, mutagenesis and computational studies (reviewed in Refs.89–90) allowed deciphering of the PTPase mechanism. The nucleophilic displacement reaction proceeds in two steps: (i) phosphoryl transfer to a functional group on the enzyme and (ii) hydrolysis of the resultant phosphoenzyme intermediate (E-P) and regeneration of the active enzyme. Upon substrate binding the flexible WPD loop (aa 350–360) moves to a closed conformation bringing Asp356 to the substrate where it acts as a general acid and transfers a proton to the scissile oxygen of the substrate initiating phosphate dissociation. Deprotonated Cys403 in the active site serves as a nucleophile to accept a phosphoryl group.89,90 Arg409 stabilizes the transition state forming a bidentate hydrogen bond with two nonbridging oxygens of the phosphate group. The phosphoryl transfer proceeds most likely via a dissociative transition state.91 During the following E-P hydrolysis step the invariant Asp356 residue acts as a general base to activate a nucleophilic water92 that is positioned by Gln446 along the S-P bond direction. The hydroxyl group of Thr410 interacts with the scissile bond and initiates E-P hydrolysis.93 Although YopH and human PTP1B follow the same catalytic mechanism, their catalytic rates vary significantly; YopH is 20-fold more active than PTPB1.94 Recent solution NMR relaxation studies revealed that the WPD loop dynamics determines the overall reaction rate, since WPD loop closure during the phosphopeptide cleavage step is tightly synchronized with the protonation of the leaving group.95
Crystal structures of YopH complexed with model peptide substrates based on an autophosphorylation site of EGFR85 revealed a second binding site within the PTPase catalytic domain.96,97 This site binds the YopH substrate p130Cas in a phosphotyrosine-dependent manner and is involved in targeting YopH to Cas in vivo.97
The N-terminal noncatalytic domain of YopH (YopH-NT) has dual properties: in bacterial cell it binds chaperone SycH, which enables its translocation,98,99 while in a host cell acquires a globular conformation and serves as a substrate binding domain.100–102 The phosphotyrosine substrate binding specificity of YopH-NT is similar to that of eukaryotic SH2 domains, which recognize pYXX(X)P amino acid sequences. However, YopH-NT shares no structural homology with eukaryotic analogs.102 The secretion segment (aa 2–17) and translocation domain (aa 18–71)103 make an integral part of the YopH-NT compact fold composed of four α-helices and two β-hairpins.100,102
Salmonella Typhimurium tyrosine phosphatase SptP
Structural aspects of SptP
SptP is a T3SS effector required for full virulence of S. Typhimurium in vivo.104 It is a modular protein with two functional domains: the C-terminal PTPase domain and the N-terminal domain similar to the Pseudomonas aeruginosa exotoxin S105 (21% sequence identity) and the Yersinia spp. cytotoxin YopE.106,107 The PTPase domain of SptP shares 30% sequence homology with YopH and is very similar structurally and mechanistically to this enzyme. SptP phosphatase domain (residues 300–543) follows the canonical tyrosine phosphatase fold, with the exception of the first helix, which is missing. Although the active site of SptP is well conserved, this phosphatase has distinct surface properties with a shape and charge distribution different from YopH, consistent with different substrates and cellular effects of these proteins.108
Upon synthesis in bacterial cell, the N-terminal domain of SptP forms a tight complex with its specific chaperone SicP.109 Since this binding is crucial for SptP stability and translocation,110,111 translation of SptP is strictly coupled to SicP expression.112
SptP function
The N-terminal domain of SptP demonstrated a potent GAP activity, inhibiting signaling through Rho GTPases Rac-1 and Cdc42. This GAP activity of SptP is responsible for downregulation of the actin-cytoskeleton rearrangements stimulated in host cells by T3SS effectors SopE and SopE2.111 However, both domains are required for another observed activity of SptP, inhibition of ERK activation.113,114 Several substrates of SptP phosphatase domain were identified, including AAA+ ATPase VCP, which when dephosphorylated by SptP, promoted membrane fusion events that might be necessary for SCV maintenance and maturation.115 SptP was also reported to suppress mast cell degranulation by dephosphorylating tyrosine kinase Syk and the vesicle fusion protein NSF.116
Phosphoserine/Phosphothreonine Lyases
Fold and catalytic mechanism
Structure
A family of homologous proteins, encompassing Shigella OspF, Salmonella SpvC (PDB: 2PIW), Chromobacterium VirA (PDB: 3BO6) and Pseudomonas syringae HopAI1, were shown to irreversibly eliminate the phospho-group from the threonine in the Thr-X-Tyr motif in the activation loop of MAP kinases.117 Although the first studies annotated OspF as a phosphatase, the subsequent analysis revealed that the action of OspF, SpvC and HopAI1 reduces molecular weight by 98 Da, corresponding to the elimination of H3PO4, and therefore resulting in a Cα–Cβ dehydroalanine/dehydrothreonine residue.117–119
The structures of OspF (PDB code 3I0U, Singer et al., unpublished), SpvC120,121 and VirA (PDB code 3BO6, Brennan et al., unpublished) are very similar and demonstrate a unique fold. The lyase domain of ∼240 residues consists of a central seven-stranded β-sheet with its convex side covered by nine α-helices [Fig. 5(a)]. A long and positively charged groove extends along the concave side of the β-sheet, necessary for binding of pT and pY residues. Activity studies revealed that OspF and SpvC have almost identical substrate specificity, with preference p38>ERK2>ERK5 and very low activity on JNK.120 Consistent with the evolutionary conserved function, the P. syringae HopAI1 lyase inactivates two plant MAP kinases, MPK3 and MPK6, that are involved in plant defense systems.122
Figure 5.
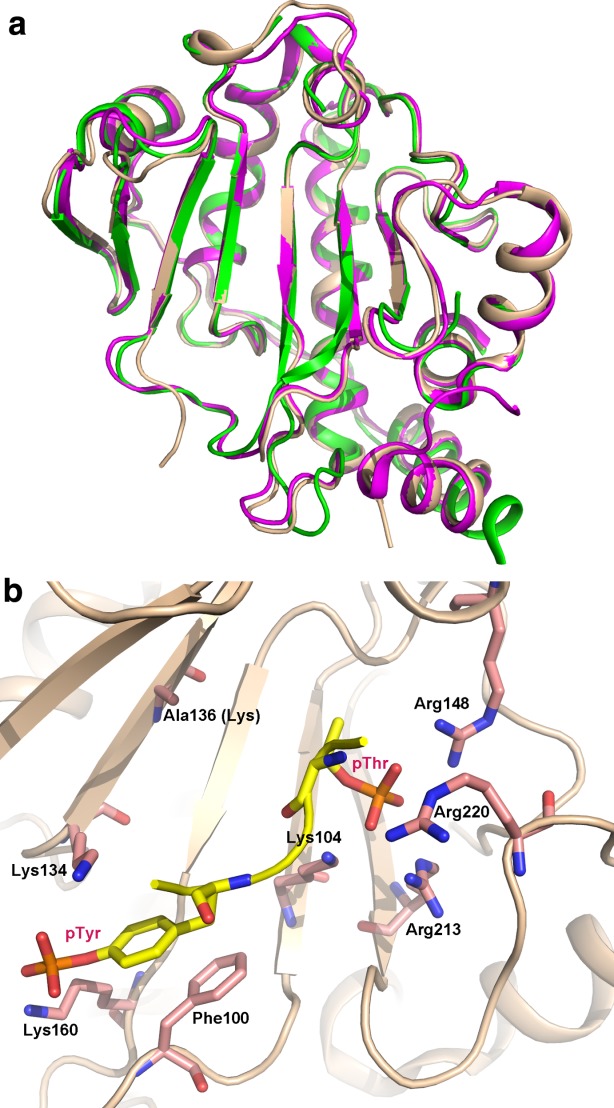
(A) Superposition of phosphothreonine lyases Salmonella SpvC (wheat), Shigella OspF (magenta) and Chromobacterium VirA (green) shown in cartoon representation. Their structures are strikingly similar; (B) the substrate binding site of SpvC with pThr and pTyr-containing peptide captured in a K136A mutant. An interactive view is available in the electronic version of the article.
Apart from the direct enzyme-substrate interaction, OspF, SpvC and VirA but not HopAI1, have mimicked a D-motif—a protein–protein interaction module, encompassing the first N-terminal 26 residues, which bind to a negatively-charged patch and a hydrophobic groove on the surface of a MAP kinase.120 The N-terminal D-motif is widely used by the MAP kinase kinases to activate MAP kinases and contains a well-established consensus sequence (R/K)2–3-X2–6-Φ-X-Φ, where Φ is a hydrophobic residue. This motif binds to a groove between α5, α6, β7, and β8 on the C-terminal lobe of ERK.123 OspF constructs with truncated or deleted D-motif lose their ability to co-purify with ERK2, suggesting its high functional importance.120
Substrate biding and catalysis
The structure of the SpvC complexed with the diphosphorylated peptide of the same sequence as the substrate's activation loop allowed the catalytic mechanism to be resolved.120,121 The peptide adopts a U-shape conformation with the pThr-Glu-pTyr residues located at the bottom of the positively charged groove in SpvC [Fig. 5(b)]. The substrate pTyr is located in the positively charged pocket mainly formed by Lys134 and Lys169, neutralizing phosphate group, and with Phe100 stacked against the tyrosine ring. Mutations of Lys160 and Phe100 affect significantly catalytic efficiency while mutation of Lys134 is less severe. The phosphate group of the substrate pThr binds in a deep cavity formed by Arg148, Arg213, Arg220, and Lys104 and is almost completely shielded from the solvent. Mutation of any of the arginines to a glutamate or to a lysine abolished SpvC activity. This phosphoresidue binding site mimics the environment of this residue in the activated form of MAP kinases where the pThr and pTyr residues are surrounded by positively charged residues.124
Lys136 and His106 were proposed to act as the catalytic residues for the β-elimination reaction of the phosphothreonine residue.120,121 Lys136 acts as a catalytic base to abstract proton from the C atom of phosphothreonine, while His106 acts as a catalytic acid to donate a proton to the leaving phosphate group. This hypothetical mechanism was substantiated by mutational studies and by QM/MM and DFT modeling calculations.125–127
Although the middle residue of the pThr-Xaa-pTyr motif is exposed to solvent, its conformational flexibility dramatically affects catalytic efficiency.120 The best substrate, p38, has a Gly in this position, ERK2 has a Glu, while the poorest substrate, JNK, has a Pro. The mutation of this Pro in the JNK peptide to Gly rendered the latter a much better substrate. The comparison of the conformation of the activation loop in the activated state of the kinase and the conformation of the substrate peptide in the SpvC active site revealed a drastic conformational change in residues N-terminal to the phosphotyrosine [Fig. 5(b)], which may explain the need for the flexible residue between the two phosphorylated residues.120
Functional role
Mice infected with the mutant Salmonella Typhimurium strain lacking the spvC gene showed pronounced colitis and inflammation compared to the wild-type bacteria.128 Deletion of the ospF gene resulted in significant changes in the differential host cell phosphoproteome.12 In addition to the expected inhibition of gene expression, the deletion affected the phosphorylation status of many proteins involved in transcription, chromatin modification and RNA processing. Additionally, proteins related to the actin cytoskeleton, microtubules, apoptosis and cell cycle were affected as well. Thus OspF massively impacts host protein phosphorylation by targeting the central MAP kinase pathway—p38 and ERK. Recently, OspF was shown to bind chromatin at the promoter region of the IL8 gene and to interact with the C-terminal chromoshadow domain of the heterochromatin protein 1 (HP1γ) involved in the recruitment of epigenetic regulators.129 Moreover, the presence of OspF led indirectly to the inhibition of phosphorylation of the regulatory Ser83 of HP1γ, which becomes phosphorylated by ERK-dependent MSK1. In addition, OspF affects chromatin remodeling through binding to the retinoblastoma protein (RB).130 OspF contains a LxCxE RB-binding motif (I178MCLE182), which is located on a long α–helix on the convex surface of OspF molecule. The mutation in this motif not only disrupted the binding to RB but also reduced the capacity of OspF to repress IL-8 secretion in vivo.130
The impact of OspF on non-MAP kinase dependent pathways remains largely unknown. So far it has been shown that OspF can activate TGF-beta activated kinase (TAK1) by disrupting the negative feedback loop regulation between p38 and TAK1, which in turn enhanced NF-κB, JNK and c-Jun signaling.131
Acetylases
The YopJ/HopZ/AvrRxv family of bacterial effector acetylases is widely distributed among animal and plant pathogens and plant symbionts. YopJ/YopP (Yersinia), AvrA (Salmonella), AvrBsT (Xantomonas campestris), YopP (Bartonella), VopJ (Vibrio), and HopZ1–3 (P. syringae) are some of the members of this family. Yersinia infection is accompanied by a profound inhibition of several MAP kinases and NF-κB pathway.132 These effects were attributed to YopJ, which was shown to downregulate ERK, p38 and JNK by decreasing the level of their phosphorylation132–134 and by preventing phosphorylation and subsequent degradation of IκB.135 Subsequently, YopJ was found to bind and inhibit activation of a number of MAP kinase kinases—MKK1, MKK3, MKK4, and MKK5, as well as IKKβ, a component of the IκB kinase complex.136
There is as yet no crystal structure for any member of this family but a limited structural insight can be derived from amino acid sequence analysis, which indicates the similarity of YopJ to adenovirus cysteine protease AVP and ubiquitin-like-specific cysteine protease 1 (ULP1), belonging to the proteolytic Clan CE.137 The deduced catalytic triad, consisting of Cys, His, and Glu, was confirmed by mutagenesis. Similar results were also demonstrated by mutations of Xantomonas campestris AvrBsT residues. Initially, YopJ was considered to be a protease/deubiquitinase.66 However, later it was shown that YopJ acetylated the Ser/Thr within the activation loop of MEK1, MEK2,138 TAK1139 as well as IKKα and IKKβ.140 Furthermore, the YopJ activity was acetyl-CoA dependent. The YopJ docking site on the kinase was localized through mutagenesis and functional studies of yeast MKK Pbs2 to α-helix G (within motif X).141
YopJ and AvrA are activated by inositol hexakisphosphate (IP6), thus indicating that IP6 is likely a cofactor common for this acetylase family.142 AvrA also inhibits the NF-κB pathway143,144 but by different means than YopJ since it does not block IKK phosphorylation. Moreover, AvrA inhibits the JNK pathway at the level of MKK4/7 but not the p38 and ERK pathways.144,145 AvrA can be downregulated by phosphorylation within the N-terminal translocation motif by one of the kinases of the ERK pathway,144 suggesting an intriguing interplay between AvrA and kinase cascades.
The substrate specificity of YopJ/HopZ/AvrRxv family members varies. The AopP effector from Aeromonas salmonicida could inhibit the NF-κB pathway downstream from IKK activation, while it did not interfere with the MAP kinase pathway,146 while VopA from Vibrio parahaemolyticus could inhibit MAK kinase pathway but not the NF-κB pathway.147 Although the acetylation of Ser and Thr residues in the activation loop was considered to be the main mechanism of inhibition, VopA could acetylate a conserved lysine residue preceding the catalytic loop in MAP kinases, important for ATP binding.148 This modification rendered MKK6 incapable of binding ATP. YopJ could only acetylate a Lys residues located in the activation loop.138 Moreover YopJ, VopA, and PopP2 (Ralstonia solanacearum) were shown to autoacetylate.140,148 The autoacetylation site for PopP2 was located at a lysine conserved in every member of the family. R. solanacearum is a plant pathogen and the PopP2 lysine mutant was not recognized by the plant defense system, suggesting a role for this Lys in acetyl-CoA recognition.149
Conclusions
Processes within eukaryotic cells are highly coordinated and are directed through a complex system of interconnected signaling pathways. Many of these signals involve adding or removing a phosphate group by dedicated enzymes, events that are recognized downstream by specialized recognition domains. Bacterial pathogens invading host cells rearrange cellular processes to adapt the initially hostile environment into one that allows bacterium to survive and prosper. To accomplish this task pathogens secrete effector proteins that modify cellular processes toward the needs of the pathogen. Since phosphorylation/dephosphorylation plays a key role in cellular signaling, pathogens have evolved sophisticated systems that affect the phosphorylation state of the key cellular signaling components thus directly and indirectly modifying the entire cellular phosphoproteome. Progress in recent years has significantly increased our understanding of the mechanisms by which pathogens subvert cellular processes and has led also at the same time to better understanding of the normal cellular processes.
Obviously, progress in our understanding of molecular mechanisms of pathogen/host warfare provides new avenues for developing new therapies. The importance of the secretion systems and in particular T3SS, T4SS as the syringes injecting bacterial effectors into the host cell and thus being the first step in infection was long recognized. These systems were targeted for drug development88,150 and progress in their development has been recently reviewed.151 Other promising pharmaceutical targets have also been identified and their usefulness for drug development is being investigated.152 Moreover, the tools evolved by bacteria represent a unique evolution–tailored set of proteins capable of modifying the desired cellular processes and the full potential of these tools is yet to be discovered. Application of bacterial effectors YopH and OspF to rewire signaling cascades was recently demonstrated in the response of to external stimuli. Similarly, Jurkat T cells were modified to mitigate off–target activities during adoptive immunotherapy.153
REFERENCES
- Christie PJ, Whitaker N, Gonzalez-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843:1578–1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diepold A, Wagner S. Assembly of the bacterial type III secretion machinery. FEMS Microbiol Rev. 2014;38:802–822. doi: 10.1111/1574-6976.12061. [DOI] [PubMed] [Google Scholar]
- Kuhle V, Hensel M. Cellular microbiology of intracellular Salmonella enterica: functions of the type III secretion system encoded by Salmonella pathogenicity island 2. Cell Mol Life Sci. 2004;61:2812–2826. doi: 10.1007/s00018-004-4248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol Rev. 2011;35:1100–1125. doi: 10.1111/j.1574-6976.2011.00271.x. [DOI] [PubMed] [Google Scholar]
- Wong ARC, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol Microbiol. 2011;80:1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x. [DOI] [PubMed] [Google Scholar]
- Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, Younis R, Matthews S, Marches O, Frankel G, Hayashi T, Pallen MJ. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci USA. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Franco IS, Shuman HA, Charpentier X. The perplexing functions and surprising origins of Legionella pneumophila type IV secretion effectors. Cell Microbiol. 2009;11:1435–1443. doi: 10.1111/j.1462-5822.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Mustelin T. A brief introduction to the protein phosphatase families. Methods Mol Biol. 2007;365:9–22. doi: 10.1385/1-59745-267-X:9. [DOI] [PubMed] [Google Scholar]
- Schmutz C, Ahrné E, Kasper CA, Tschon T, Sorg I, Dreier RF, Schmidt A, Arrieumerlou C. Systems-level overview of host protein phosphorylation during Shigella flexneri infection revealed by phosphoproteomics. Mol Cell Proteom. 2013;12:2952–2968. doi: 10.1074/mcp.M113.029918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers LD, Brown NF, Fang Y, Pelech S, Foster LJ. Phosphoproteomic analysis of Salmonella-infected cells identifies key kinase regulators and SopB-dependent host phosphorylation events. Science Signal. 2011;4:rs9. doi: 10.1126/scisignal.2001668. [DOI] [PubMed] [Google Scholar]
- Imami K, Bhavsar AP, Yu H, Brown NF, Rogers LD, Finlay BB, Foster LJ. Global impact of Salmonella pathogenicity island 2-secreted effectors on the host phosphoproteome. Mol Cell Proteom. 2013;12:1632–1643. doi: 10.1074/mcp.M112.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Hao YH, Orth K. A newly discovered post-translational modification—the acetylation of serine and threonine residues. Trends Biochem Sci. 2007;32:210–216. doi: 10.1016/j.tibs.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Cui J, Shao F. Biochemistry and cell signaling taught by bacterial effectors. Trends Biochem Sci. 2011;36:532–540. doi: 10.1016/j.tibs.2011.07.003. [DOI] [PubMed] [Google Scholar]
- Canova MJ, Molle V. Bacterial serine/threonine protein kinases in host-pathogen interactions. J Biol Chem. 2014;289:9473–9479. doi: 10.1074/jbc.R113.529917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- Wehenkel A, Bellinzoni M, Grana M, Duran R, Villarino A, Fernandez P, Andre-Leroux G, England P, Takiff H, Cervenansky C, Cole ST, Alzari PM. Mycobacterial Ser/Thr protein kinases and phosphatases: physiological roles and therapeutic potential. Biochim Biophys Acta. 2008;1784:193–202. doi: 10.1016/j.bbapap.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Poh J, Odendall C, Spanos A, Boyle C, Liu M, Freemont P, Holden DW. SteC is a Salmonella kinase required for SPI-2-dependent F-actin remodelling. Cell Microbiol. 2008;10:20–30. doi: 10.1111/j.1462-5822.2007.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley DJ, Shrestha N, Yang J, Atis N, Dayton K, Schesser K. The activities of the Yersinia protein kinase A (YpkA) and outer protein J (YopJ) virulence factors converge on an eIF2alpha kinase. J Biol Chem. 2009;284:24744–24753. doi: 10.1074/jbc.M109.010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurie-Weinberger MN, Gomez-Valero L, Merault N, Glockner G, Buchrieser C, Gophna U. The origins of eukaryotic-like proteins in Legionella pneumophila. Int J Med Microbiol. 2010;300:470–481. doi: 10.1016/j.ijmm.2010.04.016. [DOI] [PubMed] [Google Scholar]
- Hemrajani C, Berger CN, Robinson KS, Marches O, Mousnier A, Frankel G. NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc Natl Acad Sci USA. 2010;107:3129–3134. doi: 10.1073/pnas.0911609106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fookes M, Schroeder GN, Langridge GC, Blondel CJ, Mammina C, Connor TR, Seth-Smith H, Vernikos GS, Robinson KS, Sanders M, Petty NK, Kingsley RA, Baumler AJ, Nuccio SP, Contreras I, Santiviago CA, Maskell D, Barrow P, Humphrey T, Nastasi A, Roberts M, Frankel G, Parkhill J, Dougan G, Thomson NR. Salmonella bongori provides insights into the evolution of the Salmonellae. PLoS Pathog. 2011;7:e1002191. doi: 10.1371/journal.ppat.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto H, Young GM. Proteomic and functional analysis of the suite of Ysp proteins exported by the Ysa type III secretion system of Yersinia enterocolitica Biovar 1B. Mol Microbiol. 2006;59:689–706. doi: 10.1111/j.1365-2958.2005.04973.x. [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- Grishin AM, Cherney M, Anderson DH, Phanse S, Babu M, Cygler M. NleH defines a new family of bacterial effector kinases. Structure. 2014;22:250–259. doi: 10.1016/j.str.2013.11.006. [DOI] [PubMed] [Google Scholar]
- Grishin AM, Condos TE, Barber KR, Campbell-Valois FX, Parsot C, Shaw GS, Cygler M. Structural basis for the inhibition of host protein ubiquitination by Shigella effector kinase OspG. Structure. 2014;22:878–888. doi: 10.1016/j.str.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Halavaty AS, Anderson SM, Wawrzak Z, Kudritska M, Skarina T, Anderson WF, Savchenko A. Type III effector NleH2 from Escherichia coli O157:H7 str. Sakai features an atypical protein kinase domain. Biochemistry. 2014;53:2433–2435. doi: 10.1021/bi500016j. [DOI] [PubMed] [Google Scholar]
- Pruneda JN, Smith FD, Daurie A, Swaney DL, Villen J, Scott JD, Stadnyk AW, Le Trong I, Stenkamp RE, Klevit RE, Rohde JR, Brzovic PS. E2∼Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis. EMBO J. 2014;33:437–449. doi: 10.1002/embj.201386386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervet E, Charpentier X, Vianney A, Lazzaroni JC, Gilbert C, Atlan D, Doublet P. Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect Immun. 2011;79:1936–1950. doi: 10.1128/IAI.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Angulo VA, Deng W, Thomas NA, Finlay BB, Puente JL. Regulation of expression and secretion of NleH, a new non-locus of enterocyte effacement-encoded effector in Citrobacter rodentium. J Bacteriol. 2008;190:2388–2399. doi: 10.1128/JB.01602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas NA, Deng W, Puente JL, Frey EA, Yip CK, Strynadka NC, Finlay BB. CesT is a multi-effector chaperone and recruitment factor required for the efficient type III secretion of both LEE- and non-LEE-encoded effectors of enteropathogenic Escherichia coli. Mol Microbiol. 2005;57:1762–1779. doi: 10.1111/j.1365-2958.2005.04802.x. [DOI] [PubMed] [Google Scholar]
- Gao X, Wan F, Mateo K, Callegari E, Wang D, Deng W, Puente J, Li F, Chaussee MS, Finlay BB, Lenardo MJ, Hardwidge PR. Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog. 2009;5:e1000708. doi: 10.1371/journal.ppat.1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, Lenardo MJ. IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nature Immunol. 2011;12:335–343. doi: 10.1038/ni.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TH, Gao X, Tsai K, Olsen R, Wan F, Hardwidge PR. Functional differences and interactions between the Escherichia coli type III secretion system effectors NleH1 and NleH2. Infect Immun. 2012;80:2133–2140. doi: 10.1128/IAI.06358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Schroeder GN, Berger CN, Lee SF, Robinson KS, Badea L, Simpson N, Hall RA, Hartland EL, Crepin VF, Frankel G. Binding to Na(+) /H(+) exchanger regulatory factor 2 (NHERF2) affects trafficking and function of the enteropathogenic Escherichia coli type III secretion system effectors Map, EspI and NleH. Cell Microbiol. 2010;12:1718–1731. doi: 10.1111/j.1462-5822.2010.01503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Dong N, Hu L, Shao F. The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation. PLoS One. 2013;8:e57558. doi: 10.1371/journal.pone.0057558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci USA. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham TH, Gao X, Singh G, Hardwidge PR. Escherichia coli virulence protein NleH1 interaction with the v-Crk Sarcoma virus CT10 oncogene-like protein (CRKL) governs NleH1 inhibition of the ribosomal protein S3 (RPS3)/nuclear factor κB (NF-κB) pathway. J Biol Chem. 2013;288:34567–34574. doi: 10.1074/jbc.M113.512376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into ubiquitin transfer cascades from a structure of a UbcH5B<ubiquitin-HECTNEDD4L complex. Mol Cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The hydrophobic effect contributes to polyubiquitin chain recognition. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J Biol Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH. Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nature Struct Mol Biol. 2006;13:264–271. doi: 10.1038/nsmb1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJL, Timmers HTM. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24:981–993. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- Budhidarmo R, Nakatani Y, Day CL. RINGs hold the key to ubiquitin transfer. Trends Biochem Sci. 2012;37:58–65. doi: 10.1016/j.tibs.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Shin S, Case CL, Archer KA, Nogueira CV, Kobayashi KS, Flavell RA, Roy CR, Zamboni DS. Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog. 2008;4:e1000220. doi: 10.1371/journal.ppat.1000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J, Xu H, Li T, Zhou Y, Zhang Z, Li S, Liu L, Shao F. A Legionella type IV effector activates the NF-kappaB pathway by phosphorylating the IkappaB family of inhibitors. Proc Natl Acad Sci USA. 2009;106:13725–13730. doi: 10.1073/pnas.0907200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vallve S, Guzman E, Montero MA, Romeu A. HGT-DB: a database of putative horizontally transferred genes in prokaryotic complete genomes. Nucleic Acids Res. 2003;31:187–189. doi: 10.1093/nar/gkg004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK. Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 2003;4:111. doi: 10.1186/gb-2003-4-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Haenssler E, Moy MY, Isberg RR. LnaB: a Legionella pneumophila activator of NF-kappaB. Cell Microbiol. 2010;12:1083–1097. doi: 10.1111/j.1462-5822.2010.01452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meresse S, Unsworth KE, Habermann A, Griffiths G, Fang F, Martinez-Lorenzo MJ, Waterman SR, Gorvel JP, Holden DW. Remodelling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell Microbiol. 2001;3:567–577. doi: 10.1046/j.1462-5822.2001.00141.x. [DOI] [PubMed] [Google Scholar]
- Unsworth KE, Way M, McNiven M, Machesky L, Holden DW. Analysis of the mechanisms of Salmonella-induced actin assembly during invasion of host cells and intracellular replication. Cell Microbiol. 2004;6:1041–1055. doi: 10.1111/j.1462-5822.2004.00417.x. [DOI] [PubMed] [Google Scholar]
- Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- Morgan E, Campbell JD, Rowe SC, Bispham J, Stevens MP, Bowen AJ, Barrow PA, Maskell DJ, Wallis TS. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2004;54:994–1010. doi: 10.1111/j.1365-2958.2004.04323.x. [DOI] [PubMed] [Google Scholar]
- Odendall C, Rolhion N, Forster A, Poh J, Lamont DJ, Liu M, Freemont PS, Catling AD, Holden DW. The Salmonella kinase SteC targets the MAP kinase MEK to regulate the host actin cytoskeleton. Cell Host Microbe. 2012;12:657–668. doi: 10.1016/j.chom.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pinar P, Aleman A, Sondek J, Dohlman HG, Molina M, Martin H. The Salmonella Typhimurium effector SteC inhibits Cdc42-mediated signaling through binding to the exchange factor Cdc24 in Saccharomyces cerevisiae. Mol Biol Cell. 2012;23:4430–4443. doi: 10.1091/mbc.E12-03-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galyov EE, Hakansson S, Forsberg A, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- Wiley DJ, Nordfeldth R, Rosenzweig J, DaFonseca CJ, Gustin R, Wolf-Watz H, Schesser K. The Ser/Thr kinase activity of the Yersinia protein kinase A (YpkA) is necessary for full virulence in the mouse, mollifying phagocytes, and disrupting the eukaryotic cytoskeleton. Microb Pathog. 2006;40:234–243. doi: 10.1016/j.micpath.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Hakansson S, Galyov EE, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- Letzelter M, Sorg I, Mota LJ, Meyer S, Stalder J, Feldman M, Kuhn M, Callebaut I, Cornelis GR. The discovery of SycO highlights a new function for type III secretion effector chaperones. EMBO J. 2006;25:3223–3233. doi: 10.1038/sj.emboj.7601202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon D, Guo Y, Kinch LN, Grishin NV, Gardner KH, Orth K. Effectors of animal and plant pathogens use a common domain to bind host phosphoinositides. Nature Commun. 2013;4:2973. doi: 10.1038/ncomms3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juris SJ, Rudolph AE, Huddler D, Orth K, Dixon JE. A distinctive role for the Yersinia protein kinase: actin binding, kinase activation, and cytoskeleton disruption. Proc Natl Acad Sci USA. 2000;97:9431–9436. doi: 10.1073/pnas.170281997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trasak C, Zenner G, Vogel A, Yuksekdag G, Rost R, Haase I, Fischer M, Israel L, Imhof A, Linder S, Schleicher M, Aepfelbacher M. Yersinia protein kinase YopO is activated by a novel G-actin binding process. J Biol Chem. 2007;282:2268–2277. doi: 10.1074/jbc.M610071200. [DOI] [PubMed] [Google Scholar]
- Pha K, Wright ME, Barr TM, Eigenheer RA, Navarro L. Regulation of Yersinia protein kinase A (YpkA) kinase activity by multisite autophosphorylation and identification of an N-terminal substrate-binding domain in YpkA. J Biol Chem. 2014;289:26167–26177. doi: 10.1074/jbc.M114.601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Koller A, Nordfelth R, Wolf-Watz H, Taylor S, Dixon JE. Identification of a molecular target for the Yersinia protein kinase A. Mol Cell. 2007;26:465–477. doi: 10.1016/j.molcel.2007.04.025. [DOI] [PubMed] [Google Scholar]
- Ke Y, Tan Y, Wei N, Yang F, Yang H, Cao S, Wang X, Wang J, Han Y, Bi Y, Cui Y, Yan Y, Song Y, Yang X, Du Z, Yang R. Yersinia protein kinase A phosphorylates vasodilator-stimulated phosphoprotein to modify the host cytoskeleton. Cell Microbiol. 2014 doi: 10.1111/cmi.12378. [VOL:PAGE #S] [DOI] [PubMed] [Google Scholar]
- Bliska JB, Guan KL, Dixon JE, Falkow S. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc Natl Acad Sci USA. 1991;88:1187–1191. doi: 10.1073/pnas.88.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleuil F, Mogemark L, Francis MS, Wolf-Watz H, Fallman M. Interaction between the Yersinia protein tyrosine phosphatase YopH and eukaryotic Cas/Fyb is an important virulence mechanism. Cell Microbiol. 2003;5:53–64. doi: 10.1046/j.1462-5822.2003.00236.x. [DOI] [PubMed] [Google Scholar]
- Andersson K, Carballeira N, Magnusson KE, Persson C, Stendahl O, Wolf-Watz H, Fallman M. YopH of Yersinia pseudotuberculosis interrupts early phosphotyrosine signalling associated with phagocytosis. Mol Microbiol. 1996;20:1057–1069. doi: 10.1111/j.1365-2958.1996.tb02546.x. [DOI] [PubMed] [Google Scholar]
- Black DS, Bliska JB. Identification of p130Cas as a substrate of Yersinia YopH (Yop51), a bacterial protein tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. EMBO J. 1997;16:2730–2744. doi: 10.1093/emboj/16.10.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Mecsas J, Healy JI, Falkow S, Chien Y. Suppression of T and B lymphocyte activation by a Yersinia pseudotuberculosis virulence factor, yopH. J Exp Med. 1999;190:1343–1350. doi: 10.1084/jem.190.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolan HG, Durand EA, Mecsas J. Identifying Yersinia YopH-targeted signal transduction pathways that impair neutrophil responses during in vivo murine infection. Cell Host Microbe. 2013;14:306–317. doi: 10.1016/j.chom.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C, Carballeira N, Wolf-Watz H, Fallman M. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J. 1997;16:2307–2318. doi: 10.1093/emboj/16.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid N, Gustavsson A, Andersson K, McGee K, Persson C, Rudd CE, Fallman M. YopH dephosphorylates Cas and Fyn-binding protein in macrophages. Microb Pathog. 1999;27:231–242. doi: 10.1006/mpat.1999.0301. [DOI] [PubMed] [Google Scholar]
- Black DS, Marie-Cardine A, Schraven B, Bliska JB. The Yersinia tyrosine phosphatase YopH targets a novel adhesion-regulated signalling complex in macrophages. Cell Microbiol. 2000;2:401–414. doi: 10.1046/j.1462-5822.2000.00061.x. [DOI] [PubMed] [Google Scholar]
- Alonso A, Bottini N, Bruckner S, Rahmouni S, Williams S, Schoenberger SP, Mustelin T. Lck dephosphorylation at Tyr-394 and inhibition of T cell antigen receptor signaling by Yersinia phosphatase YopH. J Biol Chem. 2004;279:4922–4928. doi: 10.1074/jbc.M308978200. [DOI] [PubMed] [Google Scholar]
- Gerke C, Falkow S, Chien YH. The adaptor molecules LAT and SLP-76 are specifically targeted by Yersinia to inhibit T cell activation. J Exp Med. 2005;201:361–371. doi: 10.1084/jem.20041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Puerta ML, Trinidad AG, del Carmen Rodriguez M, Bogetz J, Sanchez Crespo M, Mustelin T, Alonso A, Bayon Y. Characterization of new substrates targeted by Yersinia tyrosine phosphatase YopH. PLoS One. 2009;4:e4431. doi: 10.1371/journal.pone.0004431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- Hooft van Huijsduijnen R. Protein tyrosine phosphatases: counting the trees in the forest. Gene. 1998;225:1–8. doi: 10.1016/s0378-1119(98)00513-7. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Wang Y, Dixon JE. Dissecting the catalytic mechanism of protein-tyrosine phosphatases. Proc Natl Acad Sci USA. 1994;91:1624–1627. doi: 10.1073/pnas.91.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey JA, Schubert HL, Fauman EB, Zhang ZY, Dixon JE, Saper MA. Crystal structure of Yersinia protein tyrosine phosphatase at 2.5 A and the complex with tungstate. Nature. 1994;370:571–575. doi: 10.1038/370571a0. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Dixon JE. Active site labeling of the Yersinia protein tyrosine phosphatase: the determination of the pKa of the active site cysteine and the function of the conserved histidine 402. Biochemistry. 1993;32:9340–9345. doi: 10.1021/bi00087a012. [DOI] [PubMed] [Google Scholar]
- Sun JP, Wu L, Fedorov AA, Almo SC, Zhang ZY. Crystal structure of the Yersinia protein-tyrosine phosphatase YopH complexed with a specific small molecule inhibitor. J Biol Chem. 2003;278:33392–33399. doi: 10.1074/jbc.M304693200. [DOI] [PubMed] [Google Scholar]
- Zhang ZY. Protein tyrosine phosphatases: prospects for therapeutics. Curr Opin Chem Biol. 2001;5:416–423. doi: 10.1016/s1367-5931(00)00223-4. [DOI] [PubMed] [Google Scholar]
- Zhang ZY. Chemical and mechanistic approaches to the study of protein tyrosine phosphatases. Acc Chem Res. 2003;36:385–392. doi: 10.1021/ar020122r. [DOI] [PubMed] [Google Scholar]
- Hengge AC, Sowa GA, Wu L, Zhang ZY. Nature of the transition state of the protein-tyrosine phosphatase-catalyzed reaction. Biochemistry. 1995;34:13982–13987. doi: 10.1021/bi00043a003. [DOI] [PubMed] [Google Scholar]
- Wu L, Zhang ZY. Probing the function of Asp128 in the lower molecular weight protein-tyrosine phosphatase-catalyzed reaction. A pre-steady-state and steady-state kinetic investigation. Biochemistry. 1996;35:5426–5434. doi: 10.1021/bi952885a. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Palfey BA, Wu L, Zhao Y. Catalytic function of the conserved hydroxyl group in the protein tyrosine phosphatase signature motif. Biochemistry. 1995;34:16389–16396. doi: 10.1021/bi00050a020. [DOI] [PubMed] [Google Scholar]
- Zhang ZY, Thieme-Sefler AM, Maclean D, McNamara DJ, Dobrusin EM, Sawyer TK, Dixon JE. Substrate specificity of the protein tyrosine phosphatases. Proc Natl Acad Sci USA. 1993;90:4446–4450. doi: 10.1073/pnas.90.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittier SK, Hengge AC, Loria JP. Conformational motions regulate phosphoryl transfer in related protein tyrosine phosphatases. Science. 2013;341:899–903. doi: 10.1126/science.1241735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan J, Lee K, Cherry S, Tropea JE, Burke TR, Jr, Waugh DS. High-resolution structure of the Yersinia pestis protein tyrosine phosphatase YopH in complex with a phosphotyrosyl mimetic-containing hexapeptide. Biochemistry. 2003;42:13113–13121. doi: 10.1021/bi030156m. [DOI] [PubMed] [Google Scholar]
- Ivanov MI, Stuckey JA, Schubert HL, Saper MA, Bliska JB. Two substrate-targeting sites in the Yersinia protein tyrosine phosphatase co-operate to promote bacterial virulence. Mol Microbiol. 2005;55:1346–1356. doi: 10.1111/j.1365-2958.2005.04477.x. [DOI] [PubMed] [Google Scholar]
- Woestyn S, Sory MP, Boland A, Lequenne O, Cornelis GR. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Microbiol. 1996;20:1261–1271. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]
- Vujanac M, Stebbins CE. Context-dependent protein folding of a virulence peptide in the bacterial and host environments: structure of an SycH-YopH chaperone-effector complex. Acta Cryst D. 2013;69:546–554. doi: 10.1107/S0907444912051086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov AG, Tropea JE, Routzahn KM, Copeland TD, Waugh DS. Structure of the N-terminal domain of Yersinia pestis YopH at 2.0 A resolution. Acta Cryst D. 2001;57:793–799. doi: 10.1107/s0907444901004875. [DOI] [PubMed] [Google Scholar]
- Montagna LG, Ivanov MI, Bliska JB. Identification of residues in the N-terminal domain of the Yersinia tyrosine phosphatase that are critical for substrate recognition. J Biol Chem. 2001;276:5005–5011. doi: 10.1074/jbc.M009045200. [DOI] [PubMed] [Google Scholar]
- Khandelwal P, Keliikuli K, Smith CL, Saper MA, Zuiderweg ER. Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure. Biochemistry. 2002;41:11425–11437. doi: 10.1021/bi026333l. [DOI] [PubMed] [Google Scholar]
- Sory MP, Boland A, Lambermont I, Cornelis GR. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K, Uralil J, Bliska JB, Galan JE. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- Kulich SM, Yahr TL, Mende-Mueller LM, Barbieri JT, Frank DW. Cloning the structural gene for the 49-kDa form of exoenzyme S (exoS) from Pseudomonas aeruginosa strain 388. J Biol Chem. 1994;269:10431–10437. [PubMed] [Google Scholar]
- Forsberg A, Wolf-Watz H. Genetic analysis of the yopE region of Yersinia spp.: identification of a novel conserved locus, yerA, regulating yopE expression. J Bacteriol. 1990;172:1547–1555. doi: 10.1128/jb.172.3.1547-1555.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels T, Wattiau P, Brasseur R, Ruysschaert JM, Cornelis G. Secretion of Yop proteins by Yersiniae. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Modulation of host signaling by a bacterial mimic: structure of the Salmonella effector SptP bound to Rac1. Mol Cell. 2000;6:1449–1460. doi: 10.1016/s1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- Fu Y, Galan JE. Identification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. J Bacteriol. 1998;180:3393–3399. doi: 10.1128/jb.180.13.3393-3399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Galan JE. A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- Button JE, Galan JE. Regulation of chaperone/effector complex synthesis in a bacterial type III secretion system. Mol Microbiol. 2011;81:1474–1483. doi: 10.1111/j.1365-2958.2011.07784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murli S, Watson RO, Galan JE. Role of tyrosine kinases and the tyrosine phosphatase SptP in the interaction of Salmonella with host cells. Cell Microbiol. 2001;3:795–810. doi: 10.1046/j.1462-5822.2001.00158.x. [DOI] [PubMed] [Google Scholar]
- Lin SL, Le TX, Cowen DS. SptP, a Salmonella typhimurium type III-secreted protein, inhibits the mitogen-activated protein kinase pathway by inhibiting Raf activation. Cell Microbiol. 2003;5:267–275. doi: 10.1046/j.1462-5822.2003.t01-1-00274.x. [DOI] [PubMed] [Google Scholar]
- Humphreys D, Hume PJ, Koronakis V. The Salmonella effector SptP dephosphorylates host AAA+ ATPase VCP to promote development of its intracellular replicative niche. Cell Host Microbe. 2009;5:225–233. doi: 10.1016/j.chom.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HW, Brooking-Dixon R, Neupane S, Lee CJ, Miao EA, Staats HF, Abraham SN. Salmonella typhimurium impedes innate immunity with a mast-cell-suppressing protein tyrosine phosphatase, SptP. Immunity. 2013;39:1108–1120. doi: 10.1016/j.immuni.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- Kim DW, Chu H, Joo DH, Jang MS, Choi JH, Park SM, Choi YJ, Han SH, Yun CH. OspF directly attenuates the activity of extracellular signal-regulated kinase during invasion by Shigella flexneri in human dendritic cells. Mol Immunol. 2008;45:3295–3301. doi: 10.1016/j.molimm.2008.02.013. [DOI] [PubMed] [Google Scholar]
- Mazurkiewicz P, Thomas J, Thompson JA, Liu M, Arbibe L, Sansonetti P, Holden DW. SpvC is a Salmonella effector with phosphothreonine lyase activity on host mitogen-activated protein kinases. Mol Microbiol. 2008;67:1371–1383. doi: 10.1111/j.1365-2958.2008.06134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li H, Long C, Hu L, Xu H, Liu L, Chen S, Wang DC, Shao F. Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase. Mol Cell. 2007;28:899–913. doi: 10.1016/j.molcel.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang H, Zhang J, Gu L, Huang N, Zhou JM, Chai J. Structural basis for the catalytic mechanism of phosphothreonine lyase. Nature Struct Mol Biol. 2008;15:101–102. doi: 10.1038/nsmb1329. [DOI] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, Chen S, Tang X, Zhou JM. A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe. 2007;1:175–185. doi: 10.1016/j.chom.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Chang CI, Xu BE, Akella R, Cobb MH, Goldsmith EJ. Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b. Mol Cell. 2002;9:1241–1249. doi: 10.1016/s1097-2765(02)00525-7. [DOI] [PubMed] [Google Scholar]
- Bellon S, Fitzgibbon MJ, Fox T, Hsiao HM, Wilson KP. The structure of phosphorylated p38gamma is monomeric and reveals a conserved activation-loop conformation. Structure. 1999;7:1057–1065. doi: 10.1016/s0969-2126(99)80173-7. [DOI] [PubMed] [Google Scholar]
- Smith GK, Ke Z, Hengge AC, Xu D, Xie D, Guo H. Active-site dynamics of SpvC virulence factor from Salmonella typhimurium and density functional theory study of phosphothreonine lyase catalysis. J Phys Chem B. 2009;113:15327–15333. doi: 10.1021/jp9052677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Z, Smith GK, Zhang Y, Guo H. Molecular mechanism for eliminylation, a newly discovered post-translational modification. J Am Chem Soc. 2011;133:11103–11105. doi: 10.1021/ja204378q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Q, Christofferson A, Zhang H, Chai J, Huang N. Computational investigation of the enzymatic mechanisms of phosphothreonine lyase. Biophys Chem. 2011;157:16–23. doi: 10.1016/j.bpc.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Haneda T, Ishii Y, Shimizu H, Ohshima K, Iida N, Danbara H, Okada N. Salmonella type III effector SpvC, a phosphothreonine lyase, contributes to reduction in inflammatory response during intestinal phase of infection. Cell Microbiol. 2012;14:485–499. doi: 10.1111/j.1462-5822.2011.01733.x. [DOI] [PubMed] [Google Scholar]
- Harouz H, Rachez C, Meijer BM, Marteyn B, Donnadieu F, Cammas F, Muchardt C, Sansonetti P, Arbibe L. Shigella flexneri targets the HP1gamma subcode through the phosphothreonine lyase OspF. EMBO J. 2014;33:2606–2622. doi: 10.15252/embj.201489244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski DV, Mumy KL, Faherty CS, McCormick BA, Maurelli AT. Shigella flexneri type III secretion system effectors OspB and OspF target the nucleus to downregulate the host inflammatory response via interactions with retinoblastoma protein. Mol Microbiol. 2009;71:350–368. doi: 10.1111/j.1365-2958.2008.06524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiterer V, Grossniklaus L, Tschon T, Kasper CA, Sorg I, Arrieumerlou C. Shigella flexneri type III secreted effector OspF reveals new crosstalks of proinflammatory signaling pathways during bacterial infection. Cell Signal. 2011;23:1188–1196. doi: 10.1016/j.cellsig.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Ruckdeschel K, Machold J, Roggenkamp A, Schubert S, Pierre J, Zumbihl R, Liautard JP, Heesemann J, Rouot B. Yersinia enterocolitica promotes deactivation of macrophage mitogen-activated protein kinases extracellular signal-regulated kinase-1/2, p38, and c-Jun NH2-terminal kinase. Correlation with its inhibitory effect on tumor necrosis factor-alpha production. J Biol Chem. 1997;272:15920–15927. doi: 10.1074/jbc.272.25.15920. [DOI] [PubMed] [Google Scholar]
- Palmer LE, Hobbie S, Galan JE, Bliska JB. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNF-alpha production and downregulation of the MAP kinases p38 and JNK. Mol Microbiol. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- Palmer LE, Pancetti AR, Greenberg S, Bliska JB. YopJ of Yersinia spp. is sufficient to cause downregulation of multiple mitogen-activated protein kinases in eukaryotic cells. Infect Immun. 1999;67:708–716. doi: 10.1128/iai.67.2.708-716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schesser K, Spiik AK, Dukuzumuremyi JM, Neurath MF, Pettersson S, Wolf-Watz H. The yopJ locus is required for Yersinia-mediated inhibition of NF-kappaB activation and cytokine expression: YopJ contains a eukaryotic SH2-like domain that is essential for its repressive activity. Mol Microbiol. 1998;28:1067–1079. doi: 10.1046/j.1365-2958.1998.00851.x. [DOI] [PubMed] [Google Scholar]
- Orth K, Palmer LE, Bao ZQ, Stewart S, Rudolph AE, Bliska JB, Dixon JE. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285:1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- Paquette N, Conlon J, Sweet C, Rus F, Wilson L, Pereira A, Rosadini CV, Goutagny N, Weber AN, Lane WS, Shaffer SA, Maniatis S, Fitzgerald KA, Stuart L, Silverman N. Serine/threonine acetylation of TGFbeta-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc Natl Acad Sci USA. 2012;109:12710–12715. doi: 10.1073/pnas.1008203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Peak-Chew SY, McMahon HT. Acetylation of MEK2 and I kappa B kinase (IKK) activation loop residues by YopJ inhibits signaling. Proc Natl Acad Sci USA. 2006;103:18574–18579. doi: 10.1073/pnas.0608995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao YH, Wang Y, Burdette D, Mukherjee S, Keitany G, Goldsmith E, Orth K. Structural requirements for Yersinia YopJ inhibition of MAP kinase pathways. PLoS One. 2008;3:e1375. doi: 10.1371/journal.pone.0001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Peak-Chew SY, Sade RS, Vallis Y, McMahon HT. The acetyltransferase activity of the bacterial toxin YopJ of Yersinia is activated by eukaryotic host cell inositol hexakisphosphate. J Biol Chem. 2010;285:19927–19934. doi: 10.1074/jbc.M110.126581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier-Hyams LS, Zeng H, Sun J, Tomlinson AD, Bao ZQ, Chen H, Madara JL, Orth K, Neish AS. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-kappa B pathway. J Immunol. 2002;169:2846–2850. doi: 10.4049/jimmunol.169.6.2846. [DOI] [PubMed] [Google Scholar]
- Du F, Galan JE. Selective inhibition of type III secretion activated signaling by the Salmonella effector AvrA. PLoS Pathog. 2009;5:e1000595. doi: 10.1371/journal.ppat.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Wu H, Wentworth C, Luo L, Collier-Hyams L, Neish AS. Salmonella AvrA coordinates suppression of host immune and apoptotic defenses via JNK pathway blockade. Cell Host Microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Fehr D, Casanova C, Liverman A, Blazkova H, Orth K, Dobbelaere D, Frey J, Burr SE. AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappaB signalling pathway. Microbiology. 2006;152:2809–2818. doi: 10.1099/mic.0.28889-0. [DOI] [PubMed] [Google Scholar]
- Trosky JE, Mukherjee S, Burdette DL, Roberts M, McCarter L, Siegel RM, Orth K. Inhibition of MAPK signaling pathways by VopA from Vibrio parahaemolyticus. J Biol Chem. 2004;279:51953–51957. doi: 10.1074/jbc.M407001200. [DOI] [PubMed] [Google Scholar]
- Trosky JE, Li Y, Mukherjee S, Keitany G, Ball H, Orth K. VopA inhibits ATP binding by acetylating the catalytic loop of MAPK kinases. J Biol Chem. 2007;282:34299–34305. doi: 10.1074/jbc.M706970200. [DOI] [PubMed] [Google Scholar]
- Tasset C, Bernoux M, Jauneau A, Pouzet C, Briere C, Kieffer-Jacquinod S, Rivas S, Marco Y, Deslandes L. Autoacetylation of the Ralstonia solanacearum effector PopP2 targets a lysine residue essential for RRS1-R-mediated immunity in Arabidopsis. PLoS Pathog. 2010;6:e1001202. doi: 10.1371/journal.ppat.1001202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi AM, Nordfelth R, Uvell H, Wolf-Watz H, Elofsson M. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem Biol. 2003;10:241–249. doi: 10.1016/s1074-5521(03)00046-2. [DOI] [PubMed] [Google Scholar]
- Marshall NC, Finlay BB. Targeting the type III secretion system to treat bacterial infections. Expert Opin Therap Targ. 2014;18:137–152. doi: 10.1517/14728222.2014.855199. [DOI] [PubMed] [Google Scholar]
- Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- Wei P, Wong WW, Park JS, Corcoran EE, Peisajovich SG, Onuffer JJ, Weiss A, Lim WA. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012;488:384–388. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E, Eargle J, Wright D, Luthey-Schulten Z. MultiSeq: unifying sequence and structure data for evolutionary analysis. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]


