Abstract
Equine lentivirus receptor 1 (ELR1) has been identified as a functional cellular receptor for equine infectious anemia virus (EIAV). Herein, recombinant ELR1 and EIAV surface glycoprotein gp90 were respectively expressed in Drosophila melanogaster S2 cells, and purified to homogeneity by Ni-NTA affinity chromatography and gel filtration chromatography. Gel filtration chromatography and analytical ultracentrifugation (AUC) analyses indicated that both ELR1 and gp90 existed as individual monomers in solution and formed a complex with a stoichiometry of 1:1 when mixed. The structure of ELR1 was first determined with the molecular replacement method, which belongs to the space group P42212 with one molecule in an asymmetric unit. It contains eight antiparallel β-sheets, of which four are in cysteine rich domain 1 (CRD1) and two are in CRD2 and CRD3, respectively. Alignment of ELR1 with HVEM and CD134 indicated that Tyr61, Leu70, and Gly72 in CRD1 of ELR1 are important residues for binding to gp90. Isothermal titration calorimetry (ITC) experiments further confirmed that Leu70 and Gly72 are the critical residues.
Keywords: equine lentivirus receptor 1, structure, gp90, analytical ultracentrifugation, isothermal titration calorimetry
Introduction
Equine infectious anemia virus (EIAV), a member of the genus lentivirus in family retroviridae, can establish a persistent infection in horses and cause a unique dynamic lentiviral disease.1 Adsorption and entry of retroviruses into target cells requires the interaction of viral envelope glycoproteins with specific cellular receptor proteins.2 The envelope proteins of EIAV comprise the surface glycoprotein (gp90) and the transmembrane glycoprotein (gp45).3 Evolution studies indicate that gp90 is the predominant site of EIAV antigenic variation, and its distinct conserved and variable domains have been defined.4–6 Currently an attenuated vaccine for EIAV has been used successfully in China, and sequence comparisons have revealed some critical mutations associated with the vaccine strain.7,8
Equine lentivirus receptor 1 (ELR1), a functional cellular receptor for both primary and cell-adapted strains of EIAV, was first identified by Zhang et al.9 In contrast to most immunodeficiency lentiviruses, such as human immunodeficiency virus 1 (HIV-1), simian immunodeficiency virus (SIV) and feline immunodeficiency virus (FIV), which generally utilize dual receptors for successful infection,10–13 EIAV appears to depend only on ELR1 for the invasion of target cells. This is similar to the single receptor usage reported for simple retroviruses, such as murine or avian leukemia viruses.9,14,15 Analysis of its deduced amino acid sequence shows that ELR1 belongs to the tumor necrosis factor receptor (TNFR) protein family, with its ectodomain containing the characteristic four cysteine rich domains designated as CRD1 to CRD4.1,16 The most homologous TNFR protein sequence to ELR1 is the herpes virus entry mediator (HVEM) protein.17
Currently no structural information on ELR1 has been reported. Here, we expressed and purified the ectodomain of ELR1 and determined its crystal structure. We demonstrated that gp90 interacts directly with ELR1 using in vitro protein binding assays and revealed that Leu70 and Gly72 in CRD1 of ELR1 are the critical residues for this interaction using isothermal titration calorimetry (ITC).
Results
ELR1 binds gp90 to form a heterodimer
Soluble ELR1 and gp90 were efficiently expressed in S2 cells and purified to homogeneity by size-exclusion chromatography [Fig. 1(A)]. Both proteins were shown to be monomeric in solution based on the elution volume in size-exclusion chromatography. The molecular masses of ELR1 and gp90 were shown to be approximately 24 kDa and 55 kDa, respectively, by SDS-PAGE analysis [Fig. 1(C)]. When the proteins were mixed at a 1:1 ratio, approximately all of the ELR1 and gp90 were eluted in fractions corresponding to a higher molecular mass [Fig. 1(A)]. AUC analysis demonstrated that the molecular masses of ELR1 and gp90 were 23.4 kDa and 66.6 kDa respectively, while the ELR1-gp90 complex was 89.2 kDa [Fig. 1(B)]. The results indicated that the two proteins formed a stable heterodimer in solution.
Figure 1.
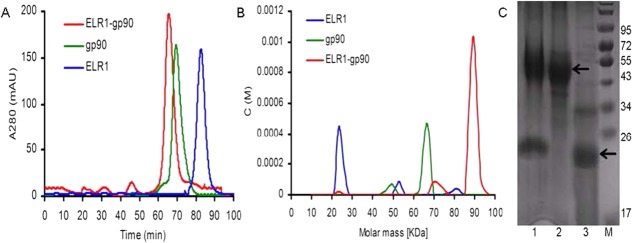
ELR1 binds gp90 to form a heterodimer. (A) ELR1, gp90, and ELR1-gp90 were subjected to gel filtration on a Superdex 200 column. (B) The masses of ELR1, gp90, and ELR1-gp90 were determined by AUC. The theoretical molecular masses of ELR1 and gp90 (without glycosylation) were 20.5 kDa and 47.7 kDa, respectively. (C) SDS-PAGE analysis. Lane 1, ELR1-gp90; Lane 2, gp90; Lane 3, ELR1; M, molecular-weight markers (kDa).
Overall structure of ELR1
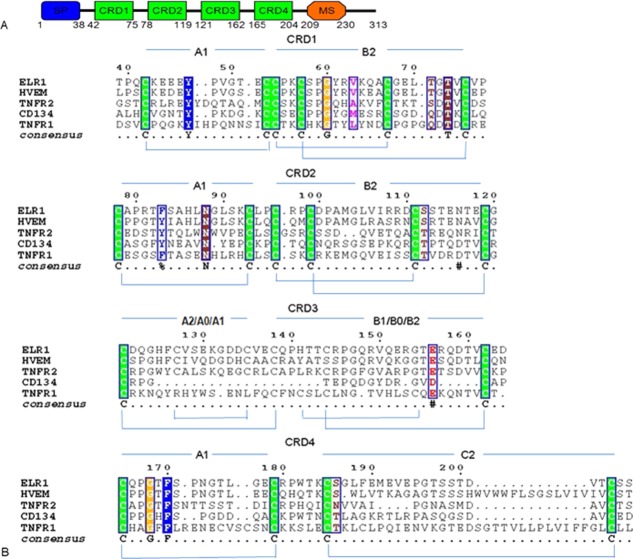
Sequence analysis showed that ELR1 adopts the canonical TNFR superfamily fold, in which CRDs 1–3 each contain three intradomain disulfide bonds and CRD4 contains two intradomain disulfide bonds (Fig. 2). However, close comparison revealed significant differences between ELR1 with other TNFR family members. CRDs 1–2 have the same disulfide connectivity as the corresponding domains of other homologs composed of A1–B2 modules, while CRD3 of ELR1 is composed of an A2–B1 module, which differs from other homologs. Compared to TNFR1, Cys127 and Cys135 contribute an additional disulfide bond in the A2 module in ELR1. Compared to HVEM, an extra disulfide bond formed by Cys144 and Cys162 is found in the B1 module of ELR1.
Figure 2.
Sequence alignment of ELR1 with other TNFRs. (A) Schematic representation of the domain organization of ELR1. Features include a signal peptide (SP, blue), four complete cysteine rich domains (CRD, green) and a membrane-spanning domain (MS, orange). (B) Sequence alignment between TNFR members by ClustalW.40 The sequences aligned are in the order: ELR1, HVEM, TNFR2, CD134 and TNFR1. Each CRD can be subdivided in two structural entities or modules and each module has been named (A1, B2, etc.) according to the definition adopted by Naismith.16 Within each CRD, cysteines forming disulfide bonds are in green.
The structure of ELR1 was determined by molecular replacement using a structural homology model based on the HVEM (PDB: 4FHQ). The crystal belongs to the space group P42212 with one molecule in an asymmetric unit (Table1). However, only residues 40–141 are clearly defined in structure and no additional electron density could be assigned to residues following 141 [Fig. 3(A)]. Due to tight packing of the crystal, we speculated that the protein was degraded during crystallization and residues following 141 were removed by residual proteases in solution. The defined structure consists of a total of eight antiparallel β-sheets, with four in CRD1 and two in CRDs 2–3, respectively [Fig. 3(B)].
Table 1.
Data Collection and Refinement Statistics
| Data collection | |
|---|---|
| Space group | P42212 |
| Wavelength (Å) | 1.00 |
| Unit cell | |
| a, b, c (Å) | 81.14, 81.14, 42.27 |
| α, β, γ (°) | 90, 90, 90 |
| Molecules/ASU | 1 |
| Resolution range (Å) | 50–1.49 (1.52–1.49) |
| Completeness (%)a | 99.69 (98.88) |
| Redundancya | 13.8(13.2) |
| No. of total reflections | 326254 |
| No. of unique reflections | 19085 |
| I/σa | 14.9(2.3) |
| Rsymab (%) | 6.6(55.1) |
| Refinement statistics | |
| Resolution (Å) | 1.50 |
| Rwork/Rfree (%)c | 19.49/20.03 |
| No. of atoms | |
| Protein | 754 |
| Water | 155 |
| B-factors (Å2) | |
| Protein | 23.0 |
| Water | 35.2 |
| r.m.s.d. | |
| Bond length (Å) | 0.006 |
| Bond angle | 1.24 |
| Ramachandran analysis | |
| Most favored (%) | 100 |
| Allowed (%) | 0 |
| Disallowed (%) | 0 |
ASU, asymmetric unit; r.m.s.d., root mean square deviation.
Values in parentheses are for the highest resolution shell.
Rsym = Σ|I − <I>|/Σ<I>, where I is the observed intensity, and <I> is the average intensity of multiple observations of symmetry-related reflections.
R = Σhkl|Fobs| − |Fcalc|/Σhkl|Fobs|. Rfree was calculated from 5% of the reflections excluded from refinement.
Figure 3.
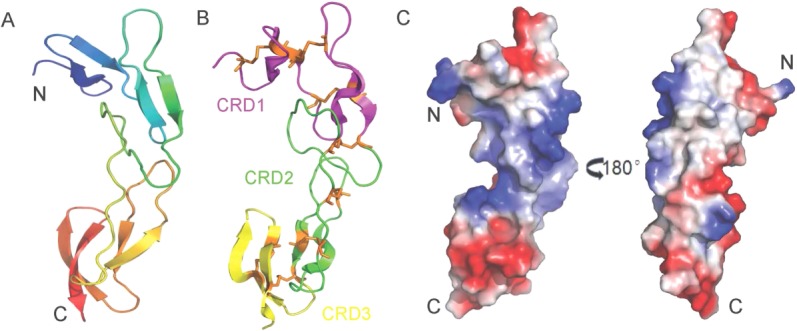
The overall structure of ELR1. (A) Cartoon representation of ELR1, showing a colored gradient from blue at the N-terminus (residue 40) to red at the C-terminus (residue 141). (B) Domain structure of ELR1. ELR1 is shown in a cartoon representation with CRDs 1–3 colored magenta, green and yellow, respectively. Disulfide bonds are shown as orange sticks. (C) The solvent-accessible electrostatic surface of ELR1. The surfaces are colored according to the electrostatic potential, ranging from deep blue (positive charge) to red (negative charge).
Table 2.
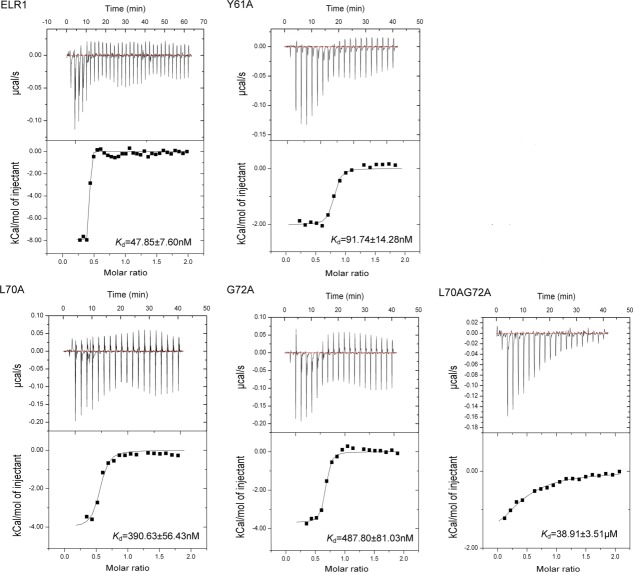
Thermodynamic Parameters of gp90 Binding to ELR1 and Its Mutants Measured by ITC
| ELR1 | Ka (M−1) | Kd | ΔH (kcal/mol) | TΔS (kcal/mol) |
|---|---|---|---|---|
| WT | 20.9 ± 1.73 E6 | 47.85 ± 7.61 nM | −5.55 ± 0.21 | 4.19 ± 0.35 |
| Y61A | 10.9 ± 0.93 E6 | 91.74 ± 14.28 nM | −2.01 ± 0.06 | 7.38 ± 0.47 |
| L70A | 2.56 ± 0.24 E6 | 390.6 ± 36.43 nM | −4.05 ± 0.29 | 4.48 ± 0.21 |
| G72A | 2.05 ± 0.22 E6 | 487.8 ± 41.03 nM | −4.62 ± 0.36 | 3.78 ± 0.19 |
| L70AG72A | 25.7 ± 2.65 E3 | 38.91 ± 3.51 μM | −2.54 ± 0.18 | 3.34 ± 0.23 |
All data were collected in the indicated buffer (50 mM Tris, 500 mM NaCl, pH 8.0) at 20°C in triplicate.
ELR1 and HVEM showed high sequence similarity of approximately 70% [Fig. 4(A)] and the crystal structures confirmed that the two TNFR family proteins adopt similar overall structures, despite some changes in their relative orientations and in the conformation of some loops [Fig. 4(B)]. Inspection of the molecular surface revealed different orientations of the N-termini in ELR1 and HVEM. The C-terminus in HVEM is highly polar with the positively charged R99 residue surrounded by hydrophobic chains, while the C-terminus in ELR1 is surrounded by negatively charged acidic residues [Fig. 4(C)].
Figure 4.
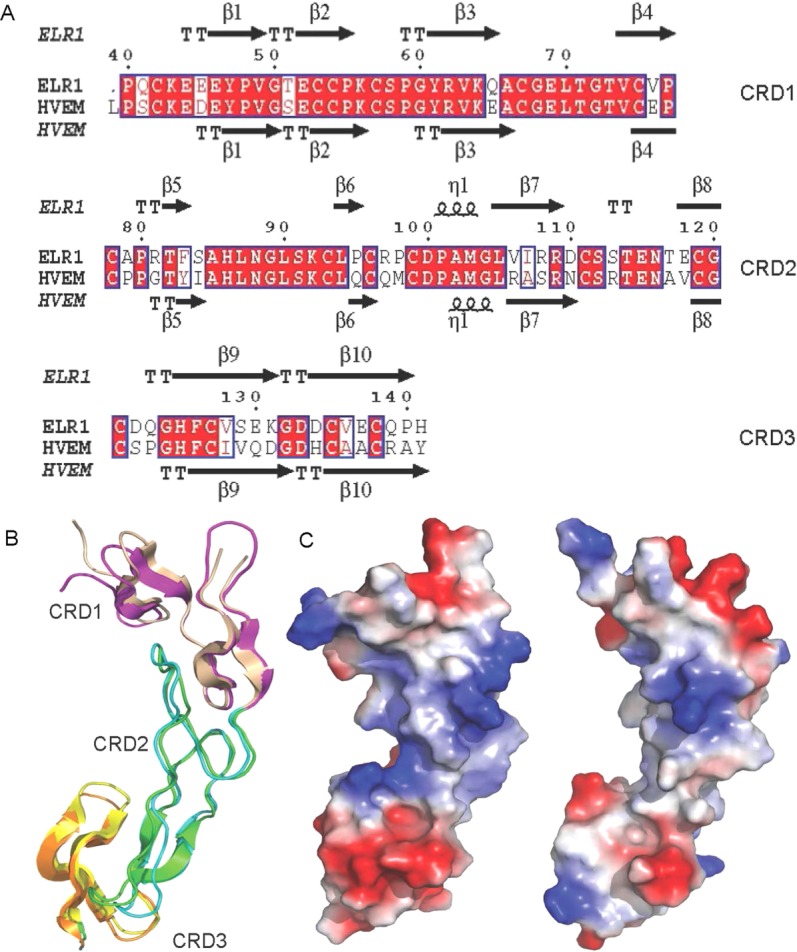
Structure comparison of ELR1 and HVEM. (A) Structure-based sequence alignment between ELR1 and HVEM (PDB: 4FHQ). Conserved residues are highlighted in red. Secondary structural elements are labeled. Numbering above the sequences corresponds to ELR1 residue numbers. (B) Superposition of the structurally conserved domains of ELR1 and HVEM. HVEM is shown in a cartoon representation with CRDs 1–3 colored yellow, cyan and orange, respectively. (C) The solvent-accessible electrostatic surface of ELR1 (left) and HVEM (right).
Putative gp90 binding sites on ELR1
Despite the lack of a precise model of the ELR1-gp90 complex, by analogy with other TNFR receptors, such as HVEM and CD134, the structure of ELR1 could provide insight into how lentivirus surface proteins interact with their receptors.
The CRD1 B2 module of ELR1 (residues 54–75) superposes very well on the equivalent region of HVEM (residues 15–37) and CD134 (residues 44–65) [Fig. 5(A)]. In the case of FIV-CD134 interactions, D60 and D62 are the critical residues for functional binding, corresponding to the position of L70 and G72 in ELR1. In the structure of the gD-HVEM complex, residues M11, A12 and L25 of gD form the core of a hydrophobic interaction with Y23 of HVEM. Superposition of ELR1 onto HVEM from the gD-HVEM (PDB: 1JMA) complex showed that Y61 of ELR1 is well positioned into the core formed by M11, A12, and L25 [Fig. 5(B)]. Furthermore, P17 and V36 of HVEM are important residues for functional binding, while S45 and K64 of CD134 at similar positions are also important residues, indicating that P55 and V74 of ELR1 located in the same position may have similar roles in the envelope–receptor interaction [Fig. 5(A)].
Figure 5.
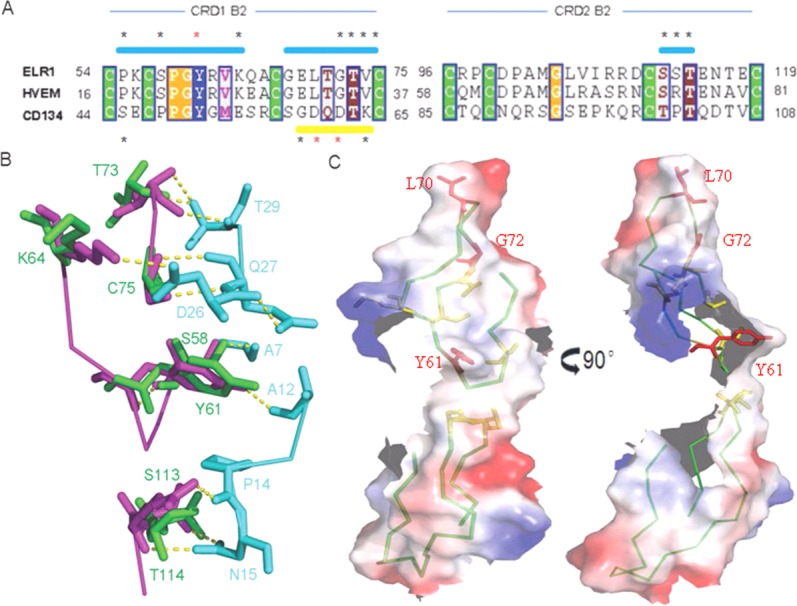
Binding sites of ELR1 to gp90. (A) Alignment of the CRDs 1–2 B2 modules of ELR1 with equivalent regions of HVEM and CD134. Identical residues are boxed and conserved cysteines are shaded green. Numbers indicate the amino acid positions. Surface binding regions are shown as a horizontal bar and colored cyan and yellow for HVEM and CD134, respectively. The important residues marked with * and the critical residues for interaction are further colored red. (B) Superposition of ELR1 onto HVEM from the gD-HVEM (PDB: 1JMA) complex to mimic the interactions. ELR1, HVEM and gD are colored green, magenta and cyan, respectively. The residues involved in the interaction are labeled. (C) Molecular surface of the ELR1 structure colored according to electrostatic potential. The critical and important residues for interaction are colored red and yellow, respectively. The critical residues are labeled.
The CRD2 B2 module of ELR1 (residues 96–119) superposes very well upon the equivalent region of HVEM (residues 58–81) and CD134 (residues 85–108) [Fig. 5(A)]. In the case of gD-HVEM interactions, S74 and T76, corresponding to S112 and T114, respectively, in ELR1, are important residues for functional binding. Superposition of ELR1 onto HVEM from the gD-HVEM complex showed that S112 and T114 of ELR1 are well positioned to form a hydrophilic interaction with N15 of gD [Fig. 5(B)]. Inspection of the molecular surface of ELR1 revealed a hydrophobic patch, composed of the critical residues L70, G72 and Y61 [Fig. 5(C)]. Other important residues, including P55, V74, S112 and T114, were all within this patch, implying that the binding site of gp90 is likely to be centered on these residues. These similarities between ELR1, CD134 and HVEM appear to reveal common themes in the use of TNFR family proteins as virus receptors, i. e., the interactions of surface proteins and their receptors are mediated by two regions on the TNFR: the CRD1 B2 module, which mediates direct contact for functional binding; and the CRD2 B2 module, which influences the surface protein binding affinity.
ITC measurements of ELR1 binding to gp90
The ELR1 residues critical for binding to gp90 were further studied by ITC. For wild-type ELR1, the heat changes corresponded best to a one-site sequential binding model that gave an apparent dissociation constant (Kd) of approximately 47.85 nM (Fig. 6). For the mutants, heat changes also corresponded best to a one-site sequential binding model. Mutant Y61A displayed a slightly higher Kd than that of the control (approximately 91.74 nM), whereas the other two mutants, L70A and G72A, yielded a much larger change (approximately 390.6 nM and 487.8 nM, respectively). Furthermore, the double mutation, L70AG72A, displayed a Kd of approximately 38.91 μM, which was significantly higher than that of the control, indicating that residues Leu70 and Gly72 are absolutely essential for strong binding of the CRD1 of ELR1 with gp90.
Figure 6.
Isothermal titration calorimetry (ITC) profiles of ELR1 binding to gp90. The observed binding isotherm is normalized as kilocalories per mole of injectant. The top panels show the raw data for heat changes (ΔH) produced by successive injections of gp90 into ELR1. The lower panel shows the integrated binding isotherms as a function of the molar ratio of gp90 to ELR1. The measured ΔH is corrected for heat changes from control experiments. The heat associated with each injection is calculated by integrating the area under the deflection of the measured signal (μcal/s).
Discussion
Studies of receptors in HIV-1, SIV and FIV infection have revealed a common theme of receptor usage, in which sequential binding of viral surface proteins to two distinct surface proteins is required for the infection of target cells.12,13,18 In contrast, the simple retroviruses, such as avian and murine oncoviruses, employ a single cell receptor to infect target cells.19–21 The general consensus has been that all lentiviruses use dual receptors for target cell infection; however, the recent finding that EIAV utilizes a single receptor protein, designated as ELR1, to infect target cells revealed an unexpected variation in the model of lentivirus receptor specificity.9 ELR1 is a member of the TNFR protein family, and TNFR-like proteins have been identified as receptors for several viruses, such as herpes simplex virus, certain avian oncoviruses and as a co-receptor component (CD134) for FIV.12,13,22,23
In contrast to HVEM, which exists as a dimer,24 the results of gel filtration experiments and AUC analysis showed that ELR1 is monomeric in solution. With the availability of soluble gp90 and ELR1, we studied their interactions in vitro and the results showed that gp90 binds specifically to ELR1 with a stoichiometry of 1:1. This is also in contrast to the gD-HVEM complex, which has a stoichiometry of 1:2.24,25 Despite both ELR1 and gp90 exist as monomers in solution, considering the trimeric state of TNFR family proteins and HIV gp120 on cell/virion surface, a trimer-trimer interaction would be plausible under physiological environment.
Lentivirus envelope proteins share similar structural features, despite a lack of amino acid sequence homology.26,27 Studies have indicated further key similarities between EIAV gp90 and HIV-1 gp120 with respect to variable-domain structure and functional properties, especially in their V3 regions.6,28–30 Comprehensive studies of the interaction between CD4 and HIV gp120 have indicated that the gp120 binding site is located at the tip of the first domain of CD4, with the critical residue F43 fitting into a large hydrophobic pocket of gp120 to initiate receptor binding.30 Alignment of ELR1 with CD134 indicated that L70 and G72 in the CRD1 B2 module are the critical resides for binding to gp90. These residues are located at the tip of CRD1 and in the β-turn projecting outward, similar to the critical residue F43 of CD4, implying that they fit into a large hydrophobic pocket of the surface protein to initiate receptor binding.30 Studies have indicated that single residues contribute a large fraction of the binding energy at protein-protein interfaces.31 These residues are located at “hot spots” of binding and are generally found at the center of the interface. Considering the structural similarities shared by EIAV gp90 and HIV-1 gp120, the structural and functional similarities between ELR1 and CD4 require further investigation.
During the past decades, ITC has been considered to be a powerful tool for investigating molecular interactions.32–34 In this study, we used this method to measure the binding affinity of ELR1 and gp90. The experiments unequivocally established that two residues, L70 and G72, in CRD1 of ELR1 are crucial to the interaction with gp90. The binding affinities of the ELR1 harboring L70A and G72A mutations for gp90 are approximately 8–10-fold weaker than that of the control, while the L70AG72A double mutation further reduces the receptor affinity by approximately 800-fold. Overall, the data support the prediction from the sequence and structure alignments that CRD1, comprising residues 54–75, plays a determining role in the interaction with gp90; this is consistent with the findings of Zhang et al.1
Taken together, these observations suggest that EIAV represents a critical transitional link between the simple oncoviruses and the more complex immunodeficiency lentiviruses in terms of genetic composition and receptor usage. The structure of the ELR1-gp90 complex is awaited to obtain a detailed characterization of the EIAV and its interactions with its receptor.
Materials and Methods
Cloning
The genes encoding full-length ELR1 and gp90 were provided by Jianhua Zhou at the Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Heilongjiang, China).7,8,35 The ectodomains of ELR1 (residues 39–206) and gp90 (residues 30–448) were amplified and cloned into the pMT/BiP/TEV-His expression vector, which was modified from the pMT/BiP/V5-His plasmid (Invitrogen, USA). To construct plasmids for the expression of ELR1 containing the mutations Y61A, L70A, G72A and L70AG72A, the pMT-ELR1 expression plasmid was mutated using the QuickChange site-directed mutagenesis kit (Stratagene, USA) according to manufacturer's instructions.
Establishment of cell lines
Recombinant S2 cell lines were generated and maintained following the manufacturer's instructions (Invitrogen, USA). Briefly, exponentially growing S2 cells were cotransfected with 20 μg of recombinant plasmid and 1 μg of the pCoBlast selection vector on the basis of the lipid-mediated transfection procedure using Cellfectin reagent (Invitrogen, USA). The cells were cultured for 2 days without drug selection before being resuspended in selection medium containing 20 μg/mL blasticidin. The selection medium was replaced every 3 days and stably transformed polyclonal cell populations were isolated after 3 weeks of selection.
Expression and purification
Stably transfected S2 cells were cultured at 27°C with constant stirring of 120 rpm until the cell density reached 6 × 106 cells/mL. Subsequently CuSO4 was added at a final concentration of 500 μM to induce production and secretion of recombinant proteins. Three days later, the medium was clarified by centrifugation at 3,000 rpm for 30 min and filtrated through a 0.22 μm Millipore filtration unit. The medium was then concentrated by ultrafiltration using a 10 kDa cutoff membrane at 4°C. After removing Cu2+ from the concentrated medium by dialysis against binding buffer (50 mM Tris, 500 mM NaCl, pH 8.0), His-tagged protein was purified using Ni-NTA resin according to the manufacturer's instruction (Qiagen, Germany). The eluted protein was then loaded onto a HiLoad 16/60 Superdex 200 size-exclusion column pre-equilibrated with the binding buffer. A sharp peak corresponding to the target protein was concentrated for further crystallization analysis.
Crystallization, data collection and structure determination
Crystallization analyses were carried out by the sitting-drop vapor-diffusion method at 293 K. Crystallization drops prepared by mixing equal volumes (1 µL) of protein solution (10 mg/mL protein, 50 mM Tris, 500 mM NaCl, pH 8.0) and precipitant solution (0.1 M citric acid, 25% w/v polyethylene glycol 3350, pH 3.5) were equilibrated against 0.1 mL reservoir solution. Diffraction data was collected at 100 K at the Shanghai Synchrotron Radiation Facility (Shanghai, China). The structure was determined by molecular replacement using the HVEM structure (PDB: 4FHQ) as a search model. The dataset was processed using the HKL2000 package36 and the preliminary model was refined using the Phenix program.37 The data collection and refinement statistics are summarized in Table1. All figures showing the structures were prepared using PyMOL.37
Analytical ultracentrifugation
Sedimentation velocity experiment was performed using a Beckman/Coulter XL-I analytical ultracentrifuge (Beckman/Coulter, USA) with double-sector or six-channel centerpieces and sapphirine windows. The experiment was conducted at 60,000 rpm and 4°C using absorbance detection and double-sector cells loaded with approximately 50 µM of all recombinant proteins. The data were analyzed using the programs SEDFIT and SEDPHAT.38,39
Isothermal titration calorimetry
ITC measurements were carried out by using VP-ITC (MicroCal, Britain). 400 µL of ELR1 (100 μM) was loaded into a sample cell and titrated with 100 µL of gp90 (250 μM) in the injection syringe. Control experiments were performed by injecting gp90 into the dialysis buffer to determine the heat change in dilution. The apparent heat change after each injection was determined by integration and corrected to remove unspecific ligand/buffer dilutions. The affinity is given as dissociation constant (Kd).
Atomic coordinates
The coordinate file and structure factors for crystal structure of ELR1 were deposited with the Protein Data Bank under accession codes 3WVT.
Glossary
- ELR1
equine lentivirus receptor 1
- TNFR
tumor necrosis factor receptor
- EIAV
equine infectious anemia virus
- AUC
analytical ultracentrifugation
- HVEM
herpes virus entry mediator
- CRD
cysteine rich domain
- HIV-1
human immunodeficiency virus 1
- SIV
simian immunodeficiency virus
- ITC
isothermal titration calorimetry
- FIV
feline immunodeficiency virus.
References
- Zhang B, Sun C, Jin S, Cascio M, Montelaro RC. Mapping of equine lentivirus receptor 1 residues critical for equine infectious anemia virus envelope binding. J Virol. 2008;82:1204–1213. doi: 10.1128/JVI.01393-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Zhang B, Jin J, Montelaro RC. Binding of equine infectious anemia virus to the equine lentivirus receptor-1 is mediated by complex discontinuous sequences in the viral envelope gp90 protein. J Gen Virol. 2008;89:2011–2019. doi: 10.1099/vir.0.83646-0. [DOI] [PubMed] [Google Scholar]
- Rushlow K, Olsen K, Stiegler G, Payne SL, Montelaro RC, Issel CJ. Lentivirus genomic organization: the complete nucleotide sequence of the env gene region of equine infectious anemia virus. Virology. 1986;155:309–321. doi: 10.1016/0042-6822(86)90195-9. [DOI] [PubMed] [Google Scholar]
- Payne SL, Fang FD, Liu CP, Dhruva BR, Rwambo P, Issel CJ, Montelaro RC. Antigenic variation and lentivirus persistence: variations in envelope gene sequences during EIAV infection resemble changes reported for sequential isolates of HIV. Virology. 1987;161:321–331. doi: 10.1016/0042-6822(87)90124-3. [DOI] [PubMed] [Google Scholar]
- Ball JM, Rushlow KE, Issel CJ, Montelaro RC. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J Virol. 1992;66:732–742. doi: 10.1128/jvi.66.2.732-742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroux C, Craigo JK, Issel CJ, Montelaro RC. Equine infectious anemia virus genomic evolution in progressor and nonprogressor ponies. J Virol. 2001;75:4570–4583. doi: 10.1128/JVI.75.10.4570-4583.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang S, Lin Y, Jiang C, Ma J, Zhao L, Lv X, Wang F, Shen R, Kong X, Zhou J. Genomic comparison between attenuated Chinese equine infectious anemia virus vaccine strains and their parental virulent strains. Arch Virol. 2011;156:353–357. doi: 10.1007/s00705-010-0877-8. [DOI] [PubMed] [Google Scholar]
- Qi X, Wang X, Wang S, Lin Y, Jiang C, Ma J, Zhao L, Lv X, Shen R, Wang F, Kong X, Su Z, Zhou J. Genomic analysis of an effective lentiviral vaccine-attenuated equine infectious anemia virus vaccine EIAV(FDDV13) Virus Genes. 2010;41:86–98. doi: 10.1007/s11262-010-0491-6. [DOI] [PubMed] [Google Scholar]
- Zhang BS, Jin S, Jin J, Li F, Montelaro RC. A tumor necrosis factor receptor family protein serves as a cellular receptor for the macrophage-tropic equine lentivirus. Proc Natl Acad Sci USA. 2005;102:9918–9923. doi: 10.1073/pnas.0501560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1-alpha, MIP-1-beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- de Parseval A, Chatterji U, Sun PQ, Elder JH. Feline immunodeficiency virus targets activated CD4(+) T cells by using CD134 as a binding receptor. Proc Natl Acad Sci USA. 2004;101:13044–13049. doi: 10.1073/pnas.0404006101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojima M, Miyazawa T, Ikeda Y, McMonagle EL, Haining H, Akashi H, Takeuchi Y, Hosie MJ, Willett BJ. Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science. 2004;303:1192–1195. doi: 10.1126/science.1092124. [DOI] [PubMed] [Google Scholar]
- Adkins HB, Brojatsch J, Naughton J, Rolls MM, Pesola JM, Young JAT. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc Natl Acad Sci USA. 1997;94:11617–11622. doi: 10.1073/pnas.94.21.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zhang BS, Weisz OA, Montelaro RC. Receptor-mediated entry by equine infectious anemia virus utilizes a pH-dependent endocytic pathway. J Virol. 2005;79:14489–14497. doi: 10.1128/JVI.79.23.14489-14497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naismith JH, Sprang SR. Modularity in the TNF-receptor family. Trends Biochem Sci. 1998;23:74–79. doi: 10.1016/s0968-0004(97)01164-x. [DOI] [PubMed] [Google Scholar]
- Montgomery RI, Warner MS, Lum BJ, Spear PG. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- de Parseval A, Chatterji U, Morris G, Sun PQ, Olson AJ, Elder JH. Structural mapping of CD134 residues critical for interaction with feline immunodeficiency virus. Nat Struct Mol Biol. 2005;12:60–66. doi: 10.1038/nsmb872. [DOI] [PubMed] [Google Scholar]
- Ott D, Rein A. Basis for receptor specificity of nonecotropic murine leukemia virus surface glycoprotein gp70SU. J Virol. 1992;66:4632–4638. doi: 10.1128/jvi.66.8.4632-4638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiden MV, Farrell K, Warsowe J, Mahan LC, Wilson CA. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J Virol. 1993;67:4056–4061. doi: 10.1128/jvi.67.7.4056-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass D, Davey RA, Hamson CA, Kim PS, Cunningham JM, Berger JM. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- Brojatsch J, Naughton J, Rolls MM, Zingler K, Young JAT. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- Adkins HB, Brojatsch J, Young JAT. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J Virol. 2000;74:3572–3578. doi: 10.1128/jvi.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitbeck JC, Peng C, Lou H, Xu R, Willis SH, Ponce De Leon M, Peng T, Nicola AV, Montgomery RI, Warner MS, Soulika AM, Spruce LA, Moore WT, Lambris JD, Spear PG, Cohen GH, Eisenberg RJ. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsters SA, Ayres TM, Skubatch M, Gray CL, Rothe M, Ashkenazi A. Herpesvirus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-kappa-B and AP-1. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- Gallaher WR, Ball JM, Garry RF, Griffin MC, Montelaro RC. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- Gallaher WR, Ball JM, Garry RF, Martin-Amedee AM, Montelaro RC. A general model for the surface glycoproteins of HIV and other retroviruses. AIDS Res Hum Retroviruses. 1995;11:191–202. doi: 10.1089/aid.1995.11.191. [DOI] [PubMed] [Google Scholar]
- Leroux C, Issel CJ, Montelaro RC. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J Virol. 1997;71:9627–9639. doi: 10.1128/jvi.71.12.9627-9639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YH, Nakaya T, Sentsui H, Kameoka M, Kishi M, Hagiwara K, Takahashi H, Kono Y, Ikuta K. Insertions, duplications and substitutions in restricted gp90 regions of equine infectious anaemia virus during febrile episodes in an experimentally infected horse. J Gen Virol. 1997;78:807–820. doi: 10.1099/0022-1317-78-4-807. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Weber PC, Salemme FR. Applications of calorimetric methods to drug discovery and the study of protein interactions. Curr Opin Struc Biol. 2003;13:115–121. doi: 10.1016/s0959-440x(03)00003-4. [DOI] [PubMed] [Google Scholar]
- Liang Y. Applications of isothermal titration calorimetry in protein science. Acta Biochem Biophys Sin. 2008;40:565–576. doi: 10.1111/j.1745-7270.2008.00437.x. [DOI] [PubMed] [Google Scholar]
- Freire E. A thermodynamic approach to the affinity optimization of drug candidates. Chem Biol Drug Des. 2009;74:468–472. doi: 10.1111/j.1747-0285.2009.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YZ, Yang F, Zhang SQ, Sun LK, Wang XF, Du C, Zhou JH. The soluble form of the EIAV receptor encoded by an alternative splicing variant inhibits EIAV infection of target cells. PLoS One. 2013;11:1–9. doi: 10.1371/journal.pone.0079299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]