Abstract
Pseudomonas aeruginosa is a Gram-negative opportunistic bacterium, synonymous with cystic fibrosis patients, which can cause chronic infection of the lungs. This pathogen is a model organism to study biofilms: a bacterial population embedded in an extracellular matrix that provide protection from environmental pressures and lead to persistence. A number of Chaperone-Usher Pathways, namely CupA-CupE, play key roles in these processes by assembling adhesive pili on the bacterial surface. One of these, encoded by the cupB operon, is unique as it contains a nonchaperone-usher gene product, CupB5. Two-partner secretion (TPS) systems are comprised of a C-terminal integral membrane β-barrel pore with tandem N-terminal POTRA (POlypeptide TRansport Associated) domains located in the periplasm (TpsB) and a secreted substrate (TpsA). Using NMR we show that TpsB4 (LepB) interacts with CupB5 and its predicted cognate partner TpsA4 (LepA), an extracellular protease. Moreover, using cellular studies we confirm that TpsB4 can translocate CupB5 across the P. aeruginosa outer membrane, which contrasts a previous observation that suggested the CupB3 P-usher secretes CupB5. In support of our findings we also demonstrate that tps4/cupB operons are coregulated by the RocS1 sensor suggesting P. aeruginosa has developed synergy between these systems. Furthermore, we have determined the solution-structure of the TpsB4-POTRA1 domain and together with restraints from NMR chemical shift mapping and in vivo mutational analysis we have calculated models for the entire TpsB4 periplasmic region in complex with both TpsA4 and CupB5 secretion motifs. The data highlight specific residues for TpsA4/CupB5 recognition by TpsB4 in the periplasm and suggest distinct roles for each POTRA domain.
Keywords: biofilm, two-partner secretion, chaperone-usher, POTRA, Pseudomonas aeruginosa, virulence factor, NMR
Introduction
Pseudomonas aeruginosa is a gram-negative opportunistic pathogen of animals, including humans, which can infect the eyes, lungs, kidneys, and urinary tract and lead to fatal consequences. P. aeruginosa is considered to be a model organism for studying bacterial biofilms1,2 and uses a large number of extracellular appendages and adhesins. Several Chaperone-Usher Pathways3–6 have been identified in P. aeruginosa (CupA to CupE)7–12 that sustain this extracellular lifestyle, although for many of them their specific targets still remain unknown. However, it has been shown that CupA pili promote adherence to solid surfaces,9 CupB/CupC pili act in synergy to support microcolony formation,12 CupD pili function to increase biofilm formation and decrease motility11,13 and CupE pili aid in colony formation during early biofilms while also shaping the three-dimensional architecture of maturing biofilms.10 Furthermore, these structures are differentially regulated,14 which supports the notion that they play distinct roles during biofilm formation and/or allow the colonization of specific environmental niches.7,8,10–12
The CupC pili form hair-like structures on the bacterial surface12 and their production is tightly controlled by the Roc systems,15 specifically the RocS1 sensor.16 CupB pili are also controlled by the Roc system, but are assembled by an atypical chaperone-usher pathway [Fig. 1(A)]. This system contains two chaperones (CupB2 and CupB4)17 and a ∼95 kDa gene product, CupB5, that is not found in other chaperone-usher pathways.18 In P. aeruginosa there are also five known two-partner secretion (TPS) systems: Tps1-Tps5,19 which secrete specific high molecular weight substrates that play distinct roles in virulence.20 The Tps3 system transports a large adhesive structure (TpsA3 or CdrA) onto the surface of P. aeruginosa that promotes interbacterial aggregation through interactions via Psl polysaccharides.21 On the other hand, the Tps4 system [Fig. 1(A)] encodes an extracellular protease (TpsA4 or LepA) that modulates host responses to infection and liberates nutrients for bacterial growth.22,23
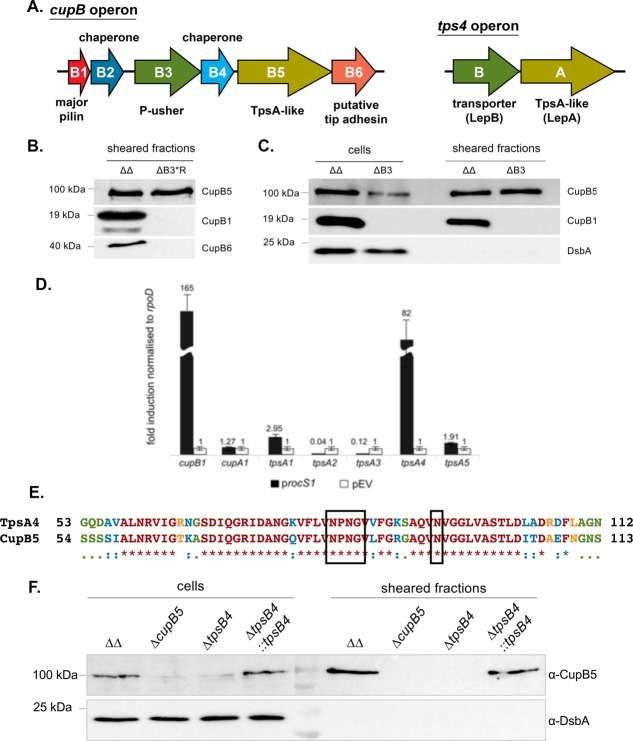
Figure 1.
CupB5 is not secreted by CupB3, but by TpsB4. (A) Schematic representation of the cupB and tps4 operons. (B) Immunoblot analysis of sheared fractions, harvested from bacteria grown on M63 agar plates, obtained from PAO1ΔΔ (ΔΔ: ΔpilA ΔfliC mutant) and PAO1ΔΔΔcupB3*R (ΔB3*R; cupB3 mutant from Ruer et al.18) conjugated with pMMBrocS1, using antibodies derived against CupB1, CupB5 and CupB6. (C) Immunoblot analysis of cell and sheared fraction, harvested from bacteria grown static in liquid M63 medium, obtained from PAO1ΔΔ (ΔΔ) and PAO1ΔΔΔcupB3 (ΔB3; cupB3 mutant from this study), conjugated with pMMBrocS1, using antibodies derived against CupB1, CupB5 and DsbA. (D) Fold induction of cupB1, cupA1, tpsA1 (PA2462), tpsA2 (PA0041), tpsA3 (PA4625), tpsA4 (PA4541) and tpsA5 (PA0690) mRNA transcription in PAO1ΔΔ in the presence (procS1) or absence (pEV) of RocS1. The fold induction was normalized to the house keeping gene rpoD, which encodes the sigma factor 70. (E) Primary sequence alignment between the N-terminus of mature P. aeruginosa TpsA4 and CupB5 using ClustalW2.66 Identical residues are red (*), highly similar residues are blue (:), weakly similar residues are green (.) and none homologous residues are orange. The conserved NPNG box and Asn residue of TpsA4 are boxed. Numbering is based on mature protein minus the N-terminal signal sequences. (F) Immunoblot of cell and sheared fraction obtained from PAO1ΔΔ (ΔΔ), PAO1ΔΔΔcupB5, PAO1ΔΔΔtpsB4 (ΔtpsB4) and PAO1ΔΔΔtpsB4::tpsB4 (ΔtpsB4::tpsB4) using antibodies derived against CupB5 and DsbA. The cupB operon was induced with RocS1.
As the name suggests, TPS systems are usually composed of just two components: a dedicated outer membrane transporter belonging to the Omp85 superfamily24 and a single passenger exoprotein generically termed TpsB and TpsA, respectively, which are usually positioned in tandem within the genome.25,26 The translocation unit is comprised of a membrane-embedded β-barrel pore with two N-terminal periplasmic POTRA (POlypeptide TRansport Associated) domains and an N-terminal α-helix that is located within the barrel.27 POTRA domains have well-defined domain orientations28 and in TpsB transporters they recognize extended conformers of the secretion domain of their TpsA passenger.29,30 In the resting state, the N-terminal helix of the transporter has been observed in the periplasm, although this compartment is believed to be sparsely populated.31 However, in the presence of cargo the equilibrium shifts toward this state, which allows the threading of the substrate through the pore and its eventual folding at the cell surface.27,31,32 The formation of β-helical architectures when outside of the cell is believed to generate the force needed for translocation.27 Although there is no direct structural evidence, periplasmic POTRA domains are thought to function by augmentation of their three-stranded β-sheet with an extended cargo conformation.33–36
Although CupB5 is predicted to form a TpsA-like structure, it was previously understood that CupB5 secretion was not dependent on a TPS system, but instead exploited the usher CupB3 from the operon in which it is embedded.18 We now show using nuclear magnetic resonance (NMR) spectroscopy that TpsB4 recognizes both CupB5 and its predicted cognate substrate TpsA4. Additional genetic manipulation of the cupB and tps4 operons coupled with cellular assays also clearly demonstrates that TpsB4 is responsible for the translocation of CupB5, rather than the P-usher CupB3. Furthermore, we provide new molecular insight into POTRA domain interactions and show that each domain plays a unique role in substrate recognition and coordinating translocation. As many other bacterial pathogens also use TPS systems to cause disease, this work highlights a general secretion mechanism that could be exploited in the fight against emerging bacterial resistance to antibiotics.
Results
CupB3-independent secretion of the TpsA-like protein CupB5
Secretion of the TpsA-like protein CupB5 was previously thought to be dependent on a P-usher,18 CupB3, suggesting a subtle integration of chaperone-usher pathways and TPS systems. In the present study and using similar growth conditions on agar plates set up in our laboratory, we observed that CupB5 could be readily recovered in the sheared fraction of this cupB3 mutant18 (PAO1ΔΔΔcupB3*R: ΔcupB3 mutant within the ΔpilA ΔfliC mutant PAO1 strain PAO1ΔΔ) and thus was still able to reach the surface [Fig. 1(A,B)]. At the same time the CupB1 and CupB6 pilin subunits were recovered in the sheared fraction of the parental strain, but not in the cupB3 mutant, confirming the expected defect in secretion for the CupB system. In addition we explored this observation using alternative conditions, in which the strains are grown in static culture instead of on agar plates. Again, using Western blot and CupB5-specific antibodies, it was found that CupB5 is systematically released in the sheared fraction. CupB5 secretion was also confirmed in a new cupB3 mutant lacking the P-usher (PAO1ΔΔΔcupB3), which was created independently for this study [Fig. 1(C)]. The periplasmic protein DsbA was used as a control and its absence in the sheared fractions indicates a lack of cell leakage. Taken together this suggests that in different laboratory environments alternative transporters to CupB3 play a role in CupB5 secretion.
TpsB4 is a genuine transporter of CupB5
TpsA proteins are usually transported by cognate TpsB transporters, therefore it is plausible that CupB5, by virtue of its similarity with TpsA-like proteins could use one of the five TpsB transporters that have been identified in P. aeruginosa.37 qRT-PCR analysis was performed to investigate whether any of the five tpsA/tpsB gene couples could be upregulated upon overexpression of rocS1 and thus be coregulated with the cupB operon, including cupB5. As a negative control we monitored the induction of cupA1, which unlike the expression of the cupC operon, is not synchronized with the cupB gene cluster.7,12 Strikingly, whereas cupB1 gene expression was readily induced upon introduction of the rocS1 gene, the tpsA4 gene also displayed an over 80-fold increase in expression [Fig. 1(D)]. The product of the tpsA4 gene has been identified as a protease (LepA)22,23 and its cognate transporter is believed to be the product of the adjacent tpsB4 (lepB) gene. None of the other tps genes, or the cupA1 gene, responded to the RocS1 control, suggesting that TpsB4 is a likely candidate for CupB5 transport.
TPS-dependent transport requires the presence of a ∼250-residue secretion or TPS domain at the N-terminus of the TpsA protein.29,30,37 This domain contains residues involved in the recognition of TpsB POTRA domains and provides the minimum sequence required to promote folding of TpsA β-helical structures on the extracellular surface and export from the cell. We hypothesized that TpsB4 is the transporter for CupB5 and should possess a similar TPS domain pattern to TpsA4. Amino acid sequence alignment between mature CupB5 (minus the N-terminal signal peptide) and mature TpsA1 (PA2462), TpsA2 (PA0041), TpsA3 (PA4625), TpsA4 (PA4541), and TpsA5 (PA0690) gave sequence identities of 14.4%, 14.4%, 34.8%, 57.2%, and 16.4%, respectively. Furthermore, in addition to CupB5 and TpsA4 sharing significant sequence identity within this domain, 40-residues between S68-D107 (TpsA4) and S69-D108 (CupB5) are either identical or highly conserved [Fig. 1(E)]. Overall, our data led us to re-evaluate previous observations that the CupB3 usher is the dedicated transporter of CupB518 and test whether TpsB4 plays a role in this process.
The tpsB4 gene was deleted from the PAO1ΔΔ chromosome and upon overexpression of RocS1 secretion of CupB5 was assessed in liquid media as described above. Our data showed that lack of tpsB4 readily abrogates CupB5 secretion, which could then be recovered by reintroducing the tpsB4 gene at the att site on the chromosome of the PAO1ΔΔΔtpsB4 strain [PAO1ΔΔΔtpsB4::tpsB4; Fig. 1(A,F)]. The lack of CupB5 in the cell fraction when the transporter is not present is expected for TPS systems, as substrates are rapidly degraded in the periplasm if not secreted efficiently.38–41 Our data indicate that CupB5 secretion is dependent on the TpsB4 transporter.
Solution structure of the TpsB4 POTRA1 domain
To address the molecular mechanism by which TpsB4 potentially recognizes two substrates, that is, both CupB5 and TpsB4, we used the multiple-threading alignment combined with fragment assembly simulation protocol of I-Tasser to model TpsB4 and facilitate the design of constructs for interactional analysis by NMR42 [Fig. 2(A)]. I-Tasser produced a TpsB4 model with a C-score of 0.58, sharing tertiary homology with the structure of the Bordetella pertussis TpsB protein, FhaC.27 The model contains a C-terminal ∼350-residue β-barrel pore and an N-terminal ∼200-residue periplasmic region (TpsB4-NT) that can be further subdivided into a ∼20-residue N-terminal plug helix (involved in gating the pore), followed by a ∼20-residue linker and two adjacent POTRA domains (POTRA1 and POTRA2, respectively). Based on these predictions we produced recombinant His6-tagged TpsB4 proteins containing the entire periplasmic region (TpsB4-NT; Residues 1–194), POTRA1–2 domains (TpsB4-P12; Residues 42–194), POTRA2 domain (TpsB4-P2; Residues 122–194), POTRA1 domain (TpsB4-P1; Residues 42–121), and the N-terminal plug helix, linker, and POTRA1 domain (TpsB4-a1P1; Residues 1–121; Fig. 2(B)). Residue numbering is based on mature TpsB4, minus the N-terminal signal sequence. These were expressed in Escherichia coli K12 strain and purified by nickel affinity and gel filtration chromatography. 1D 1H and 2D 1H-15N HSQC NMR spectra were recorded on purified proteins and showed only TpsB4-P1 and TpsB4-α1P1 to be correctly folded under these conditions. For example, NMR analyses of samples containing the POTRA2 domain showed significantly large methyl peaks in 1D spectra relative to TpsB4-α1P1 and TpsB4-P1, and although chemical shifts for the POTRA1 domain were present in 2D spectra, no dispersion was observed for the second domain (data not shown). The mis-folding of recombinant material for the second POTRA domain is likely due to lack of stabilizing interactions with the periplasmic face of the outer membrane β-barrel.
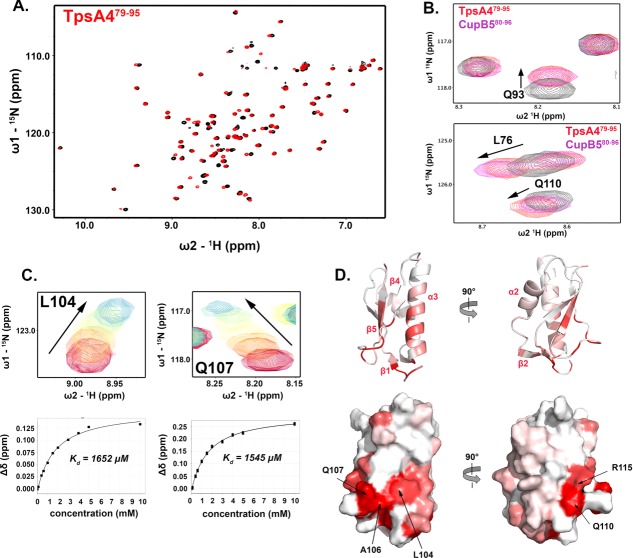
Figure 2.
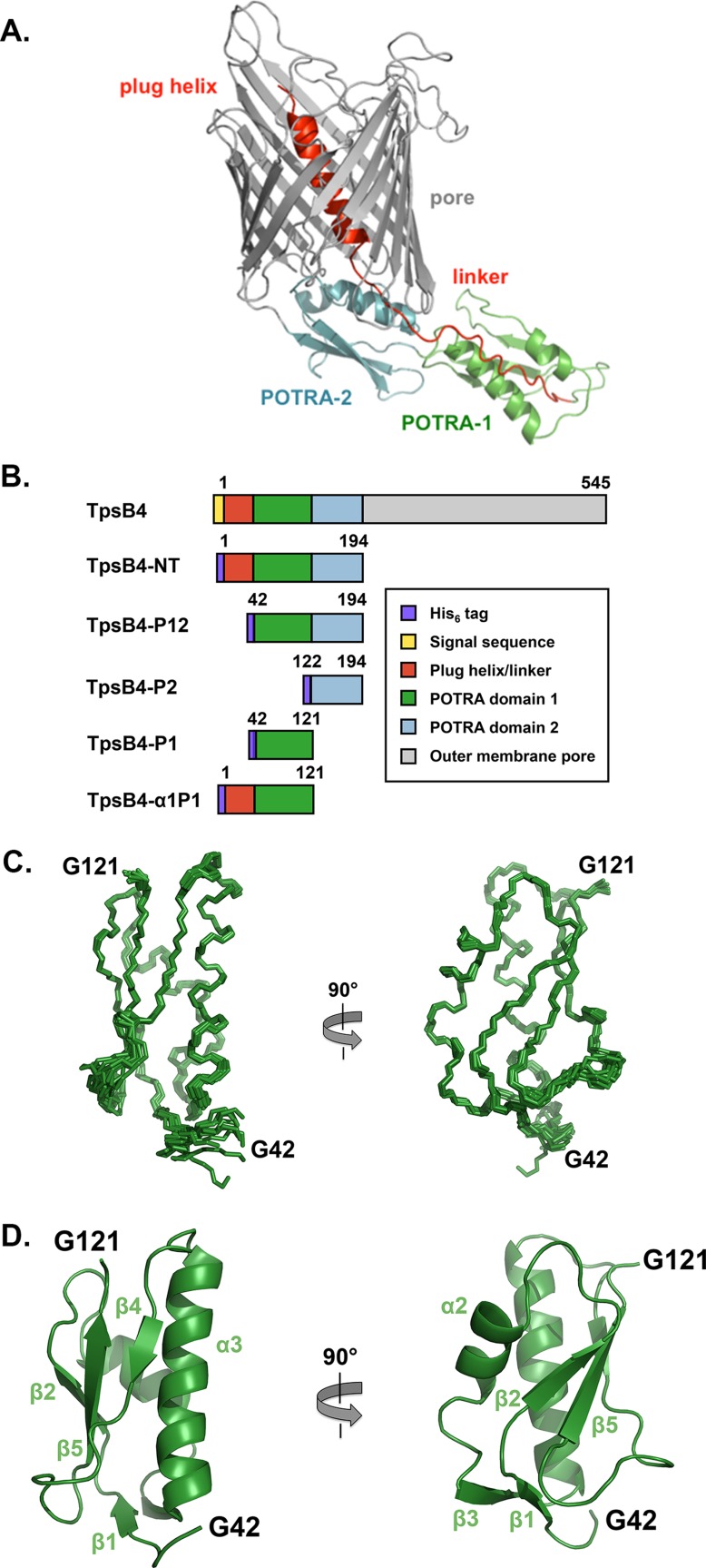
Domain organization of TpsB4 and solution NMR structure of the TpsB4 POTRA1 domain. (A) Model of full length P. aeruginosa TpsB4 generated using the I-Tasser server.42 (B) Schematic representation of the constructs used in this study and domains are colored as in (A). (C) Backbone superimposition of the ten best NMR structures of TpsB4-P1 shown in two orientations. An interactive view is available in the electronic version of the article (D) Secondary structure of one of the final TpsB4-P1 structures shown as cartoon. Numbering is based on mature TpsB4 minus the N-terminal signal sequence. An interactive view is available in the electronic version of the article
To elucidate the solution structure of the POTRA1 domain we used heteronuclear NMR methods. From both manual and ARIA (Ambiguous Restraints for Iterative Assignment) NMR assignment methods,43 a total of 1818 nuclear Overhauser effects (NOEs) were assigned in TpsB4-P1 15N/13C-edited NOESY spectra at pH 7.0. The structure determination was also supplemented with ϕ/φ dihedral angles and hydrogen bond restraints. The average pair-wise root-mean squared deviation (RMSD) for the water-refined final structures is 0.17 ± 0.04 Å for the backbone atoms and 0.49 ± 0.06 Å for the heavy atoms of residues within secondary structure. Structural statistics are shown in Table1.
Table 1.
Summary of NMR Structural Restraints and Statistics
| Distance restraints | |
|---|---|
| Total NOE | 1818 |
| Intraresidue | 480 |
| Sequential (|i − j|) = 1) | 207 |
| Short range (|i − j|) > 3) | 123 |
| Medium range (|i − j|) < 4) | 59 |
| Long range (|i − j|) > 5) | 290 |
| Ambiguous | 659 |
| Dihedral angle restraints | 106 |
| φ | 53 |
| ψ | 53 |
| Hydrogen bonds restraints | 8 |
| Structural statisticsa | |
| Violations | |
| Distance restraints (Å) | 0.014 ± 0.005 |
| Dihedral angle restraints (°) | 0.25 ± 0.016 |
| Deviation from idealized geometry | |
| Bond length (Å) | 0.0014 ± 0.000064 |
| Bond angle (°) | 0.313 ± 0.0034 |
| Impropers (°) | 0.173 ± 0.0040 |
| Average pairwise rmsd (Å) | |
| Heavy atoms | 0.17 ± 0.04 |
| Backbone atoms | 0.49 ± 0.06 |
| Structural qualityb | |
| Ramachandran statistics | |
| Favored regions (%) | 98.0 |
| Allowed regions (%) | 99.8 |
| Disallowed regions | 4 residuesc |
Average values and standard deviations over the 10 lowest energy conformers with respect to the average structure.
Percentage of residues in the Ramachandran plot regions determined by MOLPROBITY66 using an average structure from the 10 lowest energy conformers.
All outlier are situated in highly dynamic regions.
All areas of secondary structure are well defined, however, there is less precision in the β4-β5 loop and at the N-terminus preceding the β1-strand [Fig. 2(C,D)]. The increased flexibility for these regions is supported by measurement of {1H}-15N heteronuclear NOEs, which report on ns-ps timescale motions (Supporting Information Fig. S1). In addition to the canonical β-α-α-β-β POTRA-domain topology (β2-α2-α3-β4-β5),27,33 TpsB4-P1 forms an antiparallel β1-β3 sheet pairing at the N-terminal base that is not observed in FhaC27 (pdb: 2qdz), the only other published TpsB POTRA domain structures [Fig. 2(D)]. Another interesting feature is that we observe NOE correlations between the N-terminal residues of the POTRA domain and the C-terminal pole of the α3-helix. This region, which is directly adjacent to the N-terminal plug helix linker in the intact TpsB4 structure, adopts a helical conformation that creates a pseudo-continuous helix with α3, mediated by electrostatic interactions from the α3-helix dipole, the α3-helix residue S79 and the carbonyl groups of G42 and P44.
In the crystal structure of FhaC,27 electron density for the linker that connects the N-terminal plug helix and POTRA1 was not observed suggesting that it is flexible. Also, a recent biophysical study demonstrated that this linker is most likely constrained in the FhaC closed state, potentially through interactions with the POTRA domains.31 We next used NMR to probe this hypothesis in TpsB4 by comparing 1H-15N HSQC spectra of TpsB4-P1, and for a construct extended with the plug helix linker (TpsB4-α1P1). Significant chemical shift differences are observed [Fig. 3(A)]. The largest of these are mapped to the N-terminus, α3-helix and β4-β5-loop, indicating that the linker to the plug helix may fold back and interact transiently within this region [Fig. 3(B)].
Figure 3.
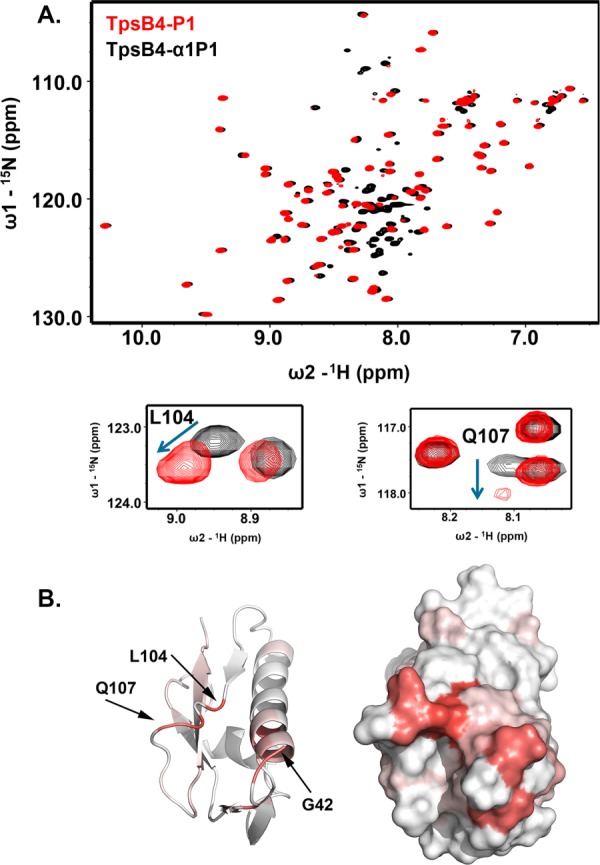
The TpsB4 N-terminal helix linker interacts with its POTRA1 domain. (A) Comparison of 1H-15N HSQC NMR spectra for 15N-labelled TpsB4-P1 (red) and TpsB4α1P1 (black), with chemical shift perturbations associated with L104 and Q107 in the β4-β5 loop expanded. (B) Chemical shift mapping on TpsB4-P1 of peak perturbation between 1H-15N HSQC NMR spectra of 15N-labelled TpsB4-P1 and TpsB4-α1P1. Deeper red areas indicate residues in closer vicinity to the TpsB4-α1P1 N-terminal plug helix/linker. Numbering is based on mature TpsB4 minus the N-terminal signal sequence. An interactive view is available in the electronic version of the article
TpsB4-P1 recognizes a conserved sequence in both TpsA4 and CupB5
We next examined how the TpsB4 POTRA1 domain recognizes its cargo. Rather than initiating these studies with CupB5, however, we focussed on TpsA4, which is believed to be its cognate substrate.23 It has been shown in other systems that these events can only be established when the secretion domain is unfolded,30 therefore a His6 tagged TpsA4 secretion domain (TpsA41–242; Residues 1–242 with numbering based on mature TpsA4 minus the N-terminal signal sequence) was purified under denaturing conditions prior to dissection. This was diluted into nondenaturing buffer in the presence of TpsB4-P1, treated with the protease trypsin and then heated to 70°C in trifluoroacetic acid to dissociate any complexes/aggregates. The TpsA41–242 peptide library was next separated by gel filtration and each peak that eluted was pooled for use in separate NMR titration experiments monitoring chemical shift perturbations (CSPs) in 1H-15N HSQC spectra of 15N-labelled TpsB4-P1 (data not shown). Samples that showed positive interactions were sent for MALDI-MS analysis. While several TpsA4 fragments were detected in these samples, repeat titrations with synthetic peptides showed that only V80FLVNPNGVVFGKSAQVNVGGLVASTLDLADR111 (TpsA480–111) bound 15N-labelled TpsB4-P1 (data not shown). Furthermore, when this was truncated at the C-terminus, the sequence K79VFLVNPNGVVFGKSAQ95 (TpsA479–95) produced identical CSPs in 1H-15N HSQC peak positions as observed for TpsA480–111 [Fig. 4(A)].
Figure 4.
NMR analysis of TpsB4 POTRA1 domain and TpsA4/CupB5 interactions. (A) 1H-15N HSQC spectra of 15N-labelled TpsB4-P1 (black) with 20 molar equivalents of TpsA479–95 (red). (B) Expanded 1H-15N HSQC spectra for 15N-labelled TpsB4-P1 with two molar equivalents of TpsA479–95 (red) or CupB580–96 (purple). (C) Two representative regions of 1H-15N HSQC spectra for 15N-labelled TpsB4-P1 titrated against TpsA479–95. Examples of the corresponding binding curves are displayed below. (D) Composite TpsA479–95 and CupB580–96 interactions mapped onto the surface of TpsB4-P1. Red areas indicate residues in close vicinity to TpsA479–95 and CupB580–96. Numbering is based on mature proteins minus N-terminal signal sequences. An interactive view is available in the electronic version of the article
The sequence of TpsA479–95 is almost indistinguishable to a region in CupB5 and NMR titrations with the corresponding peptide (CupB580–96; Q80VFLVNPNGVLFGRGAQ96 with numbering based on mature CupB5 minus the N-terminal signal sequence) show that it also interacts with TpsB4-P1 with a virtually identical pattern of CSPs [Fig. 4(B)], suggesting that these regions are functionally analogous. Although it was not possible to derive an accurate dissociation constant for CupB580–96 due to its poor solubility, it is consistent with the estimated average for TpsA479–95; Kd = 1.49 mM [±0.18 mM; Fig. 4(C)]. The affected residues are localized to a distinct, contiguous region in POTRA1 [Fig. 4(D)]: the β4-β5-loop, the adjoining β5-strand and the C-terminal region of the adjacent α3-helix and N-terminus/β1-strand. Interestingly, some of the largest CSPs lie at the extreme N-terminus of the POTRA domain (G42-T45) and into the neighboring N-terminal His6-tag. Furthermore, some of these CSPs coincide with those observed between TpsB4-α1P1 and TpsB4-P1, implying that interactions between TpsA4 or CupB5 and the POTRA domains of TpsB4 may displace its N-terminal linker connected to the plug helix.
TpsB4 POTRA domains recognize tandem TpsA4/CupB5 TPS motifs
Three motifs have been identified in TPS domains that are important for the secretion of TpsA proteins: an NPNL box, an NPNG box and a single conserved asparagine position.37,44 While the NPNL sequence is not present in TpsA4 or CupB5, the NPNG box is present in the TpsA479–95 and CupB580–96 peptides that recognize the TpsB4 POTRA1 domain [Figs. 1(E) and 4(B)]. The conserved Asp site (N97/N98 in TpsA4/CupB5) is 10-residues downstream of this NPNG box and as the entire sequence between S68–D107 in TpsA4 is almost identical in CupB5 [Fig. 1(E)] we postulated that amino acid residues C-terminal to TpsA479–95 (e.g., within residues V96–D107) may be recognized by the second TpsB4 POTRA domain (TpsB4-P2). We therefore created a series of mutants that would both validate our NMR derived observations of POTRA/TPS interactions and also examine the role of the second TpsB4 POTRA domain in the secretion of CupB5.
We first generated mutations in the conserved TPS motifs of CupB5. We introduced the double point mutation N85A/P86A, part of the conserved NPNG box, which was designed to disrupt interactions with the POTRA1 domain. We also created the double point mutation V97W/V99W, which flanks the conserved N98 residue and was intended to disrupt interactions with the POTRA2 domain [Fig. 1(E)]. In addition we created an S69A mutant outside of the expected TPS motifs as a negative control. The point mutations were generated in the CupB5-encoding plasmid, pBBR-MCS4-cupB5, introduced in the cupB5 mutant strain overexpressing RocS1 (PAO1ΔΔΔcupB5/pMMBrocS1), and secretion of CupB5 or mutant derivatives was monitored. Western blot analysis using anti-CupB5 antibodies clearly show that the N85A/P86A and V97W/V99W substitutions abrogate secretion, while S69A can still be recovered from the sheared fraction, thus supporting the role of these CupB5 TPS motifs in the interaction with TpsB4 POTRA domains [Fig. 5(A)]. However, it should be noted that levels of recovery of the S69A control is lower than PAO1ΔΔ, which may reflect its close proximity to the TPS motifs. Consistent with the effect observed for our tpsB4 mutant [Fig. 1(F)], and other TPS transporter mutants, abrogation of secretion results in CupB5 accumulation and subsequent degradation in the periplasm.
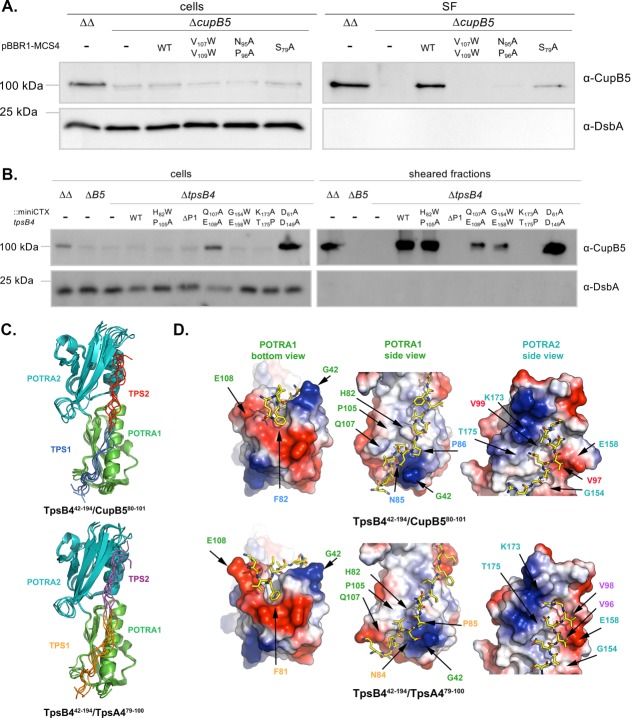
Figure 5.
Structural modelling of a TpsB4/TPS-motif complexes. (A and B) Immunoblot analysis of cell and sheared fractions obtained from PAO1ΔΔ (ΔΔ: ΔpilA ΔfliC mutant), PAO1ΔΔΔcupB5 (ΔcupB5) using antibodies derived against CupB5 and DsbA, with the cupB operon induced with RocS1. (A) PAO1ΔΔΔcupB5 was complemented with WT cupB5 and mutants cupB5S69A, cupB5N85A/P86A, and cupB5V97W/V99W. (B) PAO1ΔΔΔtpsB4 was complemented with WT tpsB4 and mutants tpsB4H82W/P105A, tpsB4G154W/E158W, tpsB4K173A/T175P and tpsB4D61A/D149A. (C) Overlay of the 5 water-refined structures with the lowest interaction energies from the top cluster of docking between TpsB442–194 and CupB580–101/TpsA479–100. TpsB4 POTRA1 is green, TpsB4 POTRA2 is teal, CupB5 TPS1 is blue, CupB5 TPS2 is red, TpsA4 TPS1 is orange and TpsA4 TPS2 is magenta. An interactive view is available in the electronic version of the article, An interactive view is available in the electronic version of the article (D) Details of TpsB442–194/CupB580–101 and TpsB442–194/TpsA479–100 interactions for the structures with the lowest interaction energy. Numbering is based on mature proteins minus N-terminal signal sequences.
We next engineered mutations in the POTRA domains of TpsB4 at positions predicted to interact with the TPS motifs of CupB5, with control mutations that should not affect binding. To confirm the importance of the POTRA1 domain in the secretion of CupB5, we created a ΔPOTRA1 mutant and a double Q107A/E108A point mutation, localized in the dynamic β4-β5 loop and targeted based on significant CSPs observed in this region during titrations against TpsA479–95 and CupB580–96. An additional double mutant was also produced (H82W/P105A), positioned at the periphery of the interaction interface based on NMR CSPs, which we expected would have negligible effect on the secretion of CupB5. Within POTRA2 we created two double point mutations, G154W/E158W in the α4-helix and K173A/T175P in the β7-strand, designed to examine whether a groove created between these secondary structure elements were important for substrate recognition. Substitutions of G154 and E158 were expected to block CupB5 binding within this groove, while mutation of K173 and T175 would prevent β-sheet augmentation between the CupB5 substrate and the TpsB4 β7-strand. Finally as a negative control, residues D61 (POTRA1) and D149 (POTRA2), which are located on opposite side of the CupB5/TpsB4 interaction surface, were each substituted by alanine. The mutations were generated in the plasmid-encoded tpsB4 gene, and the chimeric genes were recloned into the integration-proficient vector mini-CTX1. For complementation, the mini-CTX1-tpsB4 or its derivatives were conjugated into the tpsB4 mutant overexpressing rocS1, PAO1ΔΔΔtpsB4/-pMMBrocS1, thus placing the tpsB4 allele at the att site on the chromosome. Western blot analysis to monitor CupB5 secretion showed that ΔPOTRA1, and K173A/T175P mutants completely abolished, while Q107A/E108A and G154W/E158W mutants significantly impaired CupB5 transport to the cell surface [Fig. 5(B)]. At the same time and as hypothesized, the control and the H82W/P105A double mutant had no influence on CupB5 secretion [Fig. 5(B)].
Experimentally derived models of TpsB442–194/TpsA479–100 and TpsB442–194/CupB580–101 complexes
Interactions between TpsB4 POTRA1 and CupB580–96 or TpsA479–95 are in fast exchange on the NMR timescale, precluding the elucidation of NOE-based, high-resolution structures for these complexes. We therefore generated structural models that would describe a secretion complex of TpsB4 with the translocation signals from CupB5/TpsA4 by incorporating our experimental NMR and mutagenesis data. Using the HADDOCK approach,45,46 peptide sequences including both TPS motifs (CupB580–101 or TpsA479–100) were docked into the periplasmic region containing the two POTRA domains (Residues 42–194): a fusion of the NMR-derived POTRA1 structure and I-Tasser-derived POTRA2 model. Restraints were incorporated from CSPs and mutational experiments and the 200 final water-refined structures for these complexes were clustered according to a pair-wise RMSD cut-off of 3.0 Å, producing a single cluster of 15 structures for TpsB442–194/CupB580–101 and 10 structures for TpsB442–194/TpsA479–100. The average intermolecular interaction energy for these clusters was −168.4 ± 53 kcal mol−1 and −194.5 ± 82 kcal mol−1, respectively. Figure 5(C) shows an overlay of the 5 water-refined structures with the lowest interaction energies from the top cluster for each cluster. Structural statistics are shown in Table2.
Table 2.
Structural Statistics for the TpsB4 P1–2/TPS Complexes
| TpsB442–194/CupB580–101 | TpsB442–194/TpsA479–100 | |
|---|---|---|
| Number of intermolecular restraints | ||
| Total ambiguous CSP derived | 309 | 309 |
| Total ambiguous mutation derived | 71 | 71 |
| RMSD from experimental restraints and idealized covalent geometry | ||
| Bonds (Å) | 0.0037 ± 0.000072 | 0.0037 ± 0.000075 |
| Angles (°) | 0.521 ± 0.0154 | 0.523 ± 0.0159 |
| Energies (kcal mol−1) | ||
| ENOE | 19.1 ± 14.5 | 17.1 ± 11.5 |
| Ebond | 23.0 ± 0.9 | 22.9 ± 0.8 |
| Eangle | 99.6 ± 65.9 | 100.8 ± 6.1 |
| Evdwa | −652 ± 21.4 | −649.2 ± 22.3 |
| Coordinate RMSD (Å) | ||
| All backbone atoms within the interface | 1.7 ± 1.14 | 1.7 ± 1.15 |
| All backbone atoms | 1.7 ± 1.17 | 1.7 ± 1.18 |
Evdw is the Lennard–Jones van der Waals energy and is not included in the target function for simulated annealing.
These complexes share identical overall features and can be considered as an interaction between tandem POTRA domains (POTRA1 and POTRA2) and the adjacent TPS motifs [TPS1 and TPS2; Fig. 5(C,D)]. TPS1 contains the conserved NPNG box while TPS2 contains the completely conserved N97/98 residue (TpsA4/CupB5). POTRA1 is recognized by the CupB5 TPS1, kinking under the N-terminal base of this POTRA domain at position P86 (P85 in TpsA4). Here the first substantial interaction is observed between TPS1 F82 (CupB5; F81: TpsA4) and POTRA1 in a pocket created by the β4-β5-loop and β1-strand [Fig. 5(D)]. In the unbound POTRA1 domain structure, this loop is “pinned” back with the side chains of Q110 in the loop and R115 in the β5-strand (Supporting Information Fig. S2). Our models for these complexes suggest that when the peptide binds, F81/82 (TpsA4/CupB5) is accommodated by a rearrangement of this loop region.
The conserved proline in CupB5 TPS1, P86 (which is P85 in TpsA4), binds between the C-terminal pole of the α3-helix and the extreme N-terminus of POTRA1 [Fig. 5(D)], disrupting intra-POTRA1 interactions between G42-P44 and the α3-helix, while CupB5 TPS1 N85 (N84 in TpsA4) is stabilized by the dipole of the POTRA1 α3-helix. TPS2 (K92:G100 in TpsA4; R93:G101 in CupB5) binds between the β7-strand and α4-helix of POTRA2 [Fig. 5(D)], however, due to the lack of restraints that were implemented in this region, here the models are less well converged.
Discussion
Tps4 is likely a multisubstrate TPS system
In this study, we show that under various growth conditions the TpsB4 transporter is able to translocate CupB5 across the P. aeruginosa outer membrane which contradicts earlier observations in which the P-usher CupB3 was shown to secrete CupB5.18 Our new data described in this manuscript clearly identifies TpsB4 as the major CupB5 transporter and is supported by extensive mutagenesis and structural insight. While this may point to an artefact in the earlier work which did not probe the role of TpsB4 extensively, we also cannot rule out that the laboratory environment and reagents in which strains have been grown may affect the transport process and any interplay between these two systems. The cupB gene cluster is under the tight control of a complex regulatory network, which involves several sensors, including RocS1, and a yet unknown response regulator,15 so subtle environmental effects may be highly relevant in accounting for phenotypic differences observed. There may also be interplay, or at least coordination, between CupB3- and TpsB4-dependent secretion of CupB pili, CupB5 and TpsA4. For example, it was shown in the case of the Klebsiella oxytoca pullulanase (PulA) secretion that interaction with either the type II secretion machine (T2SS) or the lipoprotein sorting pathway (Lol) might be cooperating or competing events which impact on the final destination of PulA.47 Importantly, Ruer et al.18 also observed evidence for a CupB3-independent transport of CupB5, in that a deletion of the CupB3 N-terminus, a predicted POTRA-like domain, did not prevent CupB5 from reaching the surface. In light of this unexpected observation, we have further scrutinized the sequences of P. aeruginosa CupB and TPS systems and revealed here that a TPS motif is present in CupB5 that is in essence identical to the TpsA4 recognition signal, the putative cognate substrate of the TpsB4 transporter. Expression of the cupB genes is under the control of the RocS1 network and we show here that the tpsA4 gene is coregulated with the cupB genes in a RocS1-dependent manner. Recent investigations of Neisseria meningitides TPS systems have illustrated that TPS substrate recognition is defined by the recognition of specific TpsA TPS motifs by both POTRA domains of a TpsB transporter.48 We have confirmed using biophysical and genetic approaches that both TpsA4 and CupB5 TPS motifs are recognized by POTRA domains of the TpsB4 transporter, one of five TpsBs identified in P. aeruginosa.19,37 Therefore, together with the tandem organization of tpsA4/tpsB4 genes, our observations that the TpsB4 POTRA1 recognizes both TpsA4 and CupB5, and CupB5 secretion is dependent on TpsB4, this strongly supports the notion that Tps4 is a multisubstrate TPS system.
The existence of multisubstrate TPS systems, however, is not unique. For example, Bordetella bronchiseptica encodes two TpsA proteins (FhaB and FhaS) that are both secreted by FhaC.40 Although they contain 95% identity over the majority of their sequences, they display different functionalities. The bacterium N. meningitidis has three TPS systems that are scattered across its genome but are nonetheless coexpressed. Two of these, Tps1 and Tps2, each encodes a single transporter and two substrates, while Tps3 contains a single tpsA3 gene. Whereas TpsB1 secretes both of its cognate TpsA1a and TpsA1b cargo, the TpsB2 transporter exports TpsA2a, TpsA2b, and TpsA3.49,50 Usually, however, when a tpsA gene is clustered with chaperone-usher genes, it is also found with a cognate tpsB gene. Both genes can be separated by other genes in the chaperone-usher cluster as in B. pertussis51 or organized in tandem and embedded in the chaperone-usher cluster as in Pseudomonas fluorescens.52 Our observation that the product of the orphan P. aeruginosa tpsA gene, cupB5, can be transported by a distally encoded TpsB, reflects that an associated tpsB gene may have been lost due to the presence of the highly similar, alternative TpsB. In fact in P. aeruginosa, the TpsB1 and TpsB2 transporters share 99% sequence identity, as do the 250-residue TPS-domains of their predicted TpsA1 and TpsA2 substrates (71% identity over the entire TpsA1/A2 sequences) and this may represent another multisubstrate TPS system in this organism. The Tps4/CupB system we describe here reflects a coevolution between chaperone-usher pathways and two-partner secretion systems, which can be genetically linked. Although sequence differences between CupB3 and classical ushers could also suggest an alternative evolution, we have now collected clear evidence that the interaction between CupB5 and TpsB4 is effective and functional, involving key residues in the TPS translocation signal of CupB5 and the POTRA domain of TpsB4 as described here.
TpsB4 POTRA domains have distinct functions
The solution structure of TpsB4-P1 contains the usual core β-α-α-β-β structural motif, but also possesses an additional β1-β3 sheet pairing at the N-terminal base of the domain. This was not observed in FhaC,27,53 the only other TpsB structure, although it is present in one other POTRA domain containing structure: POTRA1 from a Cyanobacterial Omp8554 (pdb: 3mc8). Interestingly, significant conformational dynamics exist within the β4-β5 loop and at the N-terminus preceding the β1-strand [Fig. 2(C) and Supporting Information Fig. S1], which is where TPS1 binds and are likely important for selection of the cargo (Supporting Information Fig. S2).
A conserved proline in the NPNG box of TPS1 allows TpsA/CupB5 to be kinked around POTRA1, which would set the initial register of the TPS motifs on the POTRA domains. The main interactions between TPS1 and POTRA1 are localized to this N-terminal pole of TpsB4 and do not extend substantially along the face of the domain [Fig. 4(D)]. We also show that the linker adjoining the TpsB4 N-terminal α1 helix (involved in gating the β-barrel pore) specifically interacts with POTRA1 and is likely displaced during recognition of TpsA4/CupB5 TPS1. These observations are supported by other recent studies31 and indicate that recognition of TPS motifs by POTRA domains is coordinated with release of the N-terminal plug helix, shifting the equilibrium toward an open TpsB conformation.
Using the entire periplasmic region of FhaC and an extended 30 kDa FHA TpsA-secretion domain, low micromolar dissociation constants have been reported.44 As shown in this study, the interaction between a single POTRA domain of the TpsB4 transporter and its corresponding TPS region is significantly weaker. The presence of two POTRA domains increases the affinity between TpsB transporters and their cargo substantially, while enabling greater specificity. It has been suggested that POTRA domains recognize their substrates through β-sheet augmentation,33–36 although there is no direct structural evidence for this. Our NMR structure of POTRA1 clearly shows that β-sheet augmentation would not be possible here, as chemical shift data demonstrates that TPS1 primarily interacts with the β4-β5 loop of the domain. However, our data does support the potential for augmentation between TPS2 with the β7-strand in POTRA2. When the CupB580–101 conformation is transposed onto the POTRA domains from the FhaC crystal structure27 (pdb: 2qdz), the FHA TPS motifs follows a similar trajectory through grooves with complementary electrostatic surface potential (Supporting Information Fig. S3) and it is likely that our observations can be generalized for other TPS systems.
The G100/101 residue in TpsA4/CupB5 is positioned at the C-terminal pole of POTRA2, which in the context of intact TpsB4 would lie at the opening of the barrel. The 10 residues that follow are either identical or highly conserved between TpsA4 and CupB5 [Fig. 1(E)]. This corresponds to approximately 35 Å of extended polypeptide, which is sufficient to cross the outer membrane and to interact within the barrel of TpsB4 to facilitate transport. These observations suggest that once the TPS motifs of the cargo binds the POTRA domains (TpsA N-terminus facing into the periplasm) a hairpin could form within the TpsB4 pore and polymerization at the tip of this hairpin result in translocation of the remaining C-terminus across the outer membrane. In addition to our work here, others have shown that interactions between POTRA domains and TPS motifs are quite dynamic, which may be essential for substrate release upon folding.44 While the specific cargo release mechanism is unknown, it is interesting to note that extracellular TpsA4 is processed at its N-terminus and begins at His22, just prior to the TPS1 motif. Therefore, proteolysis of TpsA4, albeit slightly upstream of the TPS1 motif, may destabilize POTRA-TPS interactions and contribute to its release.
Tps4, CupB and CupC systems may act in synergy under specific conditions to promote virulence
TpsA4 (LepA) functions as an extracellular protease,22,23 that can activate the NF-κB pathway through cleavage of protease-activated receptors (PARs) allowing the modulation of host responses against bacterial infection.23 Furthermore, in cooperation with a P. aeruginosa hemolysin (PlcH), TpsA4 can degrade hemoglobin to liberate peptide fragments and iron as a source of nutrients for bacterial growth. In this study and in previous investigations12 we have shown that tps4, cupB, and cupC operons are induced by overexpression of RocS1. While CupC fimbriae promote the development of microcolonies during early biofilm formation, the specific role for the CupB pilus and CupB5 have yet to be determined. However, a recent study has shown that peptides released via degradation of CupB5 at the outer membrane can stimulate the overproduction of alginate, which is a major component of P. aeruginosa biofilms.55 CupB pili display a putative tip adhesin (CupB6) and it is plausible that this plays a role in adhesion to biotic surfaces, complementing the interbacterial agglomeration induced by CupC fibres. Analysis of CupB5 using the I-Tasser server42 identifies a number of β-helical structures with excellent alignment scores, including the E. coli Hbp protease (sequence coverage: 0.9, normalized Z-score: 4.0)56 and the Haemophilus influenzae Hap adhesin (sequence coverage: 0.9, normalized Z-score: 2.0)57 (Supporting Information Fig. S4). It has previously been shown by transmission electron microscopy and immunogold labelling that CupB5 is localized on the surface of P. aeruginosa alongside CupB pili.18 In enterotoxogenic E. coli, the TpsA-like molecule EtpA is secreted into the extracellular space where it associates with the flagella and mediates adhesion to host cells.58 It is tempting to speculate that CupB5 plays a similar role here by bridging or modulating adherence between the CupB pilus and specific host receptors and we are currently developing cellular adhesion assays to test these theories. While we still do not fully appreciate the implications of synergy between the cupB, cupC, and tps4 operons, it is clear that a coordination of bacterial agglutination, mechanisms of nutrient acquisition and the ability to modulate host responses to infection will provide great benefits to an organism during colonization of niche environments.
Material and Methods
P. aeruginosa strains and growth conditions
The strains and plasmids used in this study are listed in Supporting Information Table S1. Overnight cultures of P. aeruginosa strains were grown at 37°C in LB medium containing the appropriate antibiotic concentration: 80 µg/mL gentamicin (Gm), 300 µg/mL carbenicillin (Carb), 50 µg/mL tetracycline (Tet), or 2 mg/mL streptomycin (Sm). For appendage preparation, P. aeruginosa strains conjugated with pMMBrocS1 were grown at 30°C either static for 24 h in liquid M63 medium or for 4 days on M63 agar plates. Liquid medium and plates were supplemented with 0.4% l-arginine, 1 mM MgSO4 and 80 µg/mL Gm.18 Bacterial expression was induced with 0.1 mM IPTG.
Construction of PAO1ΔΔ mutants
The oligonucleotides used in this study are listed in Supporting Information Table S2. Polymerase chain reaction (PCR) was used to amplify 500-bp DNA fragments upstream of the cupB3, and tpsB4 genes from the P. aeruginosa genome using the primer pairs 950/951, and 976/977, respectively, whereas 500-bp downstream DNA fragments were generated by using the primer pairs 952/953and 978/979, respectively. The generated DNA fragments were used as a matrix in an overlapping PCR using primer pairs 950/953, and 976/979, to obtain the cupB3, and tpsB4 mutator DNA, respectively. The mutator DNA fragments were ligated into the cloning vector pCR2.1 and recloned into the suicide vector pKNG101 by restriction digest. Plasmids were conjugated in PAO1ΔΔ by three-partner conjugation.59
Construction of complementation vectors
PCR was used to amplify the tpsB4 gene with its promoter and ribosome-binding site (RBS) and the cupB5 gene and its RBS from the chromosome of PAO1ΔΔ by using primer pairs 1315/1316 and 812/813, respectively (Supporting Information Table S2). The cupB5 DNA fragment was ligated into pCR-Blunt and the tpsB4 gene into pCR2.1. tpsB4 was recloned into mini-CTX1 using the restriction enzymes SpeI/EcoRI thereby creating miniCTX-tpsB4. To obtain pBBR-MCS4-cupB5, cupB5 was transferred into pBBR1MCS-4 by using PstI/SpeI/SmaI, whereas SmaI was used to cut pCR-Blunt.
Introduction of point mutations and deletions into the plasmid-encoded cupB5 and tpsB4
To create tpsB4Q107A,E108A tpsB4G154W,E158W and tpsB4K173A,T175P the primer pairs 2039/2040, 1823/1824 and 1825/1826 were used (Supporting Information Table S2). To clone tpsB4D61A,D149A and tpsB4H82W,P105A two rounds of amplification were carried out. Use of the primer pairs 1819/1821 and 1827/1829 introduced the first point mutation H59W and D47A into tpsB4, respectively. A second PCR using the primers 1820/1822 and 1828/1830 introduced the second mutation into tpsB4H82W and tpsB4D61A, resulting in tpsB4H82W,P105A and tpsB4D61A,D149A. To create a tpsB4ΔPOTRA1 mutant bases 574–813 were deleted by PCR using the primer pair 2206/2207. Amplified plasmids were transformed into E. coli XL1-Blue and successful mutation/deletion was confirmed by sequencing. The mutated tpsB4 genes were cloned into mini-CTX1 using the restriction enzymes SpeI and EcoRI. miniCTX-tpsB4xxx or miniCTX-tpsB4ΔPOTRA1 was then conjugated into PAO1ΔΔΔtpsB4/pMMBrocS1. To engineer cupB5S69A, cupB5N85A,P86A, and cupB5V97W,V99W primer pairs 2001/2002, 1804/1805, and 1802/1803 were used, respectively. Amplified plasmids, pCR-Blunt-cupB5S69A, pCR-Blunt-cupB5N85A,P86A and pCR-Blunt-cupB5V97W,V99W, were transformed into E. coli XL1-Blue and successful mutation was confirmed by sequencing. A 600 bp cupB5 DNA fragment containing the point mutations was transferred into pBBR1-MCS4-cupB5 by using the restriction enzymes SfaAI and SgrDI. Positive clones were identified by sequencing. pBBR1-MCS4-cupB5S69A, pBBR1-MCS4-cupB5N85A,P86A and pBBR1-MCS4-cupB5V97W,V99W were conjugated into PAO1ΔΔΔcupB5/pMMBrocS1.
Quantitative real-time PCR
Total RNA was prepared from triplicate cell samples of PAO1ΔΔ and PAO1ΔΔ conjugated with pMMBrocS1, stored in RNAlater, using the RNeasy extraction kit (Qiagen) in combination with on column DNaseI treatment (Applied Biosystems). The DNase treatment was carried out twice to remove all traces of chromosomal DNA. mRNA concentration was measured with the NanoDrop 1000 spectrophotometer (NanoDrop Technologies) and normalized to 20 ng/µL. cDNA was synthesized using SuperScript III Rnase H-Reverse Transcriptase (Invitrogen), 10 mM dNTPs, RNAguard (Roche), random hexamer oligonucleotides (Amersham), 0.1 M DTT, and 1X first strand buffer (RT Superscript Kit). Negative controls without reverse transcriptase were run with all reactions. The resulting cDNA was 1:5 diluted and stored at −20°C. For quantitative real-time PCR, SYBR green PCR master mix (ABI), an upstream and downstream primer pair (Supporting Information Table S2) and water were used. Samples were analyzed by the StepOne Real-Time PCR System (Applied Biosystems). The relative quantity gene expression was determined by defining the expression of rpoD mRNA as 1.
Shearing experiments
Shearing experiments from bacteria grown on agar plates were performed as described by Ruer et al.18 Briefly, bacteria were scrapped from agar plates and an equivalent of 5 OD units was resuspended in LB. Appendages were sheared off by gentle agitation at 4°C overnight. Cells were harvested by centrifugation at 4°C: first, 4000g for 15 min, second, 4000g for 20 min and third, 13,000g for 15 min. Cell pellets, obtained after the first centrifugation step, were washed in 10 mM Tris, pH 8, sedimented by centrifugation (5 min, 13,000g, 4°C) and resuspended in 1 × SDS-loading buffer. The sheared fraction was then subjected to an ammonium sulfate precipitation at a concentration of 50% for 5 min at room temperature and sheared fraction pellets were harvested by centrifugation (75 min, 13,000g, 4°C). Precipitated proteins were harvested by ultracentrifugation (70,400g, 45 min 4°C) and pellets resuspended in 10 mM Tris, pH 8 and mixed with 5 × SDS-loading buffer. Cell and sheared fractions were heated for 10 min at 95°C. Bacteria grown in liquid medium were diluted to an equivalent of 10 units of OD600 and appendages were sheared off by gentle agitation for 3 h at 4°C. Cell and sheared fractions were separated by centrifugation (as described above) and sheared fractions subjected to ammonium sulfate precipitation. The pellet was resuspended in 10 mM Tris, pH 8 and 5 × SDS-loading buffer was added. Appendage and cell pellets were resuspended in SDS loading buffer and heated for 10 min at 95°C.
SDS-PAGE and Western blot analysis
Proteins were separated by electrophoresis on a 12% polyacrylamide gel at 160 V. The proteins were either stained with Coomassie blue or transferred to a nitrocellulose membrane (Whatman) using a semidry transfer system (Biorad). The membrane was blocked by incubation overnight at 4°C in 5% (w/v) milk containing 0.01% (v/v) Tween in PBS (PBST). The membrane was incubated for 2 h at RT with the primary antibody, diluted in 5% (w/v) milk in PBST. Membranes were washed three times for 5 min in PBST and then incubated for 45 min at RT in horseradish peroxidase (HRP)-conjugated goat-anti-rabbit/rabbit-anti-mouse secondary antibody at a dilution of 1:5000 in 5% (w/v) milk in PBST. Membranes were again washed three times for 5 min in PBST. Proteins were detected using a chemiluminescence revelation kit (Thermo Fisher Scientific).
Cloning, expression and purification
The mature (minus N-terminal signal sequence) N-terminal periplasmic region (TpsB4-NT; Residues 1–194), POTRA domains 1–2 (TpsB4-P12; Residues 42–194), PORTRA domain 2 (TpsB4-P2; Residues 122–194), PORTRA domain 1 (TpsB4-P1; Residues 42–121) and the N-terminal helix, linker, and POTRA domain 1 (TpsB4-α1P1; Residues 1–121) of TpsB4, and mature TpsA4 TPS domain (TpsA41–242; Residues 1–242) were amplified by PCR using P. aeruginosa PAO1 gDNA as template with primers B4-1/B4-2, B4-3/B4-2, B1–5/B4-4, B4-3/B4-4, B4-2/B4–5, and A4-1/A4-2, respectively (Supporting Information Table S2). Numbering is based on mature proteins. These were subsequently cloned into Ek/LIC pET46 vector (Novagen; Supporting Information Table S1) and expressed in E. coli strain BL21 (DE3) in either LB, minimal media containing 0.07% 15NH4Cl or minimal media containing both 0.07% 15NH4Cl and 0.2% 13C-glucose. TpsB4 samples were purified using nickel affinity chromatography followed by gel filtration with a Superdex-75 column. TpsA41–242 was purified in the presence of 8 M urea using first nickel affinity chromatography followed by gel filtration with a Superdex-75 column.
NMR solution structure determination
NMR measurements were performed at 310 K on a 15N13C-labelled TpsB4-P1 sample in 50 mM NaPO4 pH 7.0, 150 mM NaCl, 10% D2O. NMR experiments for backbone and side-chain assignment were performed on three different Bruker spectrometers, a DRX500, DRX600, and DRX800, equipped with TCI and TXI cryoprobes. Assignments were completed using standard triple-resonance assignment methodology60 and data were analyzed using in-house algorithms with the program NMRView.61,62 A total of 96% of the potential backbone (e.g., disregarding the N-terminal methionine and proline residues) and 76% of the potential side-chain resonances were assigned; this corresponds to 98% and 84%, respectively, when the 15-residue N-terminal histidine tag is ignored. Heteronuclear NOE cross-relaxation rates on a 15N-labelled TpsB4-P1 sample were recorded at 600 MHz. NOE data for structure calculations of TpsB4-P1 were obtained from an 800-MHz edited 1H-15N/13C nuclear Overhauser effect spectroscopy (NOESY)-heteronuclear single/multiple quantum coherence (HSQC/HMQC) experiments. However, due to spectral overlap only 63 out of a potential 91 peaks could be analyzed. The ARIA protocol for automated NOESY assignment interfaced with the CNS program was used for structure calculation.43 Secondary structure in the TpsB4-P1 POTRA domain was first identified using the chemical shift-based dihedral angle prediction software TALOS.63 For residues located in secondary structure, experimentally derived hydrogen bonds and ϕ/φ backbone dihedral angles from TALOS, were introduced as restraints in the ARIA structure calculation. The frequency window tolerances for assigning NOEs were ±0.03 ppm for direct proton dimensions, ±0.06 ppm for indirect proton dimensions, and ±0.65 ppm for nitrogen and carbon dimensions. One hundred structures were calculated in the final iteration and these had no NOE violations >0.3 Å and no dihedral angle violations >5°. The ten lowest energy structures have been deposited in the Protein Data Bank with PDB ID code 2mhj and a summary of NMR-derived restraints and statistics is reported in Table1.
Production and analysis of TpsA4 TPS domain fragments
TpsA41–242 (0.5 mM) in 20 mM Tris-HCl pH 8, 8 M urea was diluted 100-fold in 20 mM Tris-HCl pH 8, 150 mM NaCl, 2.5 mM CaCl2 and incubated with 1 μM TpsB4-P1 for 30 min at 22°C, followed by the addition of 0.5 nM trypsin for a further 30 min. This was followed by addition of 1 mM PMSF and incubation for 20 min at 70°C. This was loaded onto a Superdex-30 gel filtration column equilibrated in 25% (v/v) trifluoroacetic acid and eluted peaks were pooled and samples sent for commercial analysis by MALDI-MS (University of Leeds, UK). The remaining samples were freeze dried, washed in H2O, freeze dried again and then resuspended in 20 mM NaPO4 pH 7.0, 150 mM NaCl, 10% DMSO, 10% D2O. These were mixed with 15N-labelled TpsB4-P1 (200 μM) in the same buffer and 1H-15N HSQC NMR spectra measured.
Peptide synthesis
To increase solubility all peptides were produced with free N- and C-termini. Initial peptides used in this study (including TpsA480–111) were synthesized by ThermoScientific to >95% purity, while all other peptides (TpsA479–95 and CupB580–96) were synthesized using standard Fmoc solid phase methods, using an automated ResPepSL synthesiser (Intavis) running Multipep software, adapted from Ref.64.
NMR titrations
15N-labelled TpsB4-P1 (200 μM) in 50 mM NaPO4 pH 7.0, 150 mM NaCl, 10% DMSO, 10% D2O with the addition of 0, 0.5, 2, 3, 5, 7.5, 10, 15, 20, 25, or 50 molar equivalents of TpsA479–95 or 0, 0.5, and 2 molar equivalents of CupB580–96 peptide, were used to measure 1H-15N HSQC NMR spectra. Experiments were performed at 310 K on a Bruker DRX600. Due to peptide solubility issues saturation could not be reached. Kinetic analyses of TpsB4-P1/TpsA479–95 interactions were estimated in NMRView62 using 1H-15N CSPs from 24 separate spin systems.
Experimental driven docking
Ambiguous distance restraints from chemical shift perturbations and in vivo mutational analysis were used to drive a structural calculation of TpsB442–194/TpsA79–100 and TpsB442–194/CupB580–101 complexes using the “conformational selection and induced fit protein-peptide HADDOCK docking” approach.46 For the calculations, a composite NMR/I-Tasser42 model of TpsB442–194 and an extended conformation of either a 22 residue TpsA79–100 or CupB580–101 peptide was used with a 1:1 stoichiometry. Superposing our NMR structure of TpsB442–121 onto Residues 42–121 from the I-Tasser42 model of TpsB4 and then combining this with Residues 122–194 produced the model of TpsB442–194. PyMOL65 was used, inputting −139° for phi and −135° for psi, to generate the extended peptide conformations. Based on chemical shift perturbation, filtered for a relative solvent accessibility greater than 50%, 17 residues in TpsB4-P1 (42, 43, 44, 45, 46, 78, 79, 82, 83, 86, 104, 105, 106, 107, 109, 110, and 111) were identified as active residues. In this study we have shown that mutants in CupB5 (Residues 85, 86, 97, and 98; equivalent Positions 84, 85, 96, and 98 in TpsA4), Residues 107–108 in TpsB4 POTRA1 and Positions 154, 158, 173, and 175 in POTRA2 abrogate CupB5 secretion and these were therefore also assigned as active residues. Residues juxtaposed to these in the monomer structures that have a relative solvent accessibility greater than 50% were termed passive residues. TPS peptides were allowed to be completely flexible and the interfacial residues in TpsB442–194 were allowed to move during the simulated annealing and water refinement. 3000 initial complex structures were generated by rigid body energy minimization and the best 1000 by total energy were selected for refinement and analysis with 2500 followed by 5000 molecular dynamic cycles. The best 200 by total energy were selected for torsion angle dynamics and subsequent Cartesian dynamics in an explicit water solvent. Default scaling for energy terms was used as described previously.45 Details of the structural statistic are shown in Table2.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information Figure 1.
Supplementary Information Figure 2.
Supplementary Information Figure 3.
Supplementary Information Figure 4.
Supplementary Information Table 1.
Supplementary Information Table 2.
Supplementary Information Table 3.
Supplementary Information Table 4.
References
- Filloux A, Vallet I. Biofilm: set-up and organization of a bacterial community. Med Sci. 2003;19:77–83. doi: 10.1051/medsci/200319177. [DOI] [PubMed] [Google Scholar]
- Garnett JA, Matthews S. Interactions in bacterial biofilm development: a structural perspective. Curr Protein Pept Sci. 2012;13:739–755. doi: 10.2174/138920312804871166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillington J, Waksman G. Ordered and ushered; the assembly and translocation of the adhesive type I and P pili. Biology. 2013;2:841–860. doi: 10.3390/biology2030841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen WJ, Phan G, Waksman G. Pilus biogenesis at the outer membrane of gram-negative bacterial pathogens. Curr Opin Struct Biol. 2012;22:500–506. doi: 10.1016/j.sbi.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Zav'yalov V, Zavialov A, Zav'yalova G, Korpela T. Adhesive organelles of gram-negative pathogens assembled with the classical chaperone/usher machinery: structure and function from a clinical standpoint. Fems Microbiol Rev. 2010;34:317–378. doi: 10.1111/j.1574-6976.2009.00201.x. [DOI] [PubMed] [Google Scholar]
- Geibel S, Procko E, Hultgren SJ, Baker D, Waksman G. Structural and energetic basis of folded-protein transport by the FimD usher. Nature. 2013;496:243–+. doi: 10.1038/nature12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- Vallet I, Diggle SP, Stacey RE, Camara M, Ventre I, Lory S, Lazdunski A, Williams P, Filloux A. Biofilm formation in Pseudomonas aeruginosa: fimbrial cup gene clusters are controlled by the transcriptional regulator MvaT. J Bacteriol. 2004;186:2880–2890. doi: 10.1128/JB.186.9.2880-2890.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet I, Olson JW, Lory S, Lazdunski A, Filloux A. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc Natl Acad Sci USA. 2001;98:6911–6916. doi: 10.1073/pnas.111551898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud C, Bernard CS, Calderon V, Yang LA, Filloux A, Molin S, Fichant G, Bordi C, de Bentzmann S. The PprA-PprB two-component system activates CupE, the first non-archetypal Pseudomonas aeruginosa chaperone-usher pathway system assembling fimbriae. Environ Microbiol. 2011;13:666–683. doi: 10.1111/j.1462-2920.2010.02372.x. [DOI] [PubMed] [Google Scholar]
- Mikkelsen H, Ball G, Giraud C, Filloux A. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One. 2009;4(6) doi: 10.1371/journal.pone.0006018. ) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruer S, Stender S, Filloux A, de Bentzmann S. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J Bacteriol. 2007;189:3547–3555. doi: 10.1128/JB.00093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, Hui KL, Barraud N, Filloux A. The pathogenicity island encoded PvrSR/RcsCB regulatory network controls biofilm formation and dispersal in Pseudomonas aeruginosa PA14. Mol Microbiol. 2013;89:450–463. doi: 10.1111/mmi.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen H, Sivaneson M, Filloux A. Key two-component regulatory systems that control biofilm formation in Pseudomonas aeruginosa. Environ Microbiol. 2011;13:1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PubMed] [Google Scholar]
- Sivaneson M, Mikkelsen H, Ventre I, Bordi C, Filloux A. Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression. Mol Microbiol. 2011;79:1353–1366. doi: 10.1111/j.1365-2958.2010.07527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- Cai X, Wang R, Filloux A, Waksman G, Meng G. Structural and functional characterization of Pseudomonas aeruginosa CupB chaperones. PLoS One. 2011;6:e16583. doi: 10.1371/journal.pone.0016583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruer S, Ball G, Filloux A, de Bentzmann S. The 'P-usher', a novel protein transporter involved in fimbrial assembly and TpsA secretion. EMBO J. 2008;27:2669–2680. doi: 10.1038/emboj.2008.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A, de Bentzmann S, Aurouze M, Lazdunski A, Vallet I. Fimbrial Genes in Pseudomonas aeruginosa. New York: Springer US; 2004. and Pseudomonas putida. [Google Scholar]
- Yen MR, Peabody CR, Partovi SM, Zhai Y, Tseng YH, Saier MH. Protein-translocating outer membrane porins of gram-negative bacteria. Biochim Biophys Acta. 2002;1562:6–31. doi: 10.1016/s0005-2736(02)00359-0. [DOI] [PubMed] [Google Scholar]
- Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol Microbiol. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y, Shimizu T, Kuwano K. Cooperation between LepA and PlcH contributes to the in vivo virulence and growth of Pseudomonas aeruginosa in mice. Infect Immun. 2011;79:211–219. doi: 10.1128/IAI.01053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y, Higashimoto Y, Inoue H, Shimizu T, Kuwano K. A novel secreted protease from Pseudomonas aeruginosa activates NF-kappaB through protease-activated receptors. Cell Microbiol. 2008;10:1491–1504. doi: 10.1111/j.1462-5822.2008.01142.x. [DOI] [PubMed] [Google Scholar]
- Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- Leo JC, Grin I, Linke D. Type V secretion: mechanism(s) of autotransport through the bacterial outer membrane. Philos Trans R Soc Lond B Biol Sci. 2012;367:1088–1101. doi: 10.1098/rstb.2011.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan E, Fiedler S, Jacob-Dubuisson F, Muller M. Two-partner secretion of gram-negative bacteria: a single beta-barrel protein enables transport across the outer membrane. J Biol Chem. 2012;287:2591–2599. doi: 10.1074/jbc.M111.293068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clantin B, Delattre AS, Rucktooa P, Saint N, Meli AC, Locht C, Jacob-Dubuisson F, Villeret V. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science. 2007;317:957–961. doi: 10.1126/science.1143860. [DOI] [PubMed] [Google Scholar]
- Ward R, Zoltner M, Beer L, El Mkami H, Henderson IR, Palmer T, Norman DG. The orientation of a tandem POTRA domain pair, of the beta-barrel assembly protein BamA, determined by PELDOR spectroscopy. Structure. 2009;17:1187–1194. doi: 10.1016/j.str.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Guedin S, Willery E, Locht C, Jacob-Dubuisson F. Evidence that a globular conformation is not compatible with FhaC-mediated secretion of the Bordetella pertussis filamentous haemagglutinin. Mol Microbiol. 1998;29:763–774. doi: 10.1046/j.1365-2958.1998.00970.x. [DOI] [PubMed] [Google Scholar]
- Hodak H, Clantin B, Willery E, Villeret V, Locht C, Jacob-Dubuisson F. Secretion signal of the filamentous haemagglutinin, a model two-partner secretion substrate. Mol Microbiol. 2006;61:368–382. doi: 10.1111/j.1365-2958.2006.05242.x. [DOI] [PubMed] [Google Scholar]
- Guerin J, Baud C, Touati N, Saint N, Willery E, Locht C, Vezin H, Jacob-Dubuisson F. Conformational dynamics of protein transporter FhaC: large-scale motions of plug helix. Mol Microbiol. 2014;92:1164–1176. doi: 10.1111/mmi.12585. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Guerin J, Baelen S, Clantin B. Two-partner secretion: as simple as it sounds? Res Microbiol. 2013;164:583–595. doi: 10.1016/j.resmic.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- Gatzeva-Topalova PZ, Walton TA, Sousa MC. Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure. 2008;16:1873–1881. doi: 10.1016/j.str.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuck A, Schleiffer A, Clausen T. Augmenting beta-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J Mol Biol. 2011;406:659–666. doi: 10.1016/j.jmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Knowles TJ, Jeeves M, Bobat S, Dancea F, McClelland D, Palmer T, Overduin M, Henderson IR. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol Microbiol. 2008;68:1216–1227. doi: 10.1111/j.1365-2958.2008.06225.x. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Locht C, Antoine R. Two-partner secretion in gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol Microbiol. 2001;40:306–313. doi: 10.1046/j.1365-2958.2001.02278.x. [DOI] [PubMed] [Google Scholar]
- St Geme JW, 3rd, Grass S. Secretion of the Haemophilus influenzae HMW1 and HMW2 adhesins involves a periplasmic intermediate and requires the HMWB and HMWC proteins. Mol Microbiol. 1998;27:617–630. doi: 10.1046/j.1365-2958.1998.00711.x. [DOI] [PubMed] [Google Scholar]
- Jacob-Dubuisson F, Buisine C, Willery E, Renauld-Mongenie G, Locht C. Lack of functional complementation between Bordetella pertussis filamentous hemagglutinin and Proteus mirabilis HpmA hemolysin secretion machineries. J Bacteriol. 1997;179:775–783. doi: 10.1128/jb.179.3.775-783.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julio SM, Cotter PA. Characterization of the filamentous hemagglutinin-like protein FhaS in Bordetella bronchiseptica. Infect Immun. 2005;73:4960–4971. doi: 10.1128/IAI.73.8.4960-4971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems RJ, Geuijen C, van der Heide HG, Renauld G, Bertin P, van den Akker WM, Locht C, Mooi FR. Mutational analysis of the Bordetella pertussis fim/fha gene cluster: identification of a gene with sequence similarities to haemolysin accessory genes involved in export of FHA. Mol Microbiol. 1994;11:337–347. doi: 10.1111/j.1365-2958.1994.tb00314.x. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieping W, Habeck M, Bardiaux B, Bernard A, Malliavin TE, Nilges M. ARIA2: Automated NOE assignment and data integration in NMR structure calculation. Bioinformatics. 2007;23:381–382. doi: 10.1093/bioinformatics/btl589. [DOI] [PubMed] [Google Scholar]
- Delattre AS, Saint N, Clantin B, Willery E, Lippens G, Locht C, Villeret V, Jacob-Dubuisson F. Substrate recognition by the POTRA domains of TpsB transporter FhaC. Mol Microbiol. 2011;81:99–112. doi: 10.1111/j.1365-2958.2011.07680.x. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Trellet M, Melquiond AS, Bonvin AM. A unified conformational selection and induced fit approach to protein-peptide docking. PLoS One. 2013;8:e58769. doi: 10.1371/journal.pone.0058769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AP, Kornacker MG. Secretion of the cell surface lipoprotein pullulanase in Escherichia coli. Cooperation or competition between the specific secretion pathway and the lipoprotein sorting pathway. J Biol Chem. 1991;266:13640–13645. [PubMed] [Google Scholar]
- ur Rahman S, Arenas J, Ozturk H, Dekker N, van Ulsen P. The polypeptide transport-associated (POTRA) domains of TpsB transporters determine the system specificity of two-partner secretion systems. J Biol Chem. 2014;289:19799–19809. doi: 10.1074/jbc.M113.544627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ur Rahman S, van Ulsen P. System specificity of the TpsB transporters of coexpressed two-partner secretion systems of Neisseria meningitidis. J Bacteriol. 2013;195:788–797. doi: 10.1128/JB.01355-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ulsen P, Rutten L, Feller M, Tommassen J, van der Ende A. Two-partner secretion systems of Neisseria meningitidis associated with invasive clonal complexes. Infect Immun. 2008;76:4649–4658. doi: 10.1128/IAI.00393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spears PA, Temple LM, Miyamoto DM, Maskell DJ, Orndorff PE. Unexpected similarities between Bordetella avium and other pathogenic Bordetellae. Infect Immun. 2003;71:2591–2597. doi: 10.1128/IAI.71.5.2591-2597.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre AS, Clantin B, Saint N, Locht C, Villeret V, Jacob-Dubuisson F. Functional importance of a conserved sequence motif in FhaC, a prototypic member of the TpsB/Omp85 superfamily. FEBS J. 2010;277:4755–4765. doi: 10.1111/j.1742-4658.2010.07881.x. [DOI] [PubMed] [Google Scholar]
- Koenig P, Mirus O, Haarmann R, Sommer MS, Sinning I, Schleiff E, Tews I. Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. J Biol Chem. 2010;285:18016–18024. doi: 10.1074/jbc.M110.112649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Regt AK, Yin Y, Withers TR, Wang X, Baker TA, Sauer RT, Yu HD. Overexpression of CupB5 activates alginate overproduction in Pseudomonas aeruginosa by a novel AlgW-dependent mechanism. Mol Microbiol. 2014;93:415–425. doi: 10.1111/mmi.12665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto BR, Sijbrandi R, Luirink J, Oudega B, Heddle JG, Mizutani K, Park SY, Tame JR. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J Biol Chem. 2005;280:17339–17345. doi: 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- Meng G, Spahich N, Kenjale R, Waksman G, St Geme JW., III Crystal structure of the Haemophilus influenzae Hap adhesin reveals an intercellular oligomerization mechanism for bacterial aggregation. EMBO J. 2011;30:3864–3874. doi: 10.1038/emboj.2011.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Hilliard GM, Hamilton DJ, Luo JW, Ostmann MM, Fleckenstein JM. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature. 2009;457:594–U103. doi: 10.1038/nature07568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniga K, Delor I, Cornelis GR. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectr. 1999;34:93–158. [Google Scholar]
- Marchant J, Sawmynaden K, Saouros S, Simpson P, Matthews S. Complete resonance assignment of the first and second apple domains of MIC4 from Toxoplasma gondii, using a new NMRView-based assignment aid. Biomol NMR Assign. 2008;2:119–121. doi: 10.1007/s12104-008-9100-1. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR View—a computer-program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Thomas JC, Green JL, Howson RI, Simpson P, Moss DK, Martin SR, Holder AA, Cota E, Tate EW. Interaction and dynamics of the Plasmodium falciparum MTIP-MyoA complex, a key component of the invasion motor in the malaria parasite. Mol Biosyst. 2010;6:494–498. doi: 10.1039/b922093c. [DOI] [PubMed] [Google Scholar]
- DeLano WL. Use of PYMOL as a communications tool for molecular science. Abstracts Papers Am Chem Soc. 2004;228:U313–U314. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information Figure 1.
Supplementary Information Figure 2.
Supplementary Information Figure 3.
Supplementary Information Figure 4.
Supplementary Information Table 1.
Supplementary Information Table 2.
Supplementary Information Table 3.
Supplementary Information Table 4.