Abstract
A truly disordered protein lacks a stable fold and its backbone amide protons exchange with solvent at rates predicted from studies of unstructured peptides. We have measured the exchange rates of two model disordered proteins, FlgM and α-synuclein, in buffer and in Escherichia coli using the NMR experiment, SOLEXSY. The rates are similar in buffer and cells and are close to the rates predicted from data on small, unstructured peptides. This result indicates that true disorder can persist inside the crowded cellular interior and that weak interactions between proteins and macromolecules in cells do not necessarily affect intrinsic rates of exchange.
Keywords: amide proton exchange, intrinsically disordered proteins, in-cell NMR, SOLEXSY, macromolecular crowding
Introduction
Cellular processes occur at macromolecule concentrations of 300–400 g/L.1,2 The resulting weak, nonspecific interactions between macromolecules in the cytoplasm alter globular protein dynamics and stability.3–6 Disordered proteins are fundamentally different. Unlike many model, single domain, globular proteins, whose structures do not change until they denature,7 the properties of disordered proteins depend on solution conditions.8 Thus, the crowded nature of the cell could have large effects on protein disorder. Studies show that the cellular interior can promote structure formation in proteins that are only transiently structured in buffer.9 On the other hand, there is a growing realization from studies of globular proteins that attractive interactions favor less structure.10–16
To date, knowledge of the atomic-level structure of disordered proteins inside cells comes from crude measures: the presence or absence of crosspeaks in 15N-1H HSQC NMR spectra and chemical shifts.9,17 Absence of peaks is caused by chemical exchange and is explained in contrasting ways: (1) intramolecular exchange involving the stabilization of particular conformations (i.e., folded species) and (2) weak, transient chemical interactions between the test protein and cellular components.9,18–20 Here, we take a new approach by quantifying hydrogen exchange rates for two proteins possessing different degrees of disorder.
Structure protects backbone amide protons from solvent exchange.21 Therefore, measuring their exchange rates inside cells and comparing the values to those measured in buffer and calculated from unstructured peptides provides information about structure. Protection is quantified as the rate in an unstructured peptide (∼0.1 − 10 s−1)22 divided by the observed rate, kint/kobs. These protection factors range from >105 for backbone amide protons in the core of globular proteins to <5 for denatured proteins.5,10,23 Intrinsic rates in buffer are estimated by using the online program, SPHERE,24 which calculates kint based on data from unstructured peptides and information about the system (i.e., primary structure, pH, deuterium content, and temperature).22,25 These calculated rates remain valid in buffers containing physiologically relevant concentrations of globular proteins and cell lysates.11,26
Direct measurement of exchange in cells, however, is challenging. We investigated the disordered proteins α-synuclein and FlgM, both of which give in-cell NMR spectra,9,18,27,28 using the SOLEXSY experiment26,29 to quantify kobs in Escherichia coli and buffer. SOLEXSY uses a variable mixing time (tmix) to monitor hydrogen exchange (Supporting Information Fig. S1). Short times do not result in a buildup crosspeak. Longer times allow amide deuterons to exchange for protons and buildup of the initially deuterated amide crosspeak occurs. Similarly, a decay is observed from the initially protonated species. Rates are obtained from fits of peak volume versus mixing time.
α-Synuclein is a 140-residue protein found at the presynaptic terminals of neurons.30 Its exact role is controversial, but the protein is N-terminally acetylated and thought to play a part in dopamine trafficking.8 α-Synuclein retains the majority of its 15N-1H correlation crosspeaks inside E. coli, suggesting that the protein remains disordered in cells.17,18,27,28
FlgM is a 97-residue protein from Salmonella typhimurium that inhibits σ28, a transcription factor responsible for regulating downstream flagellar and chemotaxis genes.31,32 The protein shows characteristics of both a globular and a disordered protein. The C-terminus (residues 41–97) forms transient helices in buffer (between residues 60–73 and 83–90).33 These helices are stabilized upon binding σ28.34 The absence of crosspeaks from the 15N-1H correlation spectrum for the C-terminal region in E. coli has been interpreted as evidence that this region gains structure in cells.9 The N-terminus (residues 1–40) appears to remain disordered even in cells.9,33
Results
Amide proton exchange is base-catalyzed at physiological pH.21,22 In-cell NMR in E. coli is usually performed in dense cell slurries that are inherently low in nutrients and O2.35,36 These anaerobic conditions cause acidification of the cellular interior.17,37,38 Thus, the interior pH must be known to compare data obtained in cells to data obtained in buffer. α-Synuclein has a single histidine, H50, whose 13Cε1 proton chemical shift depends on pH.39 The C14 protons of HEPES, which is not cell permeable, were used as an external pH probe.40,41 13C-1H HSQC spectra were acquired before and after the SOLEXSY experiment.
The proton chemical shifts were compared to those from a standard curve in buffer (Supporting Information Figs. S2, S3). The cytoplasm and the extracellular medium acidified over the ∼16 h required to acquire a SOLEXSY dataset (Supporting Information Table S1), and the internal pH was ∼0.4 units lower than the external pH (Supporting Information Fig. S4). The average internal pH of all α-synuclein datasets is pH 6.7. We corrected the in-cell rates to pH 6.7 by using the second-order rate constant for base-catalyzed exchange (Supporting Information Table S2, footnotes c-e).42 We used this corrected rate to compare intracellular hydrogen exchange rates to those measured in buffer and to drive SPHERE. Since FlgM does not have a pH sensitive probe, we measured the HEPES C14 proton chemical shift before and after the SOLEXSY experiment, and applied the 0.4 pH unit correction to determine the internal pH.
During the course of an in-cell experiment the intercellular pH typically drops below 6.5 (Supporting Information Table S1). Below this pH (at 288 K) the signal to noise ratio of buildup peaks is typically too small to allow exchange rates to be quantified, because hydrogen exchange drops below 0.1 s−1 for many residues. This problem is exacerbated in-cells, where peaks are inherently broad. To estimate exchange rates from crosspeaks with weak buildup curves we used a standard curve of the ratio of buildup to decay peak volumes at a tmix of 0.3 s versus the kobs values from the fit obtained in buffer (Supporting Information Fig. S5). This procedure allows an exchange rate to be estimated by a SOLEXSY experiment with one tmix, 0.3 s. The uncertainty in this measurement was estimated as the maximum and minimum rates within 0.1 unit of the ratio.
Hydrogen exchange rates for α-synuclein and FlgM are compiled in Supporting Information Tables S3–S7. SPHERE-predicted kint values match kobs values in buffer for α-synuclein, acetylated α-synuclein, and FlgM. In addition, rates and protection factors for α-synuclein in buffer closely match those measured with CLEANEX (Supporting Information Table S3).27 The differences likely reflect differences in buffer composition and extrapolation to pH 6.7.
In buffer, 26 α-synuclein residues give quantifiable exchange rates and seven more rates can be estimated [Figs. 1(A) and 2(A), Supporting Information Table S3]. The residues are distributed throughout the primary structure. Their protection factors are all similar and less than two. The same is observed for α-synuclein in buffer supplemented with 150 mM NaCl (Supporting Information Table S3). Protection factors this small indicate a lack of structure.23
Figure 1.
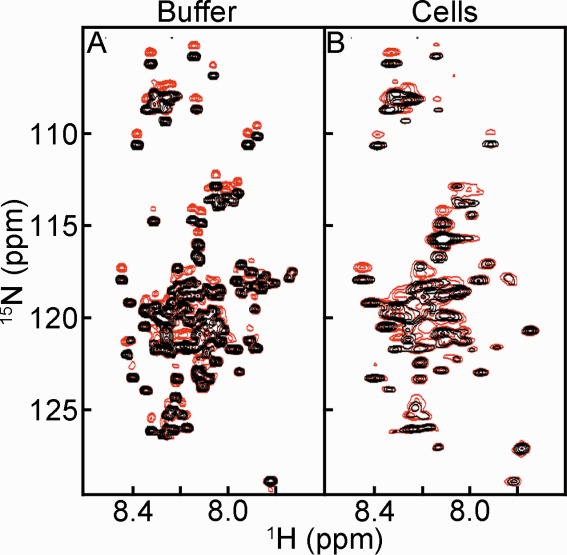
15NH/D-SOLEXSY spectra of α-synuclein in (A) buffer and (B) E. coli. Each panel is an overlay of the 0 ms (black) and 300 ms (red) mixing times. The contour levels in panels A and B are the same.
Figure 2.
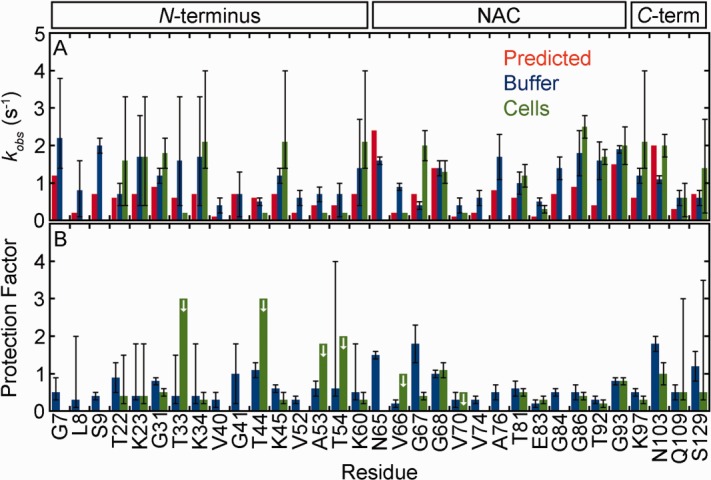
α-Synuclein exhibits similar backbone amide hydrogen exchange rates in cells and in buffer (pH 6.7, 288 K). (A) Rates from prediction (red),22,24,25 and from SOLEXSY data (blue, buffer; green, cells). Uncertainties in buffer are the standard deviation of 20 Monte Carlo noise simulations.29 Uncertainties for the in-cell data are the standard deviation of the mean from three or more trials (For T81 and G93, the uncertainties are the range of two experiments.). Asymmetric error bars are shown for rates derived from the data in Supporting Information Figure S5, as described in the text. In-cell rates without error bars are for residues that exchange too slowly for reliable fits, or have overlapped decay peaks, and were assigned a rate of ≥ 0.2 s−1, the lower limit of SOLEXSY. (B) Protection factors (kint, predicted/kobs, buffer and kint, predicted/kobs, cells). Uncertainties are from propagation of the uncertainties shown in panel A unless the rate was derived from Supporting Information Figure S5. For those residues, the uncertainty reflects a range, as described in the text. The arrows denote residues for which only a maximum value can be assigned.
In cells, nine α-synuclein residues give quantifiable rates, eight more rates can be estimated, and an additional six qualitatively show exchange [Figs. 1(B) and 2(A), Supporting Information Tables S2 and S3). Rates in cells and in buffer are in reasonable agreement with the values predicted by SPHERE. No protection factor is greater than three [Fig. 2(B)]. There is no significant difference between the protection factors, both measured and estimated, in cells and in buffer (two-tailed t-test with unequal variance,43 ρ > 0.07). The same is observed for acetylated α-synuclein. There is no significant difference between protection factors in cells and buffer (Supporting Information Fig. S6, Tables S4 and S5, two-tailed t-test with unequal variance, ρ > 0.5). These observations along with chemical shift data from the protein in cells17 indicate that α-synuclein, with or without acetylation, remains disordered in the crowded bacterial cytoplasm.
The activation energy of amide proton exchange was also measured (Supporting Information Table S8). The means and their standard deviations are 15 ± 3 kcal/mol in buffer and 17 ± 1 kcal/mol in cells. The activation energy used in SPHERE is 17 kcal/mol.22,24 These observations lend further support our to conclusion that α-synuclein has the properties of a disordered peptide in the E. coli cytoplasm.
Turning to FlgM, 24 residues give quantifiable exchange rates in buffer [Figs. 3(A) and 4(A), Supporting Information Table S7). Significantly larger protection factors (ρ < 0.002, two-tailed t-test with unequal variance) are observed for the 13 C-terminal residues compared to the 11 N-terminal residues (means and their standard deviations of 1.6 ± 0.3 and 0.5 ± 0.1, respectively). This observation suggests the existence of some structure in the C-terminal region, consistent with chemical-shift data showing that the C-terminus forms transient α-helices in buffer.33,34 However, the protection factors from the C-terminal region were all <5, indicating an absence of persistent structure.23 In summary, the agreement between the chemical-shift data and the hydrogen exchange data show that protection factors are a sensitive measure of sparsely populated secondary structure.
Figure 3.
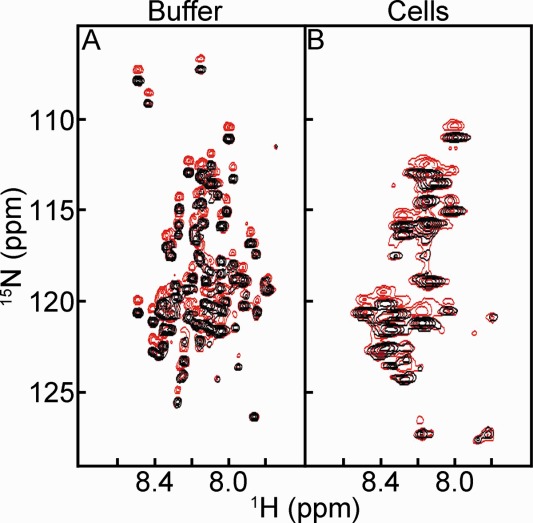
15NH/D-SOLEXSY spectra of FlgM in (A) buffer and (B) E. coli. Each panel is an overlay of the 0 ms (black) and 300 ms (red) mixing times. The contour levels in panels A and B are the same.
Figure 4.
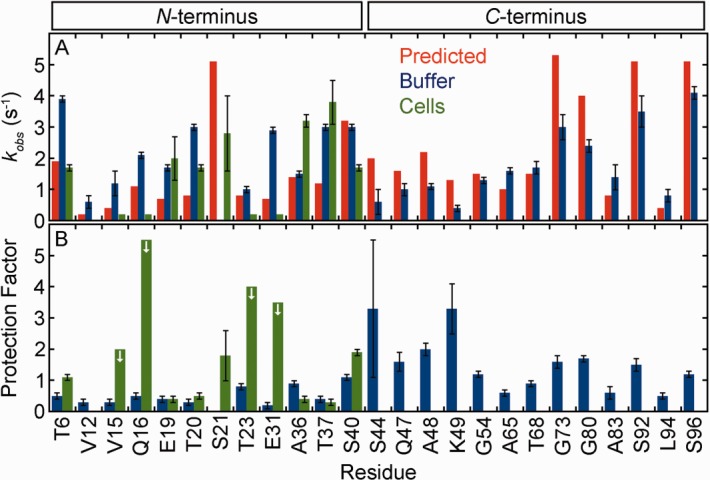
FlgM exhibits similar backbone amide hydrogen exchange rates in cells and in buffer (pH 6.7, 298 K). See the caption to Figure 2 for further information.
In cells, seven FlgM residues give quantifiable rates and four more qualitatively show exchange [Figs. 3(B) and 4(A), Supporting Information Tables S6 and S7]. The rates in cells are similar to those in buffer. For the N-terminus seven residues can be quantified in cells and 11 in buffer. Protection factors for these residues [Fig. 4(B)] are the same in cells as they are in buffer (two-tailed t-test with unequal variance, ρ > 0.1).
Discussion
Our data for α-synuclein and the N-terminal region of FlgM indicate that disorder can persist in E. coli irrespective of the existence of hard-core repulsions, i.e. macromolecular crowding.3,4 We cannot directly compare cell and buffer data from the C-terminus of FlgM because C-terminal crosspeaks disappear in cells.9 However, the smaller exchange rates in buffer and the absence of crosspeaks are both consistent with the existence of a nascent C-terminal structure. Likewise, for both acetylated and non-acetylated α-synuclein, crosspeaks from the first 12 residues and residues 37–41 show peak broadening inside cells (Supporting Information Fig. S7). This broadening is likely due to conformational exchange involving a helical form that is populated in buffer to ∼17% for acetylated α-synuclein and slightly less for the non-acetylated form.44,45 This conformational exchange is likely slowed in cells causing broadening. A similar observation has been made for a folding intermediate of the FF domain in cell lysates.6
We have shown that measured exchange rates in E. coli agree with both those obtained in buffer and those calculated based on exchange rates of small, unstructured peptides. Data for the C-terminal region of FlgM in buffer show that even small protection factors can be useful indicators of sparsely populated structured states in disordered proteins. This work also has implications for determining the stability of globular proteins in living cells. Specifically, the data indicate that predicted rates from model peptides can be applied to hydrogen exchange studies of globular proteins inside E. coli.5 Most importantly, we have shown that protein disorder can persist under physiological conditions and that the crowded cellular interior need not affect the structure of disordered proteins.
Materials and Methods
Protein expression for in-cell NMR
Plasmids harboring the gene encoding FlgM (ampicillin resistance) or α-synuclein (ampicillin resistance) were transformed into Agilent BL21 DE3 Gold cells by heat-shock. For acetylated α-synuclein, plasmids harboring pNatB (chloramphenicol resistance) and α-synuclein were co-transformed into Agilent BL21 DE3 Gold cells by electroporation. A single colony was used to inoculate a 5 mL culture of Luria-Bertani media supplemented with 100 μg/mL ampicillin (FlgM or α-synuclein) or 100 μg/mL ampicillin and 25 μg/mL chloramphenicol (pNatB/α-synuclein). The culture was grown at 37°C. After 8 h, 50 μL of the saturated culture was used to inoculate 50 mL of supplemented M9 media as described next.
For uniform labeling, M9 media (50 mM Na2HPO4, 20 mM KH2PO4, 9 mM NaCl) was supplemented with 2 mg/mL 13C-glucose, 1 mg/mL 15NH4Cl, 100 μM CaCl2, 2 mM MgSO4, 10 μg/mL thiamine and 10 μg/mL biotin, and 150 μg/mL ampicillin (100 μg/mL ampicillin and 25 μg/mL chloramphenicol for pNatB/α-synuclein). For glycine or threonine “unlabeling,” the media described above was also supplemented with 1 mg/mL natural abundance glycine or 1.5 mg/mL natural abundance threonine.46,47
For selective lysine/threonine enrichment, natural abundance glucose and NH4Cl were used in the M9 media, which was also supplemented with 0.1 mg/mL uniformly enriched 13C/15N threonine, 0.2 mg/mL uniformly 13C/15N enriched lysine and 0.2 mg/mL natural abundance glycine.48,49
The 50 mL cultures were shaken (New Brunswick Scientific Innova I26, 225 rpm) at 37°C overnight. The culture was diluted to 100 mL with supplemented M9 media and shaken until the optical density at 600 nm was at least 0.8. Isopropyl β-D-1-thiogalactopyranoside (1 mM) was used to induce expression. After 4 h cells were pelleted at 1000g and washed 3 times with 30 mL NMR buffer (100 mM HEPES, 35 mM bis-tris propane,50 50 μg/mL chloramphenicol, 100 μg/mL ampicillin, 50% D2O). For pNatB/α-synuclein the buffer contained half the amount of chloramphenicol and 15 μg/mL rifampin. Chloramphenicol or rifampin are required to halt protein expression prior to NMR. (Rifampin is required because the pNatB plasmid confers chloramphenicol resistance.) Cell pellets were resuspended in 100–200 μL of NMR buffer and loaded into a standard 5 mm NMR tube. Typical cell slurries were 70% wet cells by volume.
Protein purification
Cell pellets were frozen after in-cell NMR experiments. For α-synuclein (with or without acetylation) cells were lysed by boiling. Cell debris was removed by centrifugation at 16,000g. Using a GE AKTA FPLC, anion exchange (GE Q column, 50 mM Tris wash buffer, 50 mM Tris/1M NaCl elute buffer, 10–90% gradient) and subsequent size exclusion chromatography (GE Superdex 75 column eluted with non-supplemented M9) were used. Purified protein was dialyzed against 17 MΩ cm−1 H2O for 4 h at room temperature or overnight at 5°C. After dialysis, the sample was flash frozen in a dry-ice/ethanol bath and lyophilized.
FlgM was purified in a similar fashion except the cells were sonicated (Fisher Scientific Sonic Dismembrator Model 500, 15% amplitude, 20 s, 67% duty cycle). FlgM does not bind to anion exchange media.
LC-ESI-MS
Purified samples of α-synuclein were resuspended in 17 MΩ cm−1 H2O and subjected to LC-ESI-MS using a Restek Viva C4 column (linear gradient from 0.1% aqueous formic acid, 5% acetonitrile to 0.1% aqueous formic acid, 95% acetonitrile over 15 m, followed by 5 m in the 95% eluant) coupled to an Agilent 6520 Accurate-Mass Q-TOF running in positive ion mode. Deconvoluted masses of 15240 Da and 15280 Da were found for 13C/15N α-synuclein and 13C/15N acetylated α-synuclein, respectively. This mass difference corresponds to the addition of an acetyl group (∼40 Da). No unmodified α-synuclein was observed when the protein was co-expressed with pNatB, indicating 100% acetylation, consistent with previous work.51
pH determination
Purified α-synuclein was suspended in 50 mM citrate, 50 mM bis-tris propane, 50 mM HEPES, 50 mM borate, 5% D2O, containing 0.1% DSS at various pH values. Data were acquired at 288 K (pH: 3.0, 4.2, 5.3, 5.7, 6.0, 6.4, 6.6, 6.8, 7.0, 7.5, 7.8, 8.9) and 298 K (pH: 5.3, 6.1, 6.6, 7.0, 7.4, 7.9, 8.9) on either a 600 or 700 MHz Bruker Avance III HD spectrometer equipped with a Bruker TCI cryoprobe. 13C-HSQC spectra were acquired using sweep widths of 12 ppm in the 1H dimension and 200 ppm in the 13C dimension. Four transients were collected using 512 complex points in t2 and 64 complex increments in t1. Spectra were referenced to DSS at 0 ppm. Data were processed with Topspin 3.2. The 1H chemical shifts of H50 13Cε1-1H and 13Cδ2-1H of α-synuclein and HEPES were followed as a function of pH. The data were fit to a modified Henderson-Hasselbach equation,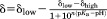 , where δlow is the low pH chemical-shift plateau and δhigh is the high plateau, n is the number of protons, and pKa is the negative logarithm of the dissociation constant.52–54 Our pKa values (6.7 for H50 and 7.5 for HEPES) are similar to literature values.39,40
, where δlow is the low pH chemical-shift plateau and δhigh is the high plateau, n is the number of protons, and pKa is the negative logarithm of the dissociation constant.52–54 Our pKa values (6.7 for H50 and 7.5 for HEPES) are similar to literature values.39,40
Using a glass electrode, buffers containing 5% and 50% D2O and α-synuclein were adjusted to a pH reading of 6.9. The solutions gave nearly identical proton chemical shifts for ε1 (7.984 ppm and 7.983 ppm) and δ2 (7.074 ppm and 7.073 ppm), for 5 and 50% D2O. Thus, the H50 13Cε1 and 13Cδ1 protons are insensitive to the H/D isotope effect and 0.2 pH units were added to the value obtained from the standard curve in solutions containing a 1:1 H2O:D2O mixture to account for the H/D isotope effect on the electrode for a 50% D2O solution.55,56 The C14 protons of HEPES, however, exhibit an isotope effect. In 5% D2O the proton chemical shift is 3.117 ppm while in 50% D2O the chemical shift is 3.134 ppm. For this reason, a correction of −0.017 ppm was applied to values obtained in 50% D2O to account for the deviation from the conditions used to acquire the standard curve, then the 0.2 pH unit correction was added to the value obtained from the standard curve.
NMR
In-cell samples were prepared as described above. For the buffer experiments purified 13C-, 15N-enriched protein (final protein concentration of ∼500 μM as judged by a Lowry assay using bovine serum albumin as the standard) was added to NMR buffer (pH 6.7, minus antibiotics). One buffer experiment with α-synuclein used NMR buffer plus 150 mM NaCl, to ensure minimal salt dependence of exchange. In-cell data were acquired at 288 K (α-synuclein, acetylated α-synuclein), and 298 K (FlgM, α-synuclein) with a 700 MHz Bruker Avance III HD spectrometer running Topspin Version 3.2 and equipped with a Bruker TCI cryoprobe. Buffer data were acquired at 288 K (α-synuclein, acetylated α-synuclein), 293 K (α-synuclein), and 298 K (FlgM, α-synuclein). An interleaved, modified26 SOLEXSY29 experiment with mixing times (tmix) of 0, 70, 140, 210, 300, 500, and 800 ms was used to measure hydrogen exchange rates. One in-cell exchange experiment with α-synuclein used tmix of 0, 70, 150, 300, 700 ms in an attempt to shorten the acquisition time and minimize pH changes. Sweep widths were 7000 Hz and 2500 Hz in the 1H and 15N dimensions, respectively. 512 complex points were collected in t2 with 128 TPPI points in t1 at each tmix. In buffer, 32 transients were acquired per increment. Forty transients were acquired for the in-cell NMR experiments. Data acquisition required ∼16 h per sample.
15N-1H and 13C-1H HSQC spectra were acquired before and after the SOLEXSY experiment to assess sample integrity and pH. Four or eight transients were collected using 512 complex points in t2 with 64 complex points in t1 for each experiment. The sweep widths were 8500 Hz in 1H and either 2500 Hz or 34500 Hz in the 15N or 13C dimension, respectively. For the in-cell samples, the cell slurry was removed after the experiment and gently pelleted. The supernatant was removed, diluted two- to three-fold, and placed in the spectrometer. A 15N-1H HSQC was acquired to assess protein leakage.57 No leakage was observed.
To ensure the pH associated with an in-cell SOLEXSY experiment was the average of the values obtained from the bracketed 13C-1H HSQC spectra, an experiment using α-synuclein was performed where 13C-1H HSQC spectra were acquired as a function of time. These HSQC spectra bracketed a short SOLEXSY (only tmix 0.3 s) experiment and an 15N-1H TROSY-HSQC experiment. This regimen not only allowed us to determine that the pH drop was linear but also allowed collection of single-plane SOLEXSY experiments where the pH drop was only averaged over ∼2.5 h instead of the ∼16 h required for a complete SOLEXSY experiment. This protocol allowed us to assess hydrogen exchange at discreet pH values, instead of exchange averaged over a ∼0.5 pH unit range.
Data processing
Data were processed with NMRPipe.58 t2 data were subjected to a cosine squared bell function (512 complex points for buffer solutions and 256 complex points for in-cell experiments) before zero-filling to 2048 points and Fourier transformation. t1 data were linear predicted to 256 points before application of a cosine squared bell function. Subsequent zero-filling to 1024 points and Fourier transform yielded the final spectra. Spectra were peak picked and integrated using the built-in nlinLS routine. Peak volumes were fitted as described.29 Published assignments were used.33,34,59,60
Acknowledgments
The authors thank Elizabeth Pielak for comments, Eric Brustad and Richard Watkins for help with mass spectroscopy and Dan Mulvihill for supplying us with the pNatB acetylation complex.
Glossary
- CLEANEX
phase-modulated CLEAN chemical exchange
- HSQC
heteronuclear single quantum correlation
- Nat
N-α-acetyltransferase
- NMR
nuclear magnetic resonance
- SOLEXSY
solvent exchange spectroscopy
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
References
- McGuffee SR, Elcock AH. Diffusion, crowding & protein stability in a dynamic molecular model of the bacterial cytoplasm. PLoS Comput Biol. 2010;6:e1000694. doi: 10.1371/journal.pcbi.1000694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SB, Trach SO. Estimation of macromolecule concentrations and excluded volume effects for the cytoplasm of Escherichia coli. J Mol Biol. 1991;222:599–620. doi: 10.1016/0022-2836(91)90499-v. [DOI] [PubMed] [Google Scholar]
- Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M, Li C, Pielak GJ. Soft interactions and crowding. Biophys Rev. 2013;5:187–194. doi: 10.1007/s12551-013-0104-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteith WB, Pielak GJ. Residue level quantification of protein stability in living cells. Proc Natl Acad Sci. 2014;111:11335–11340. doi: 10.1073/pnas.1406845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham MP, Kay LE. A similar in vitro and in cell lysate folding intermediate for the FF domain. J Mol Biol. 2014;426:3214–3220. doi: 10.1016/j.jmb.2014.07.019. [DOI] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Theillet F-X, Binolfi A, Frembgen-Kesner T, Hingorani K, Sarkar M, Kyne C, Li C, Crowley P, Gierasch LM, Pielak GJ, Elcock AH, Gershenson A, Selenko P. Physicochemical properties of cells and thier effects on intrinsically disordered proteins (IDPs) Chem Rev. 2014;114:6661–6714. doi: 10.1021/cr400695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedmon MM, Patel CN, Young GB, Pielak GJ. FlgM gains structure in living cells. Proc Natl Acad Sci USA. 2002;99:12681–12684. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M, Smith AE, Pielak GJ. Impact of reconstituted cytosol on protein stability. Proc Natl Acad Sci USA. 2013;110:19342–19347. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos AC, Sarkar M, Wang Y, Pielak GJ. Protein crowding tunes protein stability. J Am Chem Soc. 2011;133:7116–7120. doi: 10.1021/ja200067p. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sarkar M, Smith AE, Krois AS, Pielak GJ. Macromolecular crowding and protein stability. J Am Chem Soc. 2012;134:16614–16618. doi: 10.1021/ja305300m. [DOI] [PubMed] [Google Scholar]
- Minton AP. Quantitative assessment of the relative contributions of steric repulsion and chemical interactions to macromolecular crowding. Biopolymers. 2013;99:239–244. doi: 10.1002/bip.22163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H-X. Influence of crowded cellular environments on protein folding, binding, and oligomerization: biological consequences and potentials of atomistic modeling. FEBS Lett. 2013;587:1053–1061. doi: 10.1016/j.febslet.2013.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luh LM, Hänsel R, Löhr F, Kirchner DK, Krauskopf K, Pitzius S, Schäfer B, Tufar P, Corbeski I, Güntert P, Dötsch V. Molecular crowding drives active Pin1 into nonspecific complexes with endogenous proteins prior to substrate recognition. J Am Chem Soc. 2013;135:13796–13803. doi: 10.1021/ja405244v. [DOI] [PubMed] [Google Scholar]
- Phillip Y, Schreiber G. Formation of protein complexes in crowded environments - From in vitro to in vivo. FEBS Lett. 2013;587:1046–1052. doi: 10.1016/j.febslet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waudby CA, Camilloni C, Fitzpatrick AWP, Cabrita LD, Dobson CM, Vendruscolo M, Christodoulou J. In-cell NMR characterization of the secondary structure populations of a disordered conformation of α-synuclein within E. coli cells. PLoS ONE. 2013;8:e72286. doi: 10.1371/journal.pone.0072286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty BC, Young GB, Pielak GJ. Macromolecular crowding in the Escherichia coli periplasm maintains α-synuclein disorder. J Mol Biol. 2006;355:893–897. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Crowley PB, Chow E, Papkovskaia T. Protein interactions in the Escherichia coli cytosol: an impediment to in-cell NMR spectroscopy. ChemBioChem. 2011;12:1043–1048. doi: 10.1002/cbic.201100063. [DOI] [PubMed] [Google Scholar]
- Li C, Charlton LM, Lakkavaram A, Seagle C, Wang G, Young GB, Macdonald JM, Pielak GJ. Differential dynamical effects of macromolecular crowding on an intrinsically disordered protein and a globular protein: implications for in-cell NMR spectroscopy. J Am Chem Soc. 2008;130:6310–6311. doi: 10.1021/ja801020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander SW, Kallenbach NR. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17:75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, van Zijl PCM, Shortle D. Measurement of water–amide proton exchange rates in the denatured state of staphylococcal nuclease by a magnetization transfer technique. Proteins: Struct, Funct, Bioinf. 1997;28:325–332. [PubMed] [Google Scholar]
- Zhang Y-Z. Structural biology and molecular biophysics. Philadelphia: University of Pennsylvania; Protein and peptide structure and interactions studied by hydrogen exchange and NMR. [Google Scholar]
- Connelly GP, Bai Y, Jeng MF, Englander SW. Isotope effects in peptide group hydrogen exchange. Proteins. 1993;17:87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- Smith AE, Sarkar M, Young GB, Pielak GJ. Amide proton exchange of a dynamic loop in cell extracts. Protein Sci. 2013;22:1313–1319. doi: 10.1002/pro.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croke RL, Sallum CO, Watson E, Watt ED, Alexandrescu AT. Hydrogen exchange of monomeric α-synuclein shows unfolded structure persists at physiological temperature and is independent of molecular crowding in Escherichia coli. Protein Sci. 2008;17:1434–1445. doi: 10.1110/ps.033803.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CO, Monteith WB, Pielak GJ. Internal and global protein motion assessed with a fusion construct and in-cell NMR spectroscopy. ChemBioChem. 2011;12:390–391. doi: 10.1002/cbic.201000610. [DOI] [PubMed] [Google Scholar]
- Chevelkov V, Xue Y, Rao DK, Forman-Kay JD, Skrynnikov NR. 15NH/D-SOLEXSY experiment for accurate measurement of amide solvent exchange rates: application to denatured drkN SH3. J Biomol NMR. 2010;46:227–244. doi: 10.1007/s10858-010-9398-8. [DOI] [PubMed] [Google Scholar]
- Maroteaux L, Campanelli JT, Scheller RH. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8:2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K, Iino T. Role of the FliA-FlgM regulatory system on the transcriptional control of the flagellar regulon and flagellar formation in Salmonella typhimurium. J Bacteriol. 1994;176:3598–3605. doi: 10.1128/jb.176.12.3598-3605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Hanely LJ, Dahlquist FW. The C-terminal half of the anti-sigma factor FlgM contains a dynamic equilibrium solution structure favoring helical conformations. Biochemistry. 1998;37:1076–1082. doi: 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- Serber Z, Corsini L, Durst F, Dötsch V. In-cell NMR spectroscopy. Methods Enzymol. 2005;394:17–41. doi: 10.1016/S0076-6879(05)94002-0. [DOI] [PubMed] [Google Scholar]
- Serber Z, Dötsch V. In-cell NMR spectroscopy. Biochemistry. 2001;40:14317–14323. doi: 10.1021/bi011751w. [DOI] [PubMed] [Google Scholar]
- Booth IR. Regulation of cytoplasmic pH in bacteria. Microbiol Rev. 1985;49:359–378. doi: 10.1128/mr.49.4.359-378.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba N, Serber Z, Ledwidge R, Miller SM, Craik CS, Dötsch V. Quantitative identification of the protonation state of histidines in vitro and in vivo. Biochemistry. 2003;42:9227–9234. doi: 10.1021/bi0344679. [DOI] [PubMed] [Google Scholar]
- Croke RL, Patil SM, Quevreaux J, Kendall DA, Alexandrescu AT. NMR determination of pKa values in α-synuclein. Protein Sci. 2011;20:256–269. doi: 10.1002/pro.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good NE, Winget GD, Winter W, Connolly TN, Izawa S, Singh RMM. Hydrogen ion buffers for biological research. Biochemistry. 1966;5:467–477. doi: 10.1021/bi00866a011. [DOI] [PubMed] [Google Scholar]
- Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. Biomagresbank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G, Anderson JS, LeMaster DM. Polarization and polarizability assessed by protein amide acidity. Biochemistry. 2009;48:6482–6494. doi: 10.1021/bi900526z. [DOI] [PubMed] [Google Scholar]
- Harris DC. Quantitative chemical analysis. 7th ed. New York: W. H. Freeman and Company; 2007. [Google Scholar]
- Maltsev AS, Ying J, Bax A. Impact of N-terminal acetylation of α-synuclein on its random coil and lipid binding properties. Biochemistry. 2012;51:5004–5013. doi: 10.1021/bi300642h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D. Assignment of amino acid type in 1H-15N correlation spectra by labeling with 14N-amino acids. J Magn Res. 1994;105:88–90. doi: 10.1006/jmrb.1994.1106. [DOI] [PubMed] [Google Scholar]
- Krishnarjuna B, Jaipuria G, Thakur A, D'Silva P, Atreya H. Amino acid selective unlabeling for sequence specific resonance assignments in proteins. J Biomol NMR. 2011;49:39–51. doi: 10.1007/s10858-010-9459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchmore DC, McIntosh LP, Russell CB, Anderson DE, Dahlquist FW. Expression and 15N labeling of proteins for proton and 15N nuclear magnetic resonance. Methods Enzymol. 1989;177:44–73. doi: 10.1016/0076-6879(89)77005-1. [DOI] [PubMed] [Google Scholar]
- Ohki S-y, Kainosho M. Stable isotope labeling methods for protein NMR spectroscopy. Prog Nucl Magn Reson Spectrosc. 2008;53:208–226. [Google Scholar]
- Kelly AE, Ou HD, Withers R, Dötsch V. Low-conductivity buffers for high-sensitivity NMR measurements. J Am Chem Soc. 2002;124:12013–12019. doi: 10.1021/ja026121b. [DOI] [PubMed] [Google Scholar]
- Kang L, Moriarty GM, Woods LA, Ashcroft AE, Radford SE, Baum J. N-terminal acetylation of α-synuclein induces increased transient helical propensity and decreased aggregation rates in the intrinsically disordered monomer. Protein Sci. 2012;21:911–917. doi: 10.1002/pro.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, Tennant LL, Holmgren A. Proton-transfer effects in the active-site region of Escherichia coli thioredoxin using two-dimensional proton NMR. Biochemistry. 1991;30:4262–4268. doi: 10.1021/bi00231a023. [DOI] [PubMed] [Google Scholar]
- Jeng M-F, Dyson HJ. Direct measurement of the aspartic acid 26 pKa for reduced Escherichia coli thioredoxin by 13C NMR. Biochemistry. 1996;35:1–6. doi: 10.1021/bi952404n. [DOI] [PubMed] [Google Scholar]
- Markley JL. Observation of histidine residues in proteins by nuclear magnetic resonance spectroscopy. Acc Chem Res. 1975;8:70–80. [Google Scholar]
- Bundi A, Wüthrich K. 1H-NMR parameters of the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-L-Ala-OH. Biopolymers. 1979;18:285–297. [Google Scholar]
- Glasoe PK, Long FA. Use of glass electrodes to measure acidities in deuterium oxide. J Phys Chem. 1960;64:188–190. [Google Scholar]
- Barnes CO, Pielak GJ. In-cell protein NMR and protein leakage. Prot: Struct, Funct, Bioinf. 2011;79:347–351. doi: 10.1002/prot.22906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Wu KP, Baum J. Backbone assignment and dynamics of human α-synuclein in viscous 2 M glucose solution. Biomol NMR Assign. 2011;5:43–46. doi: 10.1007/s12104-010-9263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer D, Kutluay E, Bussell R, Browne G. Conformational properties of α-synuclein in its free and lipid associated states. J Mol Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


