Abstract
Using complementary approaches of potentiometry and NMR spectroscopy, we have determined that the equilibrium acid dissociation constant (pKa value) of the arginine guanidinium group is 13.8 ± 0.1. This is substantially higher than that of ∼12 often used in structure-based electrostatics calculations and cited in biochemistry textbooks. The revised intrinsic pKa value helps explains why arginine side chains in proteins are always predominantly charged, even at pH values as great as 10. The high pKa value also reinforces the observation that arginine side chains are invariably protonated under physiological conditions of near neutral pH. This occurs even when the guanidinium moiety is buried in a hydrophobic micro-environment, such as that inside a protein or a lipid membrane, thought to be incompatible with the presence of a charged group.
Keywords: protein electrostatics, pH titration, equilibrium acid dissociation constant, pKa value, guanidinium, tautomer, potentiometry, NMR spectroscopy
Introduction
The arginine side chain plays key roles in many essential biochemical processes. The planar guanidinium group often partakes in ionic and hydrogen bonding interactions critical to the structure and stability of proteins and protein complexes.1 It is also involved in proton (H+) transfer cascades in systems such as bacteriorhodopsin and cytochrome c oxidase. In these systems, arginine acts in its fully protonated state as a modulator of the pKa values of the groups serving as proton donors and acceptors.2–5 Furthermore, arginine has also been proposed to function as a general acid/base in a small number of enzymatic mechanisms,6–8 and its guanidinium moiety is a nucleophile or electrophile for a variety of post-translational modifications.9 Key to all of these functions is the charge state and hence equilibrium acid dissociation constant (pKa value) of the arginine side chain. However, within a biological context, an arginine in a measurably populated neutral state has never been unambiguously observed, even when buried in a highly hydrophobic environment such as the interior of a protein or in the middle of a lipid bilayer.10,11
The unusual ability of the arginine side chain to retain its charge under all physiological conditions is not well understood. It is especially perplexing that an arginine remains cationic even when buried in the nonpolar hydrophobic core of a folded protein.12 This is not what occurs with the ionizable side chains of lysine, histidine, tyrosine, cysteine, aspartic acid, and glutamic acid residues. For example, systematic studies have shown that the pKa values of these residues at numerous internal positions in staphylococcal nuclease shift in the direction that promotes the neutral state. The shifts can be substantial, such that lysines can have pKa values below 613 and aspartic/glutamic acids above 10.14,15 Consequently, these residues are usually neutral when buried within the interior of a protein under solution conditions with pH values near 7. This is consistent with the incompatibility between charged species and dry, hydrophobic environments. In contrast, the behavior of the arginine side chain is completely different.
Three main factors could determine the unusual ability of the arginine side chain to remain ionized, even in micro-environments that are normally considered to be incompatible with charges. First, the guanidinium moiety is large and its positive charge is delocalized over many atoms involved in a conjugated Y-π system.16 Owing to this delocalization, the charge unit of the guanidinium moiety has a large effective radius, and therefore its hydration energy, which is inversely proportional to radius, is low.17 The low hydration energy of the arginine side chain should mitigate the penalty for dehydration upon its transfer in the charged state to a hydrophobic environment. Note that this does not necessarily imply that the arginine side chain is not well hydrated. On the contrary, more crystallographic water molecules are usually found associated with the guanidinium moiety than with the primary amine of the lysine side chain.12,18 Only the carboxylic oxygen atoms of aspartate/glutamate residues appear to have higher affinities for water.19,20
The second factor that might enhance the ability of the arginine side chain to remain charged in environments, such as the protein interior or the middle of a lipid bilayer, is the conformational flexibility of its long side chain. This enables the guanidinium group to search for polar atoms, including buried water molecules, to satisfy its propensity to donate up to five hydrogen bonds.12
The third reason that arginine might always be charged under physiological conditions lies with its high intrinsic pKa value. Based on historical experiments dating back almost a century,21,22 a side chain pKa value of ∼12 is commonly cited in textbooks and used in structure-based electrostatics calculations. However, the accepted pKa values of 13.6 and 13.4 for the conjugate acids of guanidine and N-methyl guanidine, respectively,23,24 suggest strongly that the intrinsic pKa of the arginine side chain is significantly higher than 12. If so, then even when buried in the driest and most hydrophobic environments available, it may not become deprotonated to any significant level at neutral pH, or even at pH values up to 10. That is, due to an energetic penalty of ΔGo = 2.303RT(ΔpKa), the pKa of an arginine might never be perturbed to a value as low as 7 because the host proteins would unfold owing to the associated loss of net stability.17 In this respect, the properties of the lysine side chain are very different. With an intrinsic pKa value near 10.4 in the context of a random coil polypeptide,25,26 the pKa values of lysines within folded proteins can be shifted below 7. Thus, it is energetically possible to have buried lysine side chains that are completely deprotonated and neutral at physiological pH.13
In this study, we used complementary potentiometric and NMR spectroscopic methods to reinvestigate the pKa value of the arginine side chain. Our measured pKa values of 13.8 ± 0.1 are indeed almost 2 log units higher than that commonly assigned to this residue. This finding helps explain why, unlike the other ionizable amino acids, arginines remain predominantly charged in proteins under physiological conditions.
Results
Potentiometry
We used potentiometry to measure directly the release of protons by arginine in response to the addition of KOH. The results of these potentiometric titrations are summarized in Table1 and representative titration curves are shown in Figure 1. The blank used in these experiments consisted of a solution of lysine in water, allowing both calibration of the KOH titrant and control of the ionic strength without requiring the addition of supporting electrolytes. The pKa values resolved for and attributed to the α-aminium (9.4 ± 0.1) and side chain ζ-aminium (10.9 ± 0.1) groups of free lysine are consistent with accepted data.27,28
Table 1.
pKa Values Obtained from Potentiometric Titrations
| [Arg], mM | pKa values | ||
|---|---|---|---|
| Arg guanidinium | Lys α-aminium | Lys ζ-aminium | |
| 125 | 13.50 ± 0.2 | 9.39 ± 0.1 | 10.91 ± 0.1 |
| 200 | 13.61 ± 0.2 | 9.34 ± 0.1 | 10.88 ± 0.1 |
| 500 | 13.83 ± 0.2 | 9.41 ± 0.1 | 10.97 ± 0.1 |
| 500 | 13.76 ± 0.2 | 9.39 ± 0.1 | 10.93 ± 0.1 |
| 500 | 13.85 ± 0.2 | 9.37 ± 0.1 | 10.89 ± 0.1 |
| Avga | 13.8 ± 0.1 | 9.4 ± 0.1 | 10.9 ± 0.1 |
Averages and standard deviations for the three replicas with 500 mM arginine.
Figure 1.
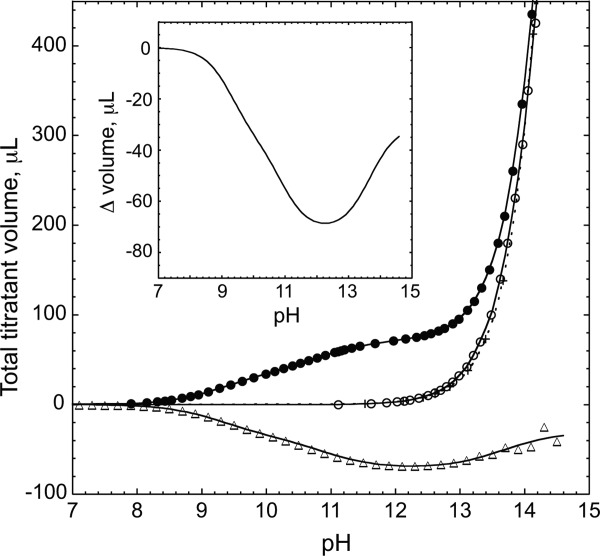
pKa measurements by potentiometry. Shown are titration curves of lysine (solid circles), arginine (open circles), and a water blank (x with dashed line). The arginine versus lysine/water difference curve is represented with open triangles. The lines are provided to guide the eye. The inset is a close up of this difference curve showing the best fit to Eq. (2). Solutions were 200 mM in each amino acid with 100 mM KCl.
The two titration events in a solution of lysine can be visualized without baseline correction. This is not the case for arginine. With charged α-carboxylate and guanidinium moieties and a neutral α-amine, the arginine titration begins at its isoionic point near pH 11 (i.e., the unadjusted pH of the initially prepared solution). Although masked by the steep rise in pH due to the necessary addition of large quantities of KOH, the guanidinium titration becomes apparent after the lysine/water blank is subtracted. As shown in the inset of Figure 1, inflection points in the difference curve can be seen for each titrating group. Fitting of this curve to a three-site binding isotherm yielded the desired pKa value of the arginine side chain.
The pKa value of the arginine guanidinium group obtained by analysis of the difference potentiometric titration curves is, on average, 13.8 ± 0.1 (Table1). This result is from multiple experiments performed over the course of five months with samples containing 500 mM arginine. Experiments were also carried out with lower concentrations of the free amino acid. Although yielding larger fitting errors due to the proportionally higher water background, a concentration-dependence was observed and a pKa value of 13.50 ± 0.2 was measured with 125 mM arginine (Table1). This small variation may be related to ionic strength differences or perhaps to the known propensity of the charged arginine side chain to associate with itself.29 However, for samples of arginine at 10 and 100 mM, similar high pKa values were obtained by NMR methods.
NMR spectroscopy
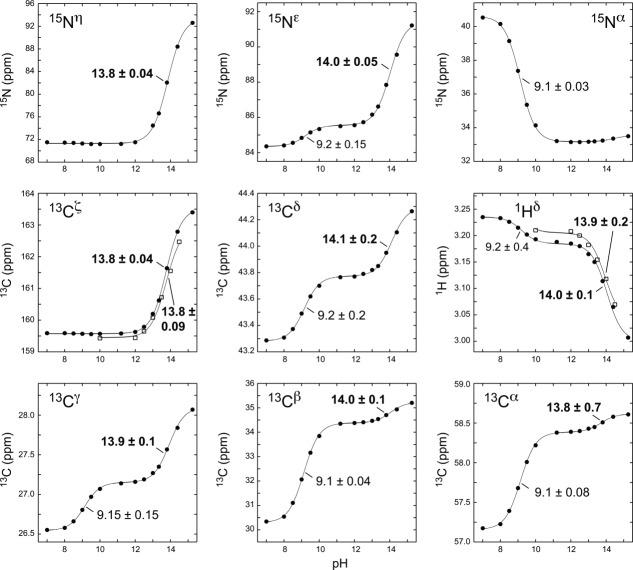
In parallel, we attempted to measure with NMR spectroscopy the pKa value of the arginine side chain in the context of a blocked tripeptide, acetyl-Gly-Arg-Gly-amide. As part of a study to define the pH-dependent chemical shifts of all the common ionizable amino acid side chains, this compound was chosen to mimic a random coil polypeptide and to avoid possible complications due to the deprotonation of the terminal α-aminium and α-carboxylic acid moieties.26 Unfortunately, under highly alkaline conditions, the tripeptide slowly hydrolyzed over the course of the titrations, which required ∼1 day due to detection of 13C at natural abundance. A similar hydrolysis problem precluded potentiometric studies of blocked arginine derivatives. As a result, reliable pH-dependent chemical shift changes could only be measured for the arginine side chain 13Cζ and 1Hδ nuclei in the intact peptide [Fig. 2(A)]. Based on these data, an average pKa value of 13.8 ± 0.1 was obtained. However, this could be an underestimation as the maximum sample pH value was only 14.5, and thus the plateau chemical shifts of the deprotonated species were not well defined. Also, according to the manufacturer's electrode specifications, interference from the ∼1 M Na+ added during the titration may have reduced the apparent sample pH by 0.1 to 0.2 log units.
Figure 2.
Arginine pKa measurements by NMR spectroscopy. Shown are titration curves of the arginine 13Cζ and 1Hδ nuclei in the 10 mM unlabeled blocked tripeptide acetyl-Gly-Arg-Gly-amide (open squares), and the side chain 13C and 15N nuclei in 100 mM 13C6/15N4-arginine (solid circles). The two 15Nη nuclei yield one degenerate signal. The curves and listed pKa values (bold for guanidinium) are the best nonlinear least squared fits to equations describing one or two sequential acid dissociation equilibria. Not shown are data for the arginine 13CO (reporting pKa 9.13 ± 0.05) and 1Hα (pKa 9.15 ± 0.04). Note that the titration data for each nuclei were fit separately and the small variations in pKa values shown in each panel of the figure reflect the sensitivity to factors such as experimental noise and the extrapolated end-point chemical shifts. Since the tautomeric forms of the neutral arginine side chain will rapidly interconvert in a pH-independent equilibrium, only a single overall macroscopic pKa value can actually be measured from the averaged chemical shift changes reported by these nuclei. Also, although arginine is following a predominantly sequential deprotonation pathway with increasing sample pH, the biphasic titration curves indicate that the chemical shifts of several reporter nuclei are sensitive to the ionization state of both the α-aminium and guanidinium groups (see Ref. 46 for further discussion).
Subsequently, we monitored via NMR spectroscopy the titration of numerous reporter nuclei in free 13C6/15N4-arginine. In this case, KOH was used to reduce possible electrode alkaline errors.30 Also, a higher final pH of 15.25 was attained. As shown in Figure 2(B), two distinct titration events were observed and could be confidently attributed to the α-aminium and guanidinium moieties. Under all pH values examined, the arginine α-carboxylic acid, with a known pKa value of ∼ 2,27,31,32 is unquestionably deprotonated. Based on the individually fit titration curves of these nuclei in data sets recorded with both 10 mM and 100 mM 13C6/15N4-arginine, the average pKa value of the α-aminium in the context of charged α-carboxylate and guanidinium moieties is 9.15 ± 0.05. This is consistent with previously reported pKa values of 9.0 to 9.3.27,31,32 Most importantly, the average of the pKa values fit individually for several guanidinium reporter nuclei in the presence of a charged α-carboxylate and neutral α-amine is 13.9 ± 0.15. The similarity of results for the acetyl-Gly-Arg-Gly-amide tripeptide and free arginine also indicates that electrode errors (K+ versus Na+) are minor and that the presence of the negatively charged carboxylate does not significantly increase the pKa value of the guanidinium group.
Discussion
Originating from pioneering studies in 192421 and 1930,22 the pKa value of the guanidinium group in free arginine is typically cited as 12 to 12.5. However, values ranging from as low as 11.4 to as high as 13.6 have been reported over the years.27,28,31–42 Although this may in part result from differences in experimental conditions, the pKa value of the arginine side chain appears to decrease only slightly with increasing temperature.28 Thus, the substantial variation in published pKa values most likely reflects the great experimental difficulty in measuring multi-step acid-base equilibria under highly alkaline conditions.
Given the importance of knowing an accurate reference acid dissociation constant for arginine, we have revisited this fundamental question using complementary potentiometric and NMR spectroscopic approaches. Our measurements were carried out independently by different personnel in two separate laboratories using different techniques and instrumentation. Collectively, we have found that the arginine guanidinium moiety has a pKa value of 13.8 to 13.9 in both the context of the free amino acid and a blocked tripeptide.
Our results confirm that the value of ∼12 commonly used for arginine in structure-based pKa calculations significantly overestimates the very weak acidity of the guanidinium group. Most importantly, a value closer to 14 is consistent with the observation that arginine side chains are always predominantly charged in proteins over a wide range of pH values spanning from physiological to as high as 10. This is the case even when the arginine side chains are buried in apparently dehydrated, hydrophobic micro-environments.12
For the arginine side chain to be neutral under physiological conditions of pH, a shift in its pKa value from 13.8 down to at least 7 would be necessary. This is equivalent to a Gibbs free energy change of almost 10 kcal/mol. Any protein bearing an arginine with such a shifted pKa would be destabilized by this full energetic penalty. Although not as large as the hydration energies of small ions, 10 kcal/mol exceeds the thermodynamic stability of most globular proteins in their folded versus unfolded states and is comparable with that of very tight ligand binding events.43 In contrast, with intrinsic pKa values of ∼10.4, 3.9, and 4.3,25,26 the side chains of lysine, aspartic acid, and glutamic acid, respectively, can be tolerated in their neutral forms within the context of a folded protein. That is, pKa values below 6 for the ζ-aminium group of lysine, and as high as 10 for the carboxylic acid groups of aspartic and glutamic acids have been measured.13–15
Even in the lipid bilayer, which in principle is more stable than most proteins, local instabilities would likely lead to water penetration and reorganization of polar head groups. This would almost certainly preclude maintenance of the strictly hydrophobic micro-environments that would be required to keep the arginine side chain in the neutral state. Thus, most biological environments are probably too unstable to tolerate the burial of an arginine side chain in its neutral form at physiological pH.
Stated conversely, the fact that charged arginine side chains are found within the interiors of stably folded proteins indicates that they do not undergo dramatic pKa shifts. Thus, proteins appear to have evolved to accommodate a buried guanidinium cation. In contrast to other ionizable side chains, the guanidinium group can participate in numerous stabilizing noncovalent interactions, including ion pairing, π-cation interactions, hydrogen bonding, and even association with another guanidinium cation. These effects could mitigate the cost of maintaining a buried side chain in the charged state despite being secluded from bulk water.29,44,45 Thus, nature appears to resort to arginine when it requires the presence of a charged species in the hydrophobic and dehydrated environment of a biological setting.
Arginine pKa measurements by potentiometry
Potentiometry is traditionally the method of choice for determining the pKa values of small molecules because it involves a direct measurement of a pH titration curve. This means that the amplitudes of the titrations determined potentiometrically reflect the number of proton binding sites. However, the resulting pKa values are macroscopic and, in the absence of any other information, not assignable to specific ionizable groups in the molecule. Furthermore, the standard potentiometry approach involves use of a glass electrode to measure pH values. At high pH, this method is fraught with difficulties caused primarily by the cation error and the sensitivity of the glass surface to these conditions.30 Fortunately, these problems were minimized in this study by using a modern glass electrode designed specifically for work under highly alkaline conditions. Also, a difference titration against a lysine/water blank enabled titrant calibration and, in effect, provided an internal reference for the data fitting. Collectively, this strongly supports the accuracy of the potentiometrically determined pKa values summarized in Table1 for the arginine side chain.
Arginine pKa measurements by NMR spectroscopy
NMR spectroscopy is often the method of choice for measuring polypeptide and protein pKa values because it provides site-specific information in the form of pH-dependent chemical shifts of nuclei associated with constituent ionizable groups.26 However, chemical shift changes may arise from additional ionization events, and the resulting pKa values are still macroscopic in the sense that they reflect the ensemble average of the pH-independent microscopic equilibria between all possible forms of a protein with a given total protonation level.46 In the case of the free amino acid arginine, the pKa values of its ionizable functional groups are sufficiently different that it follows one very dominant sequential deprontonation pathway with increasing sample pH (α-carboxylic acid then α-aminium then guanidinium). Thus, the fit pKa value of 9.15 ± 0.05 can be assigned to arise almost exclusively to the α-aminium group in the context of the charged guanidinium and α-carboxylate moieties, and that of 13.9 ± 0.15 to the guanidinium group in the context of the neutral α-amine and charged α-carboxylate.
The small difference in pKa values determined by NMR methods for the blocked acetyl-Gly-Arg-Gly-amide tripeptide (13.8 ± 0.1) and free arginine (13.9 ± 0.15) suggests that the ionization equilibrium of the guanidinium group is not substantially perturbed by the presence of the negatively charged α-carboxylate. This is expected given the length of the arginine side chain and the high ionic strength of a solution at pH ∼ 14. Consistent with this observation, a comparison of the ionization equilibria of arginine versus citrulline in solutions of 0.3 − 2 M ionic strength indicated that the pKa values of the α-carboxylic acid and α-aminium groups are perturbed by only ∼ −0.1 and +0.2 pKa units, respectively, due to electrostatic interactions with the positively charged guanidinium versus neutral carbamido moieties (i.e., in the directions expected for favorable or unfavorable electrostatic effects).32,41 Parenthetically, using internal reference compounds for pH calibration, the authors of this study also reported a pKa value of 13.54 ± 0.01 for the arginine side chain by 1H-NMR methods.32
pH-dependent chemical shifts
We are unaware of a case in which the pKa value of an arginine side chain in a protein has been measured unambiguously (although see, for example, Refs.47] and48]). Nevertheless, such studies are important to pursue for many reasons, including understanding electrostatic interactions in proteins and testing postulated roles of arginines in catalytic mechanisms and in proton transfer pathways. Therefore, the reference chemical shift data summarized in Figure 3 provide a benchmark for studying the ionization states and pKa values of arginines in polypeptides and proteins by NMR spectroscopy. In particular, the 13Cζ, 15Nε, and 15Nη nuclei should all report coincident titration curves with Δδ values similar in magnitude and sign to those listed in this figure. This is required to be reasonably confident that any pH-dependent chemical shifts result directly from the deprotonation of the arginine and not indirectly due to structural or electrostatic changes from the deprotonation of other groups, such as amino or phenol moieties, in the protein. Of course, the protein must be confirmed to remain stably folded and well behaved under highly alkaline conditions.
Figure 3.
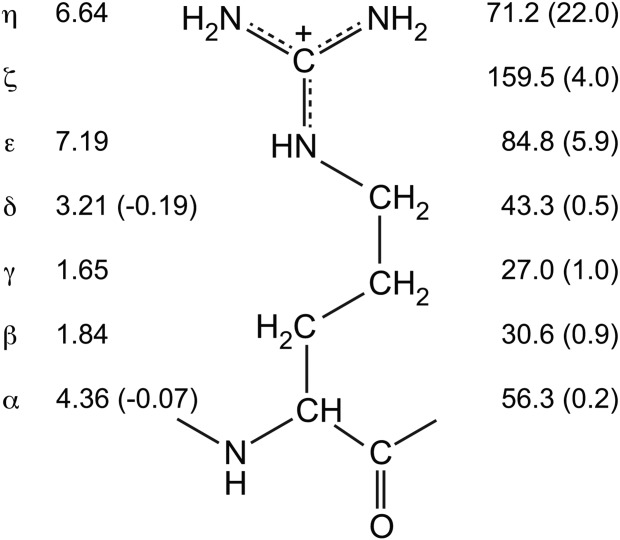
Summarized are the chemical shifts (1H left, 13C/15N right; ppm) of positively-charged arginine at neutral pH in the blocked tripeptide acetyl-Gly-Arg-Gly-amide. Shifts of prochiral 1H nuclei are averaged. Also shown in parentheses are the corresponding chemical shift changes upon deprontonation (Δδ = δhigh pH − δlow pH) of the guanidinium moiety. The latter were obtained from the pH titration curves of 13C6/15N4-arginine shown in Figure 2 using the fit limiting chemical shifts for the guanidinium moiety in the presence of the neutral α-amine, and thus assumed to be representative of a polypeptide. In the case of free arginine, the 15Nα shifts upfield by −7.5 ppm upon deprontonation (not shown). The Δδ values for the 1Hβ and 1Hγ are smaller in magnitude than 0.1 ppm, but could not be obtained reliably because of spectral overlap. Due to fast rotation about the Nε-Cζ and Cζ-Nη bond, average values are observed for the 1Hη and 15Nη nuclei.54 Although consistent with earlier studies, the Δδ values are generally larger than previously published, possibly due to better extrapolation to the high pH plateau chemical shifts.42,49,51,57–60
In the absence of any detectable pH-dependent chemical shift changes for reliable reporter nuclei (i.e., 13Cζ, 15Nε, or 15Nη), one cannot a priori conclude whether an arginine is always charged or always neutral over the conditions examined. Of course, given an intrinsic pKa value of almost 14, one expects (and finds) that the vast majority of arginines are charged in proteins. The observation of chemical shifts within the range of those shown in Figure 3 for the ionized side chain would certainly support this conclusion. In contrast, only the 15Nη nuclei with dramatically different chemical shifts of ∼71 ppm and 93 ppm in charged versus neutral arginine might serve as an indicator of its ionization state in the absence of any additional information. However, an arginine with an unusually low pKa value will most certainly be in a very unusual environment, which may also lead to strongly perturbed chemical shifts. Indeed, the guanidino moiety shows substantially smaller chemical shift differences between its charged and neutral forms within nonpolar solvents than in an aqueous environment.49 Unfortunately, this adds a further complication to consider when attempting to determine the ionization states of arginines in proteins by NMR spectroscopy.
Arginine tautomers and a route for pKa perturbations
In contrast to the planar guanidinium ion, the neutral side chain guanidine moiety is nonplanar and can adopt five possible tautomeric forms, each corresponding to the loss one of the five nitrogen-bonded protons.16 All of these neutral tautomers exist in a pH-independent equilibrium and can interconvert via bond rotations and/or proton transfer. As such, the observed acid dissociation constant of an arginine side chain is actually the sum of the five microscopic acid dissociation constants leading to each of these tautomers.
The observation of degenerate shifts for the two 15Nη nuclei, as well as similar shifts for the 15Nε and 15Nη nuclei (Figs. 2 and 3), indicates that the neutral arginine tautomers are approximately isoenergetic and interconvert rapidly on the chemical shift timescale (i.e., kex > 103 s−1). Otherwise, one would expect significantly different shifts for non-interconverting sp2 (C–N bonded) and sp3 (C–N bonded) hybridized nitrogens.50 However, the microscopic pKa values associated with each tautomer, and hence their equilibrium population distributions in the neutral form of the arginine side chain, remain uncertain. Based on an extrapolation of temperature-dependent chemical shifts recorded for neutral arginine and methylguanidine analogs in DMSO/H2O cosolvents, a distribution with ∼2/3 protonated at Nε has been estimated.51 In contrast, theoretical calculations suggested that the two lowest energy tautomers arise from deprotonation of either the Nε or one specific Nη.52 Also, the observation that the hydroxide-catalyzed rate constant for hydrogen exchange of the arginine NεH is two-fold higher than the NηH suggests a lower microscopic pKa value for Nε deprotonation and hence preferential formation of the tautomer lacking a proton at this nitrogen.53,54 A caveat to this argument is that steric factors could affect the relative rates of the diffusion-limited exchange reactions.
If a neutral arginine was indeed present in a folded protein, perhaps under highly alkaline conditions, then the relative populations of its tautomeric forms could certainly change in response to structure-dependent intermolecular interactions, such as hydrogen bonding. Moreover, it has been proposed that the pKa value of an arginine side chain could be perturbed by interactions that twist the guanidinium group towards a nonplanar conformation.6 Such structural-induced perturbations might enable the transient formation of a neutral guanidine moiety to serve, for example, as a general base in an enzymatically catalyzed reaction.
Materials and Methods
Potentiometric titrations
Solutions of the unmodified amino acids lysine and arginine (Sigma, >99.5% pure) were prepared volumetrically under an N2 atmosphere (CO2 free) with deionized water (degassed extensively by boiling under vacuum) made to 100 mM KCl. The stock solutions of lysine and arginine were initially at 500 mM and sealed under an N2 atmosphere until use. The correct volumes of sample and dilutant were withdrawn with a Hamilton syringe from the sealed containers and injected into the isolated titration vessel, also held under an N2 atmosphere. The water blanks were prepared in a similar manner. Total volumes used were in the range of 2.0 − 2.5 mL. The saturated KOH solution used as titrant was made from KOH pellets (Fisher), freshly dissolved in 100 mM KCl (prepared with degassed, deionized water).
Direct potentiometric titrations were performed using a glass electrode (pHG2HE8 from Radiometer America) that is inherently insensitive to cation error at pH values as high as 14.30,55 The electrode chain was completed with a calomel reference electrode (REF401 from Radiometer America) connected to a PHM95 pH meter (Radiometer America). The calibration of electrodes was performed with IUPAC series certified pH 9.180 and 12.45 standards (Radiometer, NIST) just prior to each experiment and rechecked after completion. Between use these buffers were stored in tightly sealed containers at 4 °C. Titrations were performed with a Dosimat E665 (Metrohm) delivering volumes as low as 1 μL. The temperature of the samples was kept at 25 ± 0.1 °C with a water-jacketed titration vessel. Titrations began immediately following initial temperature equilibration, and were monitored continuously with a thermistor probe to ensure re-equilibration after the heating that results from the exothermic dilution of highly concentrated KOH titrant.
Potentiometric titration analysis
The titration of free lysine was used to calibrate the titrants. This involved fitting the pH titration difference curve (titration of a water blank subtracted from the titration of lysine) to a two-site model describing the deprotonation of the α-aminium and side chain ζ-aminium groups. The KOH titrant concentrations were thereby determined to range from 13.7 to 14.5 M. The error in titrant concentration was estimated to be ± 0.3 M, resulting in a propagated error for the fit pKa values of ± 0.15 if > 12 and ± 0.03 if lower. Titrations were done multiple times to reduce these experimental errors.
To isolate the titration of the guanidine moiety of the arginine side chain, it was necessary to account for the water background (i.e., the steep rise in pH under highly alkaline conditions due to the increasing amounts of concentrated KOH). This requires taking the difference between the titration of the arginine sample and that of a suitable blank. The difference titration gives the volume of titrant, ΔVola, needed to achieve a given pH in the presence of the arginine side chain relative to that blank. The blank can be the aqueous solution void of arginine or, for practical reasons, a solution of lysine to help account for ionic strength effects. The difference curves were calculated by first interpolating the titration curves of both the arginine solution and the lysine solution, followed by their subtraction at equivalent pH values. The resultant difference curve is a convolution of three single-site binding isotherms: one for the guanidinium group of the arginine, one for the lysine side chain ζ-aminium, and one for the lysine α-aminium group. The arginine α-amino group, with a pKa ∼ 9 is not a factor because, unlike the case of the lysine sample, the titration of the arginine sample starts at pH ∼ 11 and thus it is essentially fully deprotonated. Each isotherm was modeled according to the fundamental equation:
| 1 |
where is the fraction of protons bound or released at each point of the titration, ΔVola is the total differential volume of base added at point a, nM is the initial number of moles of the molecule M, and [KOH] is the titrant molarity. The term m is a Hill coefficient used to evaluate the fitting, but fixed to 1 for the final analyses. The pKa values were determined from fitting the difference titration curves to the following equation:
is the fraction of protons bound or released at each point of the titration, ΔVola is the total differential volume of base added at point a, nM is the initial number of moles of the molecule M, and [KOH] is the titrant molarity. The term m is a Hill coefficient used to evaluate the fitting, but fixed to 1 for the final analyses. The pKa values were determined from fitting the difference titration curves to the following equation:
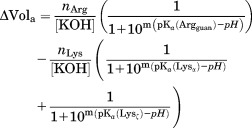 |
2 |
where Arggaun refers to the side chain guanidinium group of arginine, and Lysα and Lysζ to the α-aminium and side chain ζ-aminium groups of lysine, respectively. The parameters were resolved by fitting all simultaneously, or by first analyzing the lysine-water difference curve and then fixing those parameters while fitting the remaining terms for the arginine−lysine difference curve.
NMR-monitored titrations
The blocked tripeptide acetyl-Gly-Arg-Gly-amide (>95% purity by HPLC) was purchased from Biometik. The tripeptide was initially at ∼ 10 mM in 50 mM NaCl with 5% D2O for lock and 1 mM DSS (4,4-dimethyl-4-silapentane-1-sulfonic acid) as a pH-independent internal reference. The pH adjusted by addition of small aliquots of ∼ 0.1 M, 1 M, or solid NaOH. Uniformly 13C6/15N4-labeled l-arginine (Sigma-Aldrich) was initially at 10 mM or 100 mM in 50 mM NaCl with 5% D2O and 1 mM DSS, and the pH adjusted by addition of 0.1 M, 1 M, or solid KOH. The solid hydroxide was used to obtain the highest pH values. Sample pH values were measured at room temperature (∼20 oC) using a Thermal Scientific Orion* 3-Star pH meter and an Orion ROSS micro pH electrode (8220BNWP), calibrated with standard pH 7, 10, and 12.5 (Oakton) reference buffers. According to vendor's specifications, the electrode has a precision of ± 0.01 log units over a pH range of 0 to 14.
Spectra were recorded at 25 °C using a cryoprobe-equipped Bruker Avance III 500 MHz spectrometer. The 1H and 13C signals were referenced directly to internal DSS at 0.00 ppm, and 15N referenced indirectly via magnetogyric ratios.56 Chemical shifts for the unlabeled tripeptide were obtained from standard 1D 1H-NMR and 2D magnitude-mode 13C-HMBC spectra. Chemical shifts for the 13C6/15N4-arginine were obtained from 1D 13C-decoupled 1H-NMR, 1H/15N-decoupled 13C-NMR (with 1H NOE), and 1H/13C-decoupled 15N-NMR (with 1H NOE) spectra. Samples were in 5 mm diameter NMR tubes, except at very high pH and ionic strength conditions, for which 3 mm tubes were used to enable tuning. Also, a 100 mM sample of 13C6/15N4-arginine was required for the 15N-NMR measurements because the Bruker TCI CryoProbe is designed for direct detection of 1H and 13C, but not 15N signals.
Spectra were processed using Topspin 3. The pH-dependent chemical shifts were fit with GraphPad Prism and Matlab to published equations for one or two sequential titrations in order to obtain pKa and limiting chemical shift values.46 The standard deviations (fit precision) for the individual titrations shown in Figure 2 were obtained using a Monte Carlo approach with data points randomly varied by assumed standard deviations of 0.05 units for the sample pH, and 0.01/0.05/0.1 ppm for the 1H/13C/15N chemical shifts. The pKa values discussed in the text are the averages ± standard deviations of the means of the individually fit values for all nuclei reporting a given titration.
Acknowledgments
Instrument support was provided by the Canadian Institutes for Health Research (CIHR), the Canadian Foundation for Innovation (CFI), the British Columbia Knowledge Development Fund (BCKDF), the UBC Blusson Fund, and the Michael Smith Foundation for Health Research (MSFHR).
References
- Nandi CL, Singh J, Thornton JM. Atomic environments of arginine side chains in proteins. Protein Eng. 1993;6:247–259. doi: 10.1093/protein/6.3.247. [DOI] [PubMed] [Google Scholar]
- Petkova AT, Hu JG, Bizounok M, Simpson M, Griffin RG, Herzfeld J. Arginine activity in the proton-motive photocycle of bacteriorhodopsin: solid-state NMR studies of the wild-type and D85N proteins. Biochemistry. 1999;38:1562–1572. doi: 10.1021/bi981968z. [DOI] [PubMed] [Google Scholar]
- Hutson MS, Alexiev U, Shilov SV, Wise KJ, Braiman MS. Evidence for a perturbation of arginine-82 in the bacteriorhodopsin photocycle from time-resolved infrared spectra. Biochemistry. 2000;39:13189–13200. doi: 10.1021/bi000426q. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Hutson MS, Belenky M, Herzfeld J, Braiman MS. Role of arginine-82 in fast proton release during the bacteriorhodopsin photocycle: a time-resolved FT-IR study of purple membranes containing 15N-labeled arginine. Biochemistry. 2004;43:12809–12818. doi: 10.1021/bi049238g. [DOI] [PubMed] [Google Scholar]
- McMahon BH, Fabian M, Tomson F, Causgrove TP, Bailey JA, Rein FN, Dyer RB, Palmer G, Gennis RB, Woodruff WH. FT-IR studies of internal proton transfer reactions linked to inter-heme electron transfer in bovine cytochrome c oxidase. Biochim Biophys Acta. 2004;1655:321–331. doi: 10.1016/j.bbabio.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Schlippe YVG, Hedstrom L. A twisted base? The role of arginine in enzyme-catalyzed proton abstractions. Arch Biochem Biophys. 2005;433:266–278. doi: 10.1016/j.abb.2004.09.018. [DOI] [PubMed] [Google Scholar]
- van der Kamp MW, Perruccio F, Mulholland AJ. High-level QM/MM modelling predicts an arginine as the acid in the condensation reaction catalysed by citrate synthase. Chem Commun. 2008:1874–1876. doi: 10.1039/b800496j. [DOI] [PubMed] [Google Scholar]
- Holliday GL, Mitchell JBO, Thornton JM. Understanding the functional roles of amino acid residues in enzyme catalysis. J Mol Biol. 2009;390:560–577. doi: 10.1016/j.jmb.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Smith BC, Denu JM. Chemical mechanisms of histone lysine and arginine modifications. BBA-Gene Regul Mech. 2009;1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Vorobyov I, MacKerell AD, Allen TW. Is arginine charged in a membrane? Biophys J. 2008;94:L11–L13. doi: 10.1529/biophysj.107.121566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J, Cui Q. Does arginine remain protonated in the lipid membrane? Insights from microscopic pKa calculations. Biophys J. 2008;94:L61–L63. doi: 10.1529/biophysj.107.122945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Schlessman JL, Sue GR, Garcia-Moreno B. Arginine residues at internal positions in a protein are always charged. Proc Natl Acad Sci USA. 2011;108:18954–18959. doi: 10.1073/pnas.1104808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Castaneda CA, Cannon BR, Garcia-Moreno BE. Large shifts in pKa values of lysine residues buried inside a protein. Proc Natl Acad Sci USA. 2011;108:5260–5265. doi: 10.1073/pnas.1010750108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Cannon BR, Castaneda CA, Robinson A, Garcia-Moreno B. High tolerance for ionizable residues in the hydrophobic interior of proteins. Proc Natl Acad Sci USA. 2008;105:17784–17788. doi: 10.1073/pnas.0805113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isom DG, Castaneda CA, Cannon BR, Velu PD, Garcia-Moreno EB. Charges in the hydrophobic interior of proteins. Proc Natl Acad Sci USA. 2010;107:16096–16100. doi: 10.1073/pnas.1004213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raczynska ED, Cyranski MK, Gutowski M, Rak J, Gal JF, Maria PC, Darowska M, Duczmal K. Consequences of proton transfer in guanidine. J Phys Org Chem. 2003;16:91–106. [Google Scholar]
- Dwyer JJ, Gittis AG, Karp DA, Lattman EE, Spencer DS, Stites WE, Garcia-Moreno EB. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys J. 2000;79:1610–1620. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessman JL, Abe C, Gittis A, Karp DA, Dolan MA, Garcia-Moreno EB. Crystallographic study of hydration of an internal cavity in engineered proteins with buried polar or ionizable groups. Biophys J. 2008;94:3208–3216. doi: 10.1529/biophysj.107.122473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PE, Neilson GW, Dempsey CE, Barnes AC, Cruickshank JM. The hydration structure of guanidinium and thiocyanate ions: Implications for protein stability in aqueous solution. Proc Natl Acad Sci USA. 2003;100:4557–4561. doi: 10.1073/pnas.0735920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach T, Bowers MT. Hydration of biomolecules. Chem Phys Lett. 2009;480:1–16. [Google Scholar]
- Hunter A, Borsook H. The dissociation constants of arginine. Biochem J. 1924;18:883–890. doi: 10.1042/bj0180883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CLA, Kirk PL, Appleman K. The apparent dissociation constants of arginine and of lysine and the apparent heats of ionization of certain amino acids. J Biol Chem. 1930;88:285–293. [Google Scholar]
- Hall NF, Sprinkle MR. Relations between the structure and strength of certain organic bases in aqueous solution. J Am Chem Soc. 1932;54:3469–3485. [Google Scholar]
- Angyal SJ, Warburton WK. The basic strengths of methylated guanidines. J Chem Soc. 1951:2492–2494. [Google Scholar]
- Grimsley GR, Scholtz JM, Pace CN. A summary of the measured pKa values of the ionizable groups in folded proteins. Protein Sci. 2009;18:247–251. doi: 10.1002/pro.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platzer G, Okon M, McIntosh LP. pH-dependent random coil 1H, 13C, and 15N chemical shifts of the ionizable amino acids: a guide for protein pKa measurements. J Biomol NMR. 2014;60:109–129. doi: 10.1007/s10858-014-9862-y. [DOI] [PubMed] [Google Scholar]
- Perrin DD. Dissociation constants of organic bases in aqueous solution. London: Butterworths; 1965. [Google Scholar]
- Nagai H, Kuwabara K, Carta G. Temperature dependence of the dissociation constants of several amino acids. J Chem Eng Data. 2008;53:619–627. [Google Scholar]
- Vondrasek J, Mason PE, Heyda J, Collins KD, Jungwirth P. The molecular origin of like-charge arginine-arginine pairing in water. J Phys Chem B. 2009;113:9041–9045. doi: 10.1021/jp902377q. [DOI] [PubMed] [Google Scholar]
- Licht S. pH measurement in concentrated alkaline-solutions. Anal Chem. 1985;57:514–519. [Google Scholar]
- Dawson RMC, Elliott DC, Elliott WH, KJones KM. Data for biochemical research. Oxford: Clarendon Press; 1986. [Google Scholar]
- Orgovan G, Noszal B. The complete microspeciation of arginine and citrulline. J Pharmaceut Biomed. 2011;54:965–971. doi: 10.1016/j.jpba.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Birch TW, Harris LJ. A redetermination of the titration dissociation constants of arginine and histidine with a demonstration of the zwitterion constitution of these molecules. Biochem J. 1930;24:564–575. doi: 10.1042/bj0240564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein JP. Studies of the peptides of trivalent amino acids - iii. The apparent dissociation constants, free energy changes, and heats of ionization of peptides involving arginine, histidine, lysine, tyrosine, and aspartic and glutamic acids, and the behavior of lysine peptides toward nitrous acid. J Biol Chem. 1933;101:603–621. [Google Scholar]
- Melville J, Richardson GM. The titration constants of some amides and dipeptides in relation to alcohol and formaldehyde titrations of amino-N. Biochem J. 1935;29:187–195. doi: 10.1042/bj0290187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert A. Quantitative studies of the avidity of naturally occurring substances for trace metals. II. Amino acids having 3 ionizing groups. Biochem J. 1952;50:690–698. doi: 10.1042/bj0500690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke ER, Matrtell AE. Metal chelates of arginine and related ligands. J Inorg Nucl Chem. 1970;32:911–926. [Google Scholar]
- Nair MS, Santappa M. Equilibrium studies on copper(II) mixed-ligand complexes containing L-arginine and L-histidine, histamine or imidazole. Indian J Chem A. 1982;21:58–62. [Google Scholar]
- Bajpai AK, Yadava H. Ionophoretic technique in the study of dissociation-constants of amino acids. Afinidad. 1988;45:149–151. [Google Scholar]
- Albourine A, Petitramel M, Thomasdavid G, Vallon JJ. Potentiometry, visible-spectrophotometry and circular-dichroism of binary and ternary copper(II) complexes with 1-canavanine and 1-arginine. Can J Chem. 1989;67:959–966. [Google Scholar]
- Noszal B, Kassaitanczos R. Microscopic acid-base equilibria of arginine. Talanta. 1991;38:1439–1444. doi: 10.1016/0039-9140(91)80293-9. [DOI] [PubMed] [Google Scholar]
- Baillargeon MW, Laskowski M, Neves DE, Porubcan MA, Santini RE, Markley JL. Soybean trypsin-inhibitor (kunitz) and its complex with trypsin—13C nuclear magnetic-resonance studies of the reactive site arginine. Biochemistry. 1980;19:5703–5710. doi: 10.1021/bi00566a006. [DOI] [PubMed] [Google Scholar]
- Creighton TE. The biophysical chemistry of nucleic acids and proteins. London: Helvetian Press; 2010. [Google Scholar]
- Tissot AC, Vuilleumier S, Fersht AR. Importance of two buried salt bridges in the stability and folding pathway of barnase. Biochemistry. 1996;35:6786–6794. doi: 10.1021/bi952930e. [DOI] [PubMed] [Google Scholar]
- Dougherty DA. The cation-π interaction. Acc Chem Res. 2013;46:885–893. doi: 10.1021/ar300265y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh LP, Naito D, Baturin SJ, Okon M, Joshi MD, Nielsen JE. Dissecting electrostatic interactions in Bacillus circulans xylanase through NMR-monitored pH titrations. J Biomol NMR. 2011;51:5–19. doi: 10.1007/s10858-011-9537-x. [DOI] [PubMed] [Google Scholar]
- Niemeyer MI, Gonzalez-Nilo FD, Zuniga L, Gonzalez W, Cid LP, Sepulveda FV. Neutralization of a single arginine residue gates open a two-pore domain, alkali-activated K+ channel. Proc Natl Acad Sci USA. 2007;104:666–671. doi: 10.1073/pnas.0606173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cymes GD, Ni Y, Grosman C. Probing ion-channel pores one proton at a time. Nature. 2005;438:975–980. doi: 10.1038/nature04293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao YW, Braiman M. Modeling amino acid side chains in proteins: 15N NMR spectra of guanidino groups in nonpolar environments. J Phys Chem B. 2005;109:16953–16958. doi: 10.1021/jp051279e. [DOI] [PubMed] [Google Scholar]
- Witanowski M, Stefaniak L, Szymanski S, Webb GA. 14N NMR study of isomeric structures of urea and its analogs. Tetrahedron. 1976;32:2127–2129. [Google Scholar]
- Kanamori K, Roberts JD. A 15N NMR study of the barriers to isomerization about guanidinium and guanidino carbon nitrogen bonds in L-arginine. J Am Chem Soc. 1983;105:4698–4701. [Google Scholar]
- Norberg J, Foloppe N, Nilsson L. Intrinsic relative stabilities of the neutral tautomers of arginine side-chain models. J Chem Theory Comput. 2005;1:986–993. doi: 10.1021/ct049849m. [DOI] [PubMed] [Google Scholar]
- Yavari I, Roberts JD. Differential rates of proton-exchange for guanidinium nitrogens of L-arginine determined by natural-abundance 15N nuclear magnetic-resonance spectroscopy. Biochem Bioph Res Co. 1978;83:635–640. doi: 10.1016/0006-291x(78)91037-9. [DOI] [PubMed] [Google Scholar]
- Henry GD, Sykes BD. Determination of the rotational-dynamics and ph-dependence of the hydrogen-exchange rates of the arginine guanidino group using NMR spectroscopy. J Biomol NMR. 1995;6:59–66. doi: 10.1007/BF00417492. [DOI] [PubMed] [Google Scholar]
- Popov K, Ronkkomaki H, Lajunen LHJ. Guidelines for NMR measurements for determination of high and low pKa values. Pure Appl Chem. 2006;78:663–675. [Google Scholar]
- Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1H, 13C, and 15N chemical-shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Yamaguch T, Imanari M. 15N FT-NMR spectra of amino-acids in natural abundance - pH-dependence of 15N chemical-shifts for L-arginine. Tetrahedron Lett. 1974:1809–1812. [Google Scholar]
- Blomberg F, Maurer W, Ruterjans H. 15N nuclear magnetic-resonance investigations on amino acids. Proc Natl Acad Sci USA. 1976;73:1409–1413. doi: 10.1073/pnas.73.5.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London RE, Walker TE, Whaley TW, Matwiyoff NA. 15N nmr-studies of 13C, 15N labeled arginine. Org Magn Resonance. 1977;9:598–600. [Google Scholar]
- Kanamori K, Cain AH, Roberts JD. Studies of pH and anion complexation effects on L-arginine by natural abundance 15N nuclear magnetic resonance spectroscopy. J Am Chem Soc. 1978;100:4979–4981. [Google Scholar]