Abstract
Notch is an intercellular signaling pathway that is highly conserved in metazoans and is essential for proper cellular specification during development and in the adult organism. Misregulated Notch signaling underlies or contributes to the pathogenesis of many human diseases, most notably cancer. Signaling through the Notch pathway ultimately results in changes in gene expression, which is regulated by the transcription factor CSL. Upon pathway activation, CSL forms a ternary complex with the intracellular domain of the Notch receptor (NICD) and the transcriptional coactivator Mastermind (MAM) that activates transcription from Notch target genes. While detailed in vitro studies have been conducted with mammalian and worm orthologous proteins, less is known regarding the molecular details of the Notch ternary complex in Drosophila. Here we thermodynamically characterize the assembly of the fly ternary complex using isothermal titration calorimetry. Our data reveal striking differences in the way the RAM (RBP-J associated molecule) and ANK (ankyrin) domains of NICD interact with CSL that is specific to the fly. Additional analysis using cross-species experiments suggest that these differences are primarily due to fly CSL, while experiments using point mutants show that the interface between fly CSL and ANK is likely similar to the mammalian and worm interface. Finally, we show that the binding of the fly RAM domain to CSL does not affect interactions of the corepressor Hairless with CSL. Taken together, our data suggest species-specific differences in ternary complex assembly that may be significant in understanding how CSL regulates transcription in different organisms.
Keywords: notch signaling, CSL, RBP-J, isothermal titration calorimetry, X-ray crystallography, transcription, protein−protein interactions
Introduction
From the model organisms Drosophila melanogaster and Caenorhabditis elegans to more complex metazoans, such as mammals, the highly conserved Notch pathway serves as a cell-to-cell communication mechanism to regulate the transcription of numerous target genes.1 Genes controlled by the Notch pathway play a critical role in cell fate specification, thereby making the pathway essential for a number of developmental and homeostatic processes, including embryogenesis, organogenesis, hematopoiesis, and stem cell maintenance.2–4 Emphasizing its important and highly pleiotropic role in multicellular organisms is the fact that aberrant Notch signaling has been implicated in a wide variety of diseases, including cerebrovascular disease, as well as a diverse array of cancers and developmental disorders.2,5,6
Genetic studies in flies and worms identified the central components of Notch signaling, which consist of the receptor Notch, the ligand DSL (Delta, Serrate, Lag-2), and the nuclear effector CSL (CBF1/RBP-J, Su(H), Lag-1).1,7 Notch pathway activation occurs when a DSL ligand on a signal-sending cell interacts with the Notch receptor on an adjacent signal-receiving cell.8 This interaction triggers proteolytic cleavage of the Notch receptor, generating the NICD (Notch intracellular domain), which translocates to the nucleus and interacts with the DNA binding transcription factor CSL. A third protein, Mastermind (MAM), also binds to the complex, forming the ternary complex (CSL-NICD-MAM) necessary for transcriptional activation of target genes regulated by the pathway. In the absence of an activating signal, the Notch pathway also functions to repress the transcription of some, but not all, target genes.9,10 This is achieved when a corepressor protein, such as Hairless,11 interacts with CSL present on the DNA of a Notch target gene. Corepressors mediate interactions with histone remodeling complexes, e.g. histone deacetylase and methyltransferase, which convert the local chromatin to a repressive environment.9 The ability of CSL to differentially regulate gene expression is determined by its interaction with coregulatory proteins (coactivators or corepressors), placing CSL at the center of a transcriptional switch [Fig. 1(A)].
Figure 1.
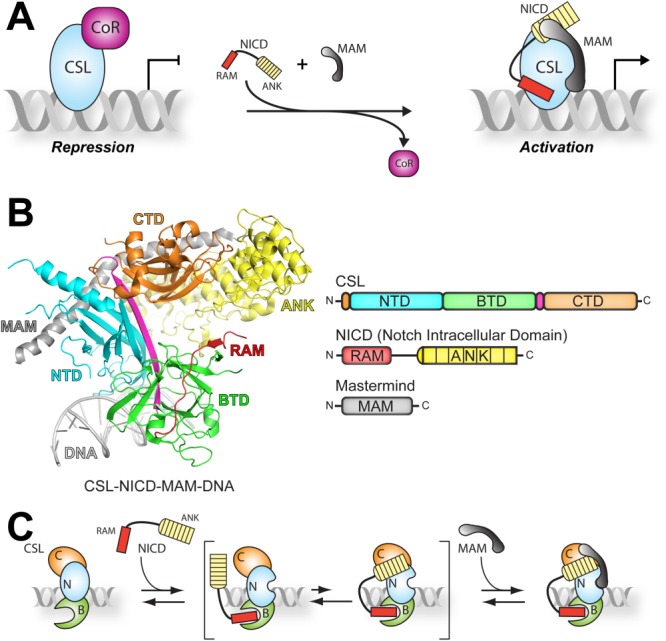
Overview of CSL-mediated transcription regulation. A: Model of CSL functioning as a transcriptional switch. Left, pathway inactivity allows corepressors (CoR, magenta) to interact with CSL present on DNA in the regulatory regions of target genes, and thereby repress gene transcription. Right, when the pathway is active, the corepressor complex is exchanged for two coactivators, Notch intracellular domain (NICD, red and yellow) and Mastermind (Mam, gray) to activate transcription from Notch target genes. B: Ribbon diagram (left) and domain schematics (right) of the CSL-NICD-MAM ternary complex bound to DNA.17 Coloring is consistent in both images. CSL consists of three domains—NTD (cyan), BTD (green), and CTD (orange). A beta-strand that bridges all three domains of CSL is colored magenta. The NTD and BTD of CSL make contacts with the DNA (gray). The RAM domain (red) of NICD interacts solely with the BTD of CSL while the ANK domain (yellow) interacts with both the NTD and CTD of CSL. Mastermind (gray) binds as a long helix across a composite surface created by the ANK domain bound to the NTD and CTD of CSL. C: Model of ternary complex assembly.12 According to this model, the RAM domain (red) of NICD binds to the BTD of CSL (green) in a high affinity interaction. The ANK domain (yellow) of NICD interacts very weakly with CSL until the second coactivator, MAM (gray), is present, locking the complex into an active conformation.
As shown in Figure 1(B), CSL is a DNA binding protein consisting of three domains—the N-terminal domain (NTD), the beta-trefoil domain (BTD), and the C-terminal domain (CTD).12,13 The BTD and NTD make both specific and nonspecific contacts to DNA, allowing CSL to bind DNA sequences present in genes regulated by the Notch pathway.13 Two domains of NICD mediate its interaction with CSL: the RAM (RBP-J associated molecule) and ANK (ankyrin) domains.14,15 RAM binds solely to the BTD of CSL, whereas ANK binds the CTD and NTD of CSL.16,17 The third protein of the CSL-NICD-MAM ternary complex, Mastermind, binds as a long α-helix with a distinctive bend, allowing it to make contacts with ANK as well as the CTD and NTD of CSL.16,17
Detailed biochemical and biophysical studies have defined a step-wise assembly mechanism for the CSL-NICD-MAM ternary complex [Fig. 1(C)].12,18 These studies showed that RAM forms a high affinity interaction with the BTD of CSL, initiating complex formation between CSL and NICD.19–21 These studies also showed that isolated constructs of ANK or MAM do not appreciably interact with CSL; conversely, when ANK and MAM are both present, formation of the CSL-NICD-MAM ternary complex occurs.19–21 It should be mentioned that these binding studies were performed with mammalian (human and mouse) and C. elegans proteins, and given the high degree of sequence conservation between orthologous Notch proteins, it has been assumed that the assembly mechanism of the CSL-NICD-MAM ternary complex is conserved for all organisms.
However, previous studies from our group using Notch proteins from D. melanogaster have compelled us to re-examine this assumption. In these studies, we demonstrated that the corepressor Hairless binds exclusively to the CTD of Su(H) (the fly ortholog of CSL).22 We also showed using EMSA that NICD (RAMANK) from Drosophila could efficiently displace Hairless from CSL in the absence of MAM.22 Given that previous studies demonstrated ANK interacts very weakly or not at all with the CTD of CSL, this suggests two possible mechanisms: one, RAM binding to the BTD induces a dramatic long-range conformational change in the CTD, which inhibits Hairless binding; and/or two, unlike the mammalian or worm ANK domain, the fly ANK domain interacts with the CTD of CSL, in the absence of MAM, and therefore can compete with Hairless for binding Su(H).
To address these two possible mechanisms, we used isothermal titration calorimetry to describe the binding interactions between Drosophila NICD and Su(H). Unexpectedly, we show that the ANK domain of Drosophila NICD is able to bind to Su(H) in the absence of MAM, which does not occur with the mammalian or worm orthologous proteins. To determine the molecular basis of this difference, we conducted a series of cross-species binding experiments using Drosophila and mammalian Notch proteins that suggest Su(H) is the primary factor that mediates this phenomenon. Additionally, point mutations were introduced into Su(H) and Drosophila NICD, based on the CSL-NICD-MAM X-ray structures, to disrupt the CTD-ANK interface. While single mutations do not appreciably affect binding, a quadruple ANK domain mutant significantly reduced binding to Su(H), which suggests that the molecular interactions of the Drosophila CSL-NICD complex are similar to those observed in the CSL-NICD-MAM ternary complex structures.16,17 Moreover, EMSA and ITC studies demonstrate that RAM binding does not affect Hairless interactions with the CTD of CSL. Taken together, our data define the assembly mechanism for Notch transcription complexes from D. melanogaster, which suggests that the molecular details of assembly are not strictly conserved in all metazoans.
Results
Analysis of Su(H)–NICD interactions
To define the thermodynamic binding parameters that underlie complexes formed between Su(H) and the Notch intracellular domain from Drosophila, we used ITC with highly purified preparations of recombinant Su(H) and NICD from bacteria. As shown in Table1 and Figure 2(A), a construct corresponding to the RAM and ANK domains of Drosophila NICD (dRAMANK) binds Su(H) with 60 nM affinity. This is slightly weaker than the affinity we previously measured between mouse CSL and NICD proteins (Kd ∼ 20 nM) and stronger than the binding we measured between the C. elegans orthologous proteins (Kd ∼ 3 μM) under identical conditions.20 For the mouse and worm NICD proteins, RAM contributes almost entirely to the observed binding to CSL; however, when we examined the individual contributions of the RAM and ANK domains of Drosophila NICD to Su(H) binding, we saw a distinct difference from the mouse and worm orthologs. The binding affinity between Su(H) and dRAM is 345 nM and the binding affinity between Su(H) and dANK is 668 nM [Fig. 2(C,B)]. We also analyzed the binding between dANK and a construct that corresponds to the CTD of Su(H) [dCTD, Fig. 2(D)]. In this case, we observed weaker binding between dCTD and dANK (Kd 21 μM) than between Su(H) and dANK (Kd 668 nM). Remarkably, these data suggest that the fly Notch proteins are behaving in a much different manner than the previously characterized worm and mammalian proteins.19–21
Table 1.
Calorimetric Data for the Binding of Drosophila NICD to Su(H)
| Cell | Syringe | K (M−1) | Kd (µM) | ΔG° (kcal/mol) | ΔH° (kcal/mol) | −TΔS° (kcal/mol) |
|---|---|---|---|---|---|---|
| dRAMANK | Su(H) | 1.9 ± 0.9 × 107 | 0.060 | −9.9 ± 0.2 | −23.8 ± 0.5 | 13.9 ± 0.5 |
| Su(H) | dRAM | 3.0 ± 0.7 × 106 | 0.345 | −8.8 ± 0.1 | −17.4 ± 0.7 | 8.6 ± 0.9 |
| Su(H) | dANK | 1.5 ± 0.4 × 106 | 0.668 | −8.4 ± 0.1 | −6.1 ± 0.4 | −2.2 ± 0.5 |
| dCTD | dANK | 4.7 ± 0.4 × 104 | 20.9 | −6.3 ± 0.05 | −9.3 ± 1.0 | 2.9 ± 1.0 |
All experiments were performed at 25°C. Values are the mean of at least three independent experiments and errors represent the standard deviation of multiple experiments.
Figure 2.
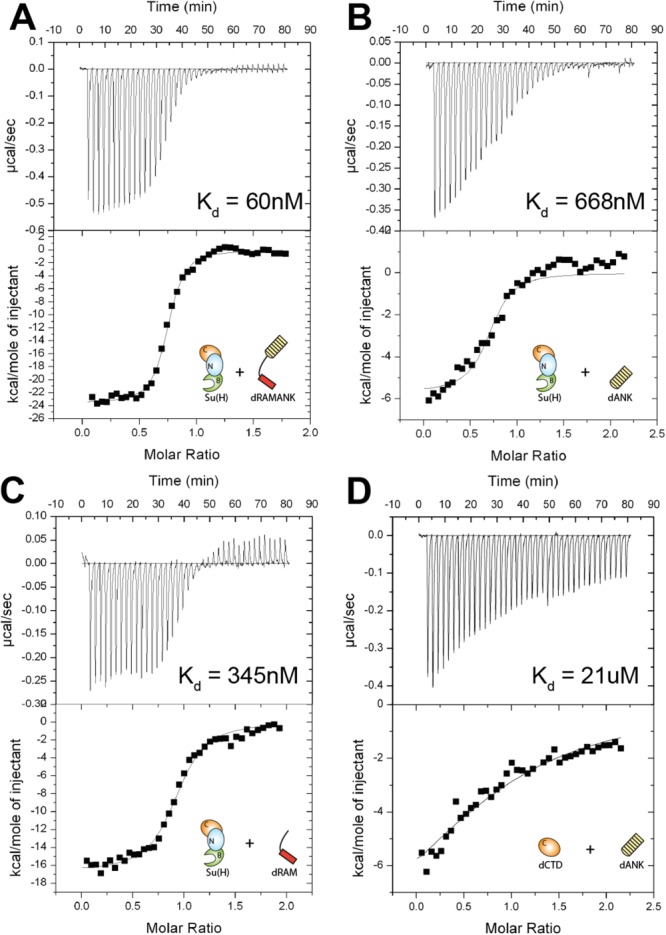
Thermodynamic binding analysis of Notch proteins from Drosophila. Figure shows representative thermograms (raw heat signal and nonlinear least squares fit to the integrated data) for Su(H) binding Drosophila NICD. Each experiment was performed at 25°C, with 40 titrations of 7 μL injections spaced 120 s apart. The experimentally determined dissociation constant (Kd) is shown for each experiment. A: Su(H) binding dRAMANK. B: Su(H) binding dANK. C: Su(H) binding dRAM. D: CTD of Su(H) binding dANK.
Cross-species binding studies of CSL-NICD interactions
To define where the difference(s) in CSL-NICD binding resides in flies, we performed cross-species ITC experiments with Notch components from mouse and Drosophila (Table2 and Figs. 3 and 4). In the initial set of experiments (Fig. 3), we assessed the interaction between RBP-J (mouse CSL) and NICD from Drosophila (dRAMANK). The binding affinity between RBP-J and dRAMANK is 50 nM [Fig. 3(A)], which is identical to the binding observed between Su(H) and dRAMANK (Kd 60 nM) within error. Similar to previous binding studies of the mammalian Notch proteins,19–21 RBP-J bound dRAM with 40 nM affinity [Fig. 3(B)], suggesting that dANK does not interact with the CTD of RBP-J.20 We confirmed this by measuring the binding between (1) RBP-J and dANK and (2) the CTD of RBP-J (mCTD) and dANK, and in both cases we could not detect any binding by ITC [Table2 and Fig. 3(C,D)]. It should also be mentioned that we measured the binding between mCTD and the ANK domain of mouse NICD (mANK), since it had not been measured previously, and as expected, we saw no interaction (Table2). Taken together, these data suggest that the binding profile of RBP-J with dRAMANK resembles the binding profile for the mouse orthologous proteins.
Table 2.
Calorimetric Data for NICD–CSL Binding Between Mouse and Drosophila Components
| Cell | Syringe | K (M−1) | Kd (µM) | ΔG° (kcal/mol) | ΔH° (kcal/mol) | −TΔS° (kcal/mol) | |
|---|---|---|---|---|---|---|---|
| Su(H) + mNICD | mRAMANK | Su(H) | 4.9 ± 0.8 × 106 | 0.206 | −9.1 ± 0.09 | −17.7 ± 0.8 | 8.6 ± 0.8 |
| mRAM | Su(H) | 2.5 ± 1.1 × 106 | 0.437 | −8.7 ± 0.2 | −14.6 ± 0.7 | 5.9 ± 0.9 | |
| mANK | Su(H) | NBD | NBD | NBD | NBD | NBD | |
| Su(H) | mANK | NBD | NBD | NBD | NBD | NBD | |
| dCTD | mANK | NBD | NBD | NBD | NBD | NBD | |
| RBP-J + dNICD | dRAMANK | RBP-J | 2.8 ± 1.6 × 107 | 0.050 | −10.0 ± 0.4 | −16.6 ± 0.5 | 6.5 ± 0.9 |
| dRAM | RBP-J | 2.9 ± 0.5 × 107 | 0.040 | −10.1 ± 0.1 | −14.9 ± 0.2 | 4.7 ± 0.3 | |
| dANK | RBP-J | NBD | NBD | NBD | NBD | NBD | |
| RBPJ | dANK | NBD | NBD | NBD | NBD | NBD | |
| mANK | mCTD | NBD | NBD | NBD | NBD | NBD | |
| mCTD | dANK | NBD | NBD | NBD | NBD | NBD |
All experiments were performed at 25°C. Values are the mean of at least three independent experiments and errors represent the standard deviation of multiple experiments. NBD, no binding detected.
Figure 3.

Cross-species binding experiments (RBP-J + dNICD). Figure shows representative thermograms (raw heat signal and nonlinear least squares fit to the integrated data) for RBP-J binding Drosophila NICD. Each experiment was performed at 25°C, with 40 titrations of 7 μL injections spaced 120 s apart. The experimentally determined dissociation constant (Kd) is shown for each experiment. A: RBP-J binding dRAMANK. B: RBP-J binding dRAM. C: RBP-J does not bind dANK. D: CTD of RBP-J does not bind dANK. NBD, no binding detected.
Figure 4.

Cross-species binding experiments (Su(H) + mNICD). Figure shows representative thermograms (raw heat signal and nonlinear least squares fit to the integrated data) for Su(H) binding mouse NICD. Each experiment was performed at 25°C, with forty titrations of 7 μL injections spaced 120 s apart. The experimentally determined dissociation constant (Kd) is shown for each experiment. A: Su(H) binding mRAMANK. B: Su(H) binding mRAM. C: Su(H) does not bind mANK. D: CTD of Su(H) does not bind mANK. NBD, no binding detected.
The second set of cross-species ITC experiments assessed the interaction between Su(H) and the mouse NICD (mRAMANK) (Fig. 4). With an affinity of 206 nM [Fig. 4(A)], the interaction between Su(H) and mRAMANK is approximately three-fold weaker than the Su(H)-dRAMANK complex and approximately ten-fold weaker than the RBPJ-mRAMANK complex.20 The interaction of Su(H) and mRAM yielded an affinity of 437 nM [Fig. 4(B)], which is similar to the affinity for Su(H)-mRAMANK (Kd 206 nM), suggesting that the mouse ANK domain (mANK) does not interact with Su(H). We confirmed this by binding experiments with Su(H) and mANK, as well as dCTD and mANK, which in both cases displayed no observable binding by ITC [Fig. 4(C,D)]. From these experiments, we conclude that the difference in CSL-NICD binding between mouse and fly proteins likely lies primarily with Su(H) and not dNICD.
Binding analysis of Su(H) – dRAMANK point mutations
Using the CSL-NICD-MAM ternary complex structures as a guide,16,17 point mutations were made to Su(H) and to dRAMANK targeting the CTD-ANK interface (Supporting Information Figs. S1 and S2).17 These mutations focused on a conserved Glu-Arg ion pair buried at the CTD-ANK interface previously shown to have a deleterious effect on complex formation and transcription when mutations produce like charges.19,23,24 Additionally, we tested a quadruple mutant in dRAMANK (R1985E/R2027E/R2093E/E2094R), hereafter termed dRAMANK4xMUT, that was shown to affect binding and abrogate ternary complex formation with the human Notch proteins.19 We first tested each point mutant with a wild-type partner. As shown in Table3, the combinations of Su(H) with dRAMANKR1985E or dRAMANKR2027E showed only small differences in affinity, but were not statistically significant difference in binding when compared to wild-type Su(H) with wild-type dRAMANK. Similarly, the combination of Su(H)E446R with dRAMANK or dRAMANKR1985E or dRAMANKR2027E also showed no statistically significant difference in binding affinity when compared to wild-type Su(H) with wild-type dRAMANK (Table3). However, when we tested the binding of dRAMANK4xMUT with Su(H) we observed a significant four-fold reduction in binding (Kd 261 nM) compared with wild-type, which is similar, but not identical, to the affinity of Su(H) for dRAM (Kd 345 nM). Altogether, binding analysis of Su(H) and dRAMANK mutants suggest that the contacts at the interface between the CTD of Su(H) and the ANK domain of fly NICD are similar to what was observed in the human and worm CSL-NICD-MAM ternary complex structures.
Table 3.
Calorimetric Data for Su(H) and dNICD Point Mutants
| Cell | Syringe | K (M−1) | Kd (µM) | ΔG° (kcal/mol) | ΔH° (kcal/mol) | −TΔS° (kcal/mol) | ΔΔG° (kcal/mol) |
|---|---|---|---|---|---|---|---|
| dRAMANKR1985E | Su(H) | 5.8 ± 1.8 × 106 | 0.180 | −9.2 ± 0.1 | −18.3 ± 1.5 | 9.1 ± 1.6 | 0.7 |
| dRAMANKR2027E | Su(H) | 1.5 ± 0.2 × 107 | 0.068 | −9.7 ± 0.1 | −28.3 ± 0.7 | 18.5 ± 0.6 | 0.2 |
| dRAMANK | Su(H)E446R | 6.4 ± 2.1 × 106 | 0.170 | −9.2 ± 0.2 | −18.5 ± 1.4 | 9.2 ± 1.4 | 0.7 |
| dRAMANKR1985E | Su(H)E446R | 6.3 ± 1.9 × 106 | 0.167 | −9.2 ± 0.1 | −21.6 ± 0.6 | 12.3 ± 0.8 | 0.7 |
| dRAMANKR2027E | Su(H)E446R | 8.9 ± 2.4 × 106 | 0.118 | −9.4 ± 0.1 | −20.0 ± 0.7 | 10.6 ± 0.9 | 0.5 |
| GST-dRAMANK4xMUT | Su(H) | 3.9 ± 0.8 × 106 | 0.261 | −8.9 ± 0.1 | −16.8 ± 0.9 | 7.8 ± 1.0 | 1.0 |
All experiments were performed at 25°C. Values are the mean of at least three independent experiments and errors represent the standard deviation of multiple experiments. 4xMUT=R1985E/R2027E/R2093E/E2094R.
Characterizing the effect RAM binding has on Su(H)-Hairless interactions
Previous work from our lab using Notch proteins from worm and mammals demonstrated that RAM binding to the BTD of CSL promotes ternary complex formation by inducing a distal conformational change in the NTD of CSL, thereby creating a binding site for MAM.20 In other work, we showed that the corepressor Hairless binds exclusively to the CTD of Su(H); however, in EMSA experiments dRAMANK was able to efficiently displace Hairless from Su(H).22 To determine whether RAM binding affects Hairless interactions with the CTD of Su(H), we employed two methods: competition ITC and EMSA. For our competition ITC binding experiments, we preformed complexes of Su(H) and RAM before titrating with Hairless. The experiment yielded an affinity of 1 nM, which is essentially identical to the affinity we previously measured for Su(H) and Hairless (Table4).22 For the EMSAs, we preformed complexes of Su(H) and Hairless before adding either RAM, ANK, and MAM or ANK and MAM (Fig. 5). Both gels showed nearly identical results—the Su(H)-Hairless complexes persisted, as ANK and MAM [Fig. 5(A)] or RAM, ANK, and MAM [Fig. 5(B)] were very ineffective at displacing Hairless from Su(H). Taken together, these results suggest that RAM binding to Su(H) does not affect Su(H)-Hairless interactions.
Table 4.
Calorimetric Data for Competition ITC Between Su(H)-RAM and Hairless
| Cell | Syringe | K (M−1) | Kd (µM) | ΔG° (kcal/mol) | ΔH° (kcal/mol) | −TΔS° (kcal/mol) |
|---|---|---|---|---|---|---|
| Su(H) | Hairless | 9.1 ± 2.4 × 108 | 0.001 | −12.2 ± 0.2 | −16.1 ± 0.7 | 3.9 ± 0.8 |
| Su(H) - RAM | Hairless | 7.8 ± 4.3 × 108 | 0.001 | −12.0 ± 0.3 | −9.5 ± 0.7 | −2.5 ± 1.0 |
All experiments were performed at 25°C. Values are the mean of at least three independent experiments and errors represent the standard deviation of multiple experiments. Values for Su(H)-Hairless were taken from our publication Maier et al., 2011.
Figure 5.
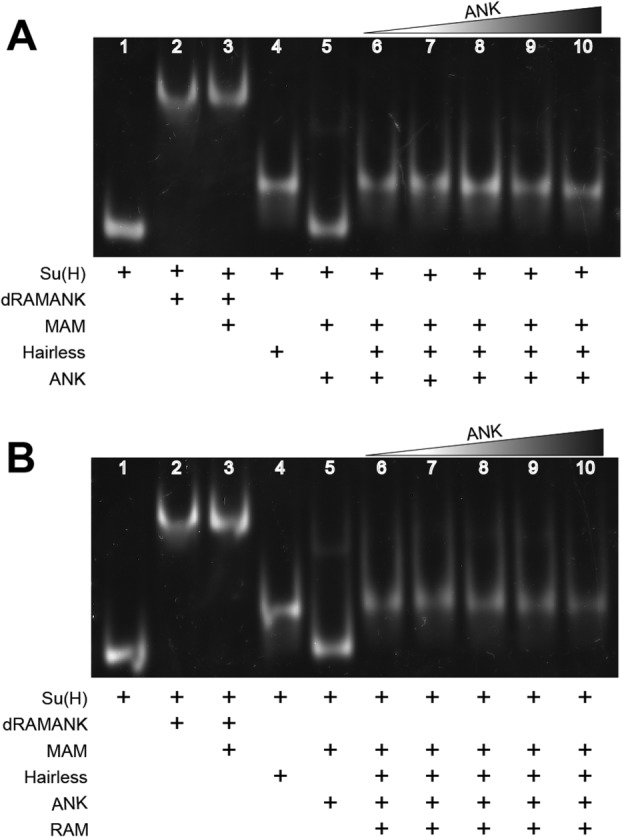
Characterizing the effect RAM has on Su(H)-Hairless interactions. Figure shows representative EMSAs in which ANK + MAM (A) or RAM + ANK + MAM (B) compete for binding to the preformed Su(H)-Hairless-DNA complex. The control lanes (1−5) for both EMSAs contain Su(H)-DNA, Su(H)-dRAMANK-DNA, Su(H)-dRAMANK-MAM-DNA, Su(H)-Hairless-DNA, and Su(H)-dANK-MAM-DNA, respectively. ANK was added in increasing amounts (Lanes 6−10) either with (B) or without (A) RAM. In both cases, ANK or RAM + ANK compete poorly for the Su(H)-Hairless complex.
Discussion
Canonical Notch signaling ultimately results in changes in gene expression, which is regulated by the DNA binding transcription factor CSL.7,8,25 Upon pathway activation, CSL forms a ternary complex with the intracellular domain of the Notch receptor (NICD) and the transcriptional coactivator Mastermind (MAM) to activate gene expression from Notch targets.12 CSL also interacts with corepressors, such as Hairless, to repress transcription from some, but not all, Notch responsive genes.10,11 Both the mechanism of signal transduction and the individual components of the Notch pathway are highly conserved, for example fly and mouse CSL proteins share ∼78% sequence identity within their structural core (Supporting Information Figs. S1 and S2).12 Previously, extensive structural, biophysical, and biochemical/cellular studies were performed on Notch proteins, primarily from mammals and worms, resulting in a detailed model of CSL-NICD-MAM ternary complex formation.12 Given the high degree of conservation between orthologous components, it has been widely assumed that the assembly mechanism of the ternary complex would also be strictly conserved between organisms. However, previous studies from our group prompted us to reassess whether this assumption held true for Notch proteins from Drosophila.22
A hallmark of the assembly mechanism is that RAM forms a high affinity interaction with the BTD of CSL (for the mouse proteins Kd ∼ 20 nM).19–21 This serves to tether ANK to CSL, greatly increasing its local concentration for subsequent interactions with the CTD of CSL and MAM [Fig. 1(C)].26 Despite this dramatic increase in local concentration, in the absence of MAM, the binding of ANK to CSL is nearly immeasurable.19–21,27 Here, we show that the fly proteins behave quite differently. In this case, both RAM and ANK bind to Su(H) (fly CSL) with sub-micromolar affinity (Table1 and Fig. 2). Interestingly, yeast two-hybrid studies performed 20 years ago also observed significant interactions between Su(H) and the isolated ANK domain of NICD.14,15 Additionally, two other points are worth mentioning: one, while ANK also binds the isolated CTD of Su(H), it does so with 30-fold less affinity. This may be due to the interactions ANK makes with the NTD of CSL, as observed in the CSL-NICD-MAM-DNA X-ray structures,16,17 as well as an entropic penalty that may result from folding coupled to binding for the isolated CTD construct. And two, due to the chelate effect, the Gibb's free energy of binding (ΔG°) for RAMANK interacting with Su(H) is greater than it is for the isolated constructs of RAM or ANK, but the free energies are not strictly additive, which is commonly seen for small molecules binding to macromolecules.28 This may be due to the ∼55 Å distance between where RAM binds the BTD and ANK binds the CTD of CSL.
Given this striking difference in the binding interactions between fly and mammalian Notch proteins, we sought to identify the molecular basis for this observation. As there are no major sequence differences between mammalian and fly orthologs of CSL and NICD (Supporting Information Figs. S1 and S2), in particular at the CTD-ANK interface, there is no obvious reason as to why dANK binds CTD, whereas mANK does not. In an effort to discern which component, dANK or Su(H), is largely responsible for this effect, we performed cross-species ITC experiments using mouse and fly Notch proteins. These studies convincingly showed that RBP-J interacts with dRAMANK in a very similar manner as it does with mRAMANK, that is both mouse and fly RAM form a high affinity interaction with the BTD of RBP-J, and neither dANK nor mANK interact with the CTD of RBP-J. However, the results of the cross-species experiments with Su(H) and mRAMANK were not as clear-cut. In this case, Su(H) bound both mRAM and mRAMANK with roughly similar affinities, as the two-fold difference in binding was not statistically significant. Consistent with this, mANK did not bind Su(H). However, mRAMANK bound Su(H) with three-fold less affinity than dRAMANK, which was statistically significant and comparable to the affinity between dRAM and Su(H). Taken together, these data seem to suggest that Su(H) is the factor playing the largest role in the difference between mammalian and fly Notch proteins. Future binding studies will focus on the approximately 30 residues different between the CTDs of mouse and fly (Supporting Information Fig. S1) to better understand how these changes allow Su(H) to bind ANK.
To further scrutinize Su(H)-dRAMANK interactions, we designed point mutations based on the CSL-NICD-MAM-DNA X-ray structures that focused on a Glu-Arg salt bridge buried at the CTD-ANK interface (Supporting Information Figs. S1 and S2).16,17,23 we tested the binding of both dRAMANKR1985E and dRAMANKR2027E, which correspond to the arginines observed in the worm and human X-ray structures, respectively, that would pair with Glu446 on Su(H), as well as the Su(H)E446R mutant.16,17,23 Interestingly, none of the single mutants had a dramatic effect on binding (Table3); however, the quadruple mutant dRAM ANK4xMUT did significantly reduce affinity almost to the level observed for Su(H)-dRAM binding. Similar results have been seen previously with the human Notch proteins19, that is single mutants in RAMANK had little to no effect on ternary complex formation in EMSA and FRET assays, but the corresponding quadruple did. This suggests that the molecular contacts at the dCTD-dANK interface are similar to what was observed in the human and worm structures when MAM was bound to the complex. Certainly, future studies of the fly Notch proteins will prove useful for characterizing interactions between the CTD of Su(H) and the ANK domain of NICD in the absence of MAM, which may provide additional insights into ternary complex assembly.
Previously, we showed that the corepressor Hairless binds solely to the CTD of Su(H); however, we also showed in competitive binding assays that dRAMANK could efficiently displace Hairless from Su(H) in the absence of MAM.22 In light of herein described binding experiments, in which dANK was shown to bind Su(H), provide a molecular explanation for why dRAMANK is an effective competitor for Su(H)-Hairless complexes. Consistent with this reasoning, we demonstrated via ITC and EMSA that RAM does not affect Hairless binding to Su(H) (Table4 and Fig. 5). Together, these results indicate RAM binding to the BTD does not cause a long-range conformational change in the CTD of Su(H), but is important for tethering ANK to Su(H).
Finally, we present a revised model of CSL-NICD-MAM ternary complex formation that is fly-specific (Fig. 6). In this case, when NICD binds Su(H) both RAM and ANK have appreciable interactions with Su(H). We suspect that when MAM binds Su(H)-dRAMANK, it forms a ternary complex very similar to what was observed in the human and worm X-ray structures. While the biological significance of a fly-specific model is not immediately obvious, it is interesting to speculate that perhaps the difference in dRAMANK binding to Su(H) is necessary for displacement of Hairless from Su(H), but this will require further study. Nonetheless, it will be important for future studies to take into consideration possible species-specific differences in Notch signaling, which may impact interpretation of results and phenotypes.
Figure 6.

Revised model of ternary complex assembly for Drosophila Notch proteins. In contrast to the mammalian and worm Notch proteins, our binding data suggest that the binding of Drosophila NICD to Su(H) is partitioned between its RAM and ANK domains, such that ANK has appreciable interactions with the CTD of Su(H) in the absence of MAM.
Materials and Methods
Cloning, expression, and protein purification
The cloning, expression, and purification of constructs that correspond to Mus musculus RBP-J (53-474), as well as the RAMANK (1744-2113), RAM (1744-1771), and ANK (1827-2133) constructs from mouse Notch1 were described previously.20 Additionally, the cloning, expression, and purification of Drosophila melanogaster Su(H) (98–523) and the CTD (101-119 + 415-523) domain of Su(H), as well as the RAMANK (1762-2142) and ANK (1858-2142) domains of fly Notch were previously described.22 The construct corresponding to the RAM (1762-1790) domain of fly Notch was cloned, expressed, and purified similar to the RAM domain from mouse Notch1.
Isothermal titration calorimetry
Proteins for use in isothermal titration calorimetry (ITC) experiments were degassed and buffer-matched using either size exclusion chromatography or dialysis. Protein concentrations were determined by UV absorbance at 280 nm. ITC experiments were performed with a MicroCal VP-ITC microcalorimeter. All experiments were conducted at 25°C in a buffer of 50 mM sodium phosphate, pH 6.5, and 150 mM sodium chloride. A typical experiment consisted of 10 µM macromolecule in the cell and 100 µM ligand in the syringe. Data were analyzed with the ORIGIN software package and fit to a one-site binding model. The reported binding data are the average of at least three individual experiments (n = 3). For the competition ITC experiment, proteins were prepared separately as described above. The Hairless construct (232-358) retained an N-terminal SMT3 fusion tag from purification; however, no binding was detected between SMT3 and Su(H) (data not shown). Purified Su(H) and dRAM were combined in a 1 : 1 ratio and placed in the microcalorimeter cell and then titrated with Hairless.
Electrophoretic mobility shift assays
EMSAs were performed as described previously.20,22 Briefly, purified constructs of Su(H) and Hairless (232-269) were incubated for 15 minat room temperature with a 19-mer duplex DNA (-GTTACTGTGG GAAAGAAAG-) containing a single CSL-binding site (in bold type) from the Hes-1 gene. Various combinations of purified Drosophila RAMANK, RAM, ANK, and MAM proteins were added to the preformed DNA-Su(H)-Hairless complexes and incubated for an additional 15 min at room temperature. The complexes were separated on a 7% polyacrylamide gel containing 0.5x Tris-borate buffer, pH 7.0, for 3 h at 4°C and visualized using SYBR-GOLD stain (Invitrogen).
Acknowledgments
The authors thank members of the Kovall lab for their support and helpful comments for the manuscript.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
Supplementary Information
References
- Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140. doi: 10.1242/jcs.127308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME. Introduction—notch in development and disease. Semin Cell Dev Biol. 2012;23:419–420. doi: 10.1016/j.semcdb.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Liu J, Sato C, Cerletti M, Wagers A. Notch signaling in the regulation of stem cell self-renewal and differentiation. Curr Top Dev Biol. 2010;92:367–409. doi: 10.1016/S0070-2153(10)92012-7. [DOI] [PubMed] [Google Scholar]
- Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch and disease: a growing field. Semin Cell Dev Biol. 2012;23:473–480. doi: 10.1016/j.semcdb.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25:318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray S, Furriols M. Notch pathway: making sense of suppressor of hairless. Curr Biol. 2001;11:R217–221. doi: 10.1016/s0960-9822(01)00109-9. [DOI] [PubMed] [Google Scholar]
- Maier D. Hairless: the ignored antagonist of the Notch signalling pathway. Hereditas. 2006;143:212–221. doi: 10.1111/j.2007.0018-0661.01971.x. [DOI] [PubMed] [Google Scholar]
- Kovall RA, Blacklow SC. Mechanistic insights into Notch receptor signaling from structural and biochemical studies. Curr Top Dev Biol. 2010;92:31–71. doi: 10.1016/S0070-2153(10)92002-4. [DOI] [PubMed] [Google Scholar]
- Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004;23:3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J Biol Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- Del Bianco C, Aster JC, Blacklow SC. Mutational and energetic studies of Notch 1 transcription complexes. J Mol Biol. 2008;376:131–140. doi: 10.1016/j.jmb.2007.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann DR, Wilson JJ, Kovall RA. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J Biol Chem. 2008;283:14781–14791. doi: 10.1074/jbc.M709501200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubman OY, Ilagan MX, Kopan R, Barrick D. Quantitative dissection of the Notch:CSL interaction: insights into the Notch-mediated transcriptional switch. J Mol Biol. 2007;365:577–589. doi: 10.1016/j.jmb.2006.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier D, Kurth P, Schulz A, Russell A, Yuan Z, Gruber K, Kovall RA, Preiss A. Structural and functional analysis of the repressor complex in the Notch signaling pathway of Drosophila melanogaster. Mol Biol Cell. 2011;22:3242–3252. doi: 10.1091/mbc.E11-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA. Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr Opin Struct Biol. 2007;17:117–127. doi: 10.1016/j.sbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Yuan Z, Friedmann DR, VanderWielen BD, Collins KJ, Kovall RA. Characterization of CSL (CBF-1, Su(H), Lag-1) mutants reveals differences in signaling mediated by Notch1 and Notch2. J Biol Chem. 2012;287:34904–34916. doi: 10.1074/jbc.M112.403287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Bertagna A, Toptygin D, Brand L, Barrick D. The effects of conformational heterogeneity on the binding of the Notch intracellular domain to effector proteins: a case of biologically tuned disorder. Biochem Soc Trans. 2008;36:157–166. doi: 10.1042/BST0360157. [DOI] [PubMed] [Google Scholar]
- VanderWielen BD, Yuan Z, Friedmann DR, Kovall RA. Transcriptional repression in the Notch pathway: thermodynamic characterization of CSL-MINT (Msx2-interacting nuclear target protein) complexes. J Biol Chem. 2011;286:14892–14902. doi: 10.1074/jbc.M110.181156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks WP. On the attribution and additivity of binding energies. Proc Natl Acad Sci USA. 1981;78:4046–4050. doi: 10.1073/pnas.78.7.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Supplementary Information


