Abstract
Heme coordination state determines the functional diversity of heme proteins. Using myoglobin as a model protein, we designed a distal hydrogen-bonding network by introducing both distal glutamic acid (Glu29) and histidine (His43) residues and regulated the heme into a bis-His coordination state with native ligands His64 and His93. This resembles the heme site in natural bis-His coordinated heme proteins such as cytoglobin and neuroglobin. A single mutation of L29E or F43H was found to form a distinct hydrogen-bonding network involving distal water molecules, instead of the bis-His heme coordination, which highlights the importance of the combination of multiple hydrogen-bonding interactions to regulate the heme coordination state. Kinetic studies further revealed that direct coordination of distal His64 to the heme iron negatively regulates fluoride binding and hydrogen peroxide activation by competing with the exogenous ligands. The new approach developed in this study can be generally applicable for fine-tuning the structure and function of heme proteins.
Keywords: crystal structures, heme proteins, hydrogen bonding, non-native states, protein design
Heme proteins perform a wide array of biological functions such as O2 carrying, electron transfer, catalysis, and signaling through a diverse group of heme coordination states.1 For example, cytochrome c (cyt c) functions as an electron transfer protein with a heme coordinated by both His and Met, and exhibits an enhanced peroxidase activity upon cleavage of the axial Met–heme ligation.2 Myoglobin (Mb), an O2 carrier, has a six-coordinated heme by axial His93 and water in its met form, where a distal His64 stabilizes the axial water through hydrogen bonding (Figure 1 a).3 Differently, two newly discovered members of the globin family, cytoglobin (Cgb)4 and neuroglobin (Ngb),5 have a heme with two axial histidine residues, His81/His113 and His64/His96, respectively. In addition, nonsymbiotic plant hemoglobin (Hb) such as rice Hb also contains a heme with bis-His coordination, His73/His108.6 It was shown that bis-His heme coordination has a novel role in regulating ligand binding and reactivity,7 whereas the structural basis and molecular mechanism underlying its formation are not well understood. In the case of Ngb, although an intramolecular disulfide bond of Cys46–Cys55 was found to regulate ligand binding, the X-ray structure showed that both WT Ngb and its mutant without the disulfide bond, C46G/C55S/C120S Ngb, are in a bis-His heme coordination state.5 Therefore, it is enticing to develop a strategy to regulate the heme coordination state and fine-tune the diverse functions of heme proteins.
Figure 1.
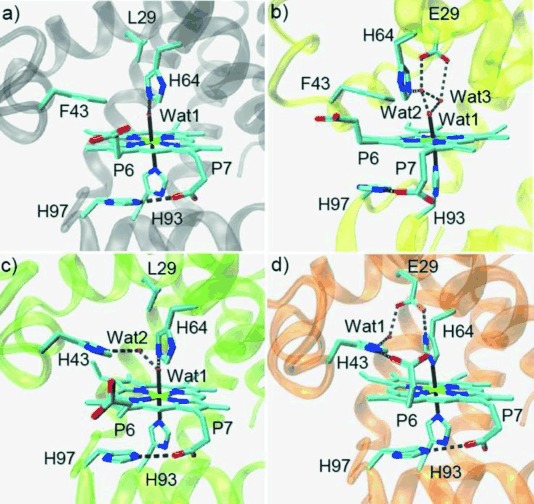
Crystal structures of the met form of WT Mb (A, PDB code 1 JP63), L29E Mb (B, PDB code 4PQ6), F43H Mb (C, PDB code 4PQC), and L29E/F43H Mb (D, PDB code 4PQB), showing the heme coordination state and distal hydrogen-bonding network.
To this end, it is pivotal to develop such a strategy for reconstructing a model heme protein such as Mb with different heme coordination states. Mb has been shown to be an ideal model for rational protein design.8 As shown previously,9 a straightforward approach to constructing a bis-His coordination state is to introduce a proximal ligand, such as moving the distal histidine from position 64 to 68 in Mb, resulting in His68/His93 coordination. Meanwhile, sequence alignment showed that the highly conserved ligands in natural globins with bis-His heme coordination are in positions corresponding to 64 and 93 of Mb.4b Up to now, there has not been an X-ray or NMR structure reported for Mb or its mutants adopting a non-native state of bis-His coordination with native ligands His64/His93. To regulate the heme coordination state and tune the distal His64 to be an axial ligand, we developed an approach to design a distal hydrogen-bonding network, by which a bis-His coordinated sate of Mb was successfully achieved with the native ligands.
Based on X-ray crystallography studies of globins with bis-His coordination states,4–6 we found that there are hydrogen-bonding interactions between the heme and the polypeptide chain. For example, there is a distal water molecule in the heme pocket of Ngb forming two hydrogen bonds with His64 (Nδ atom) and the backbone carbonyl group of Phe61. Moreover, two heme propionate groups, P7 and P6, form hydrogen-bonding interactions with two lysine residues, Lys67 and Lys95, respectively, on both sides of the heme plane (Figure S1).5a These interactions likely play key roles in the formation of bis-His heme coordination of Ngb, thus providing us with valuable clues for rational design to tune the heme coordination state of Mb.
Moreover, by inspections of the X-ray structures of heme proteins, we noticed that there is a conserved His-Asp pair in the proximal pocket of peroxidase, and the hydrogen bond is crucial for protein stability and catalysis.10 To test whether a similar His–Asp/Glu pair could be introduced in the distal pocket of Mb to construct a distal hydrogen-bonding network, we engineered a single mutant of L29E Mb. As shown in Figure 1 b, the X-ray crystal structure revealed that, instead of interacting with His64, Glu29 interacts directly with two distal water molecules (wat2 and wat3), where wat2 also forms a hydrogen bond with His64, and both wat2 and wat3 interact with the axial wat1. Consequently, the interaction between His64 and axial wat1 was weakened by this unique hydrogen-bonding network, and the distance between His64 and heme iron slightly increased from 4.40 Å in WT Mb to 4.64 Å in L29E Mb.
On the other hand, it was found that heme P7 interacts with a lysine at position 67 in Ngb, which likely facilitates bis-His coordination. We thus attempted to construct a similar hydrogen-bonding interaction in Mb. Meanwhile, a previous study showed that the T67R Mb mutant was not in a bis-His coordination state,11 and thus we did not test the T67K Mb mutant. Alternatively, the X-ray structure of Mb shows that on the heme proximal side, heme P7 interacts with His97, with the imidazole plane in parallel with the heme plane (Figure 1 a). We thus became interested in whether a similar interaction could be engineered on the distal side for the heme P6. We found that Phe43 is a potential target due to its suitable conformation with respect to the heme plane and then constructed a F43H Mb mutant. Unexpectedly, the X-ray structure showed that His43 does not interact directly with heme P6. Instead, it forms a hydrogen bond with an additional distal water (wat2) that also interacts with the axial wat1 (Figure 1 c).
With the structural information of L29E Mb and F43H Mb, we envisaged that the combination of two mutations may provide stronger interactions to achieve a bis-His heme coordination state, compared with that by a single mutation. Therefore, we engineered a double mutant of L29E/F43H Mb and crystallized it successfully. The structure showed that, as expected, the heme group is coordinated by both proximal His93 and distal His64 through the Nɛ atom with a distance of 2.06 and 2.12 Å (Figure 1 d), respectively. The bis-His heme coordination thus confirms the spectroscopic observations as well as computer modeling in previous studies.12 Moreover, one O atom of Glu29 interacts with Nδ atom of His64 by forming a strong hydrogen bond (2.49 Å), which resembles the conserved His–Asp pair in the proximal pocket of peroxidase.10 The other O atom of Glu29 is bridged with the Nɛ atom of His43 by a water molecule, and the Nδ atom of His43 further interacts with heme P6, similar to the interaction between His97 and heme P7. These observations suggest that the combination of both His64–Glu29 and Glu29–wat–His43–heme–P6 interactions facilitates the formation of a bis-His heme coordination state.
To probe conformational alterations induced by bis-His heme coordination, we compared the overall structure of L29E/F43H Mb with WT Mb, as well as Cgb, Ngb, and rice Hb with globin folding but possessing a bis-His coordinated heme. As shown in Figure 2 a, when heme planes are superimposed, the helix E of L29E/F43H Mb shifts toward the heme plane with respect to that of WT Mb, resulting in movement of distal His64 by 2.28 Å for the Nɛ atom and direct coordination to the heme iron. The resultant conformations of helix E and distal His64, together with helices C and D, overlap well with that of Cgb (Figure 2 b), although Mb and Cgb share a low sequence identity of 25 %.4 The helix E and distal His64 of L29E/F43H Mb also overlap well with that of both Ngb (Figure 2 c) and rice Hb (Figure 2 d). The comparison indicates that not only the heme coordination state but also the overall conformation is regulated by the designed distal hydrogen-bonding network.
Figure 2.
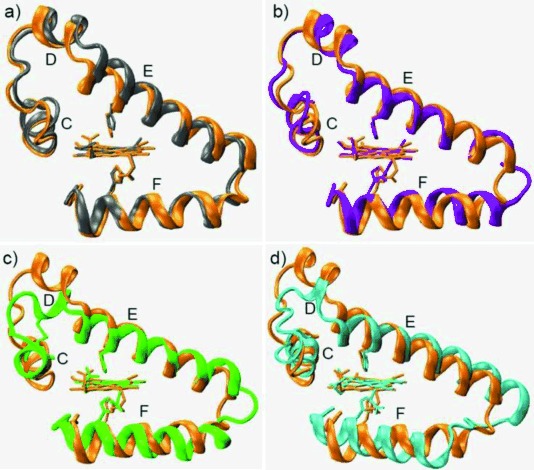
Comparison of helix C–F regions by overlapping the crystal structure of L29E/F43H Mb (orange) with WT Mb (gray, PDB code 1 JP63) (a), cytoglobin (purple, PDB code 1 V5H4b) (b), neuroglobin (green, PDB code 4 MPM5b) (c), and rice Hb (cyan, PDB code 1D8U6) (d).
To examine the coordination of distal His64 and interactions of the distal hydrogen-bonding network, we used fluoride as a probe, as applied previously for heme proteins.13 The stopped-flow spectra showed that the Soret band rapidly shifts from 415 to 407 nm upon mixing L29E/F43H Mb with fluoride, with an increase of a charge-transfer (CT1) band at 610 nm (Figure 3 a). The resultant new spectrum (407 and 610 nm) resembles that of the fluoride–Mb complex (406 and 607 nm),13e suggesting that His64 dissociates from the heme iron and fluoride binds alternatively. The 3 nm red shift of the CT1 band from 607 to 610 nm indicates the presence of multiple hydrogen-bonding interactions between fluoride and distal residues in L29E/F43H Mb, likely donated by His64, His43, Glu29, and distal water, instead of only distal His64 and a water molecule in WT Mb.13a As shown in Figure 3 a, inset, the absorbance change of the Soret band was fitted well to a single exponential decay equation. By plotting the rate constants (kobs) versus fluoride concentrations (Figure 3 b), we obtained the association rate constants, ka (kon), and the dissociation rate constants, kd (koff), summarized in Table 1. The kon and koff values determined for WT Mb are similar to that reported for horse Mb (6.0±0.5 m−1 s−1 and 0.10±0.01 s−1).13c L29E Mb has a kon value approximately twofold greater than that of WT Mb, with a koff value decreased by approximately threefold, which suggests that distal Glu29 facilitates the binding of fluoride and stabilizes it by hydrogen-bonding interactions. Meanwhile, F43H Mb exhibits slightly increased kon and koff values compared with WT Mb, suggesting little contribution from the second distal His43. The double mutant L29E/F43H Mb shows a similar kon value to that of F43H Mb, whereas it shows an approximate fivefold increased koff value compared with that of F43H Mb and WT Mb. These observations indicate that the direct coordination of distal His64 competes with fluoride for binding and promotes its dissociation from the heme iron. The role of His64 in bis-His coordinated L29E/F43H Mb thus resembles that in Cgb and Ngb, where the axial ligand plays a key role in tuning the binding affinity of exogenous ligands.7
Figure 3.
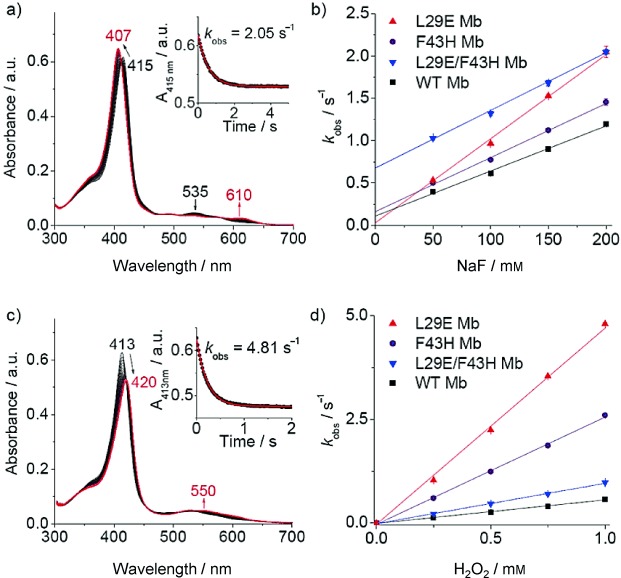
a) Stopped-flow spectra upon mixing L29E/F43H Mb and fluoride (5 μm and 100 mm, respectively, final concentration) in 100 mm KH2PO4 buffer (pH 7.0) for 5 s. b) Plots of observed rate constants versus fluoride concentrations. c) Stopped-flow spectra upon mixing L29E Mb and H2O2 (5 μm and 1 mm, respectively, final concentration) in the same buffer for 2 s. d) Plots of observed rate constants versus H2O2 concentrations. Insets in (a) and (c) show the single-exponential fit of the decay of the Soret band signal. For (b) and (d), data represent the mean ± SD for n=3 replicates.
Table 1.
Comparisons of the association and dissociation rate constants (kon and koff) of fluoride binding, and the rate constant (k1) of compound II formation for wild-type (WT) myoglobin (Mb) and its mutants.[a]
| Mbs | kon (F−) [m−1 s−1] | koff (F−) [s−1] | k1 (H2O2) [mm−1 s−1] |
|---|---|---|---|
| WT Mb | 5.14±0.15 | 0.13±0.01 | 0.57±0.01 |
| L29E Mb | 9.77±0.17 | 0.04±0.01 | 4.85±0.12 |
| F43H Mb | 6.42±0.16 | 0.15±0.02 | 2.59±0.05 |
| L29E/F43H Mb | 6.80±0.23 | 0.67±0.02 | 0.97±0.02 |
[a] Data represent the mean ±SD for n=3 replicates.
To further probe the effect of the distal hydrogen-bonding network and bis-His heme coordination on protein reactivity, we evaluated the ability of activation of hydrogen peroxide by L29E Mb, F43H Mb, and L29E/F43H Mb. Stopped-flow kinetic studies showed that, ferric L29E Mb readily reacts with hydrogen peroxide and yields an oxoferryl heme (Compound II), as indicated by a shift of the Soret band from 413 nm to 420 nm and an increase of the visible band at 550 nm (Figure 3 c). Similar to fluoride binding, the decay curve of A413 nm obeyed pseudo first-order kinetics (Figure 3 c, inset). Plots of kobs values versus hydrogen peroxide concentrations showed good linearity for both Mb mutants and WT Mb (Figure 3 d). The rate constants (k1) of Compound II formation for L29E Mb and F43H Mb are approximately 8.5-fold and 4.5-fold higher than that for WT Mb, respectively (Table 1). The value of 0.57±0.01 mm−1 s−1 determined for WT Mb agrees with that reported previously by Watanabe and co-workers (0.51 mm−1 s−1 at 20 °C).14 The enhanced peroxide activation ability of L29E Mb is attributed to the distal Glu29, as in chloroperoxidase where a distal glutamate rather than a histidine is responsible for the cleavage of the peroxide O−O bond.15 On the other hand, the enhanced ability of F43H Mb is due to the suitable distance of distal His43 to the heme iron (5.67 Å) for activation of hydrogen peroxide, similar to that in native peroxidases, such as cyt c peroxidase (CcP) (5.6 Å)16 and horseradish peroxidase (HRP) (6.0 Å).17 Notably, the ability of hydrogen peroxide activation of L29E/F43H Mb was greatly decreased compared with two single mutants, indicating that the direct coordination of distal His64 blocks the binding of hydrogen peroxide and thus inhibits the formation of Compound II. In comparison to WT Mb, the activity of L29E/F43H Mb is slightly higher (∼1.7-fold), which suggests that hydrogen peroxide can compete with the heme axial ligand, His64, where both distal Glu29 and His43 likely participate in activation of heme-bound hydrogen peroxide.
In summary, inspired by the structural information of globins with bis-His heme coordination, we designed a distal hydrogen-bonding network in Mb by introducing both a distal Glu29 and His43, and regulated the heme into a bis-His coordination state with native ligands, His64 and His93. A single mutation of L29E or F43H was found to form a distinct hydrogen-bonding network involving distal water molecules, instead of the bis-His heme coordination, which highlights the importance of the combination of multiple hydrogen-bonding interactions to regulate the heme coordination state. Kinetic studies further revealed that direct coordination of distal His64 to the heme iron negatively regulates fluoride binding and hydrogen peroxide activation by competing with the exogenous ligands. Distinctly, Glu29 promotes fluoride binding by multiple hydrogen-bonding interactions. Moreover, both Glu29 and His43 facilitate the activation of hydrogen peroxide, which resembles the active site of native peroxidases such as chloroperoxidase and CcP, respectively. As developed in this study, the design of a distal hydrogen-bonding network enables us to regulate the heme coordination state that determines protein functions. This design can be generally applied to other heme proteins for fine-tuning their structure and function.
Experimental Section
Protein preparation: WT sperm whale Mb was expressed in BL21(DE3) cells using the Mb gene of pMbt7–7 and purified using a procedure described previously.18 L29E Mb, F43H Mb, and L29E/F43H Mb were constructed by using the QuickChange Site Directed Mutagenesis Kit (Stratagene). The mutations were confirmed by DNA sequencing. L29E Mb and L29E/F43H Mb were expressed in inclusion bodies and purified using a procedure described previously.12a F43H Mb was expressed and purified using the same procedure as that for WT Mb.
X-ray crystallography: L29E Mb, F43H Mb and L29E/F43H Mb with a high purity were exchanged into 20 mm KH2PO4 (pH 7.0) and concentrated to ∼1.5 mm. The vapor diffusion hanging drop technique was used to crystallize the protein. The well buffer contained 0.2 m sodium acetate trihydrate, 0.1 m sodium cacodylate trihydrate pH 6.5, and 30 % w/v polyethylene glycol (PEG) 8 000. Crystal trays were set up by transferring well buffer (250 μL) into each well. Then well buffer (2 μL) and protein (2 μL) were mixed and placed on a siliconized glass slide. Crystallization was achieved at 10 °C after ∼3 weeks. Diffractable crystals were soaked in a cryoprotectant solution of 30 % PEG 400, mounted onto cryogenic loops, and frozen quickly in liquid nitrogen. Diffraction data were collected from a single crystal at the Shanghai Synchrotron Radiation Facility (SSRF) BL17U beamline, China, using a MAR mosaic 225 CCD detector with a wavelength of 0.9793 Å at 100 K. The diffraction data were processed and scaled with HKL-2000.19 The structure was solved by the molecular replacement method, and the 1.6 Å structure of WT Mb (PDB entry 1 JP63) was used as the starting model. Manual adjustment of the model was carried out using the program COOT,20 and the models were refined by PHENIX21 and Refmac5.22 Stereochemical quality of the structures was also checked by using PROCHECK.23 All residues locate in the favored and allowed region and none in the disallowed region (Table S1 in the Supporting Information).
Fluoride binding kinetics: Fluoride binding to the heme center of WT Mb, L29E Mb, and L29E/F43H Mb was carried out with a SF-61DX2 Hi-Tech KinetAsyst™ dual mixing stopped-flow spectrophotometer (TgK Scientific, Bradford-on-Avon, UK). The binding kinetics was measured by mixing the protein (10 μm in 100 mm KH2PO4 buffer, pH 7.0) in one syringe with increasing concentrations of NaF (0.1–0.4 m) in the second syringe, with an equal volume of solutions. The observed rate constant (kobs) was obtained by fitting the change in the protein Soret band intensity to a mono-exponential decay equation. The association rate constant, ka (kon), and the dissociation rate constant, kd (koff), were determined from a plot of kobs versus the fluoride concentration,13c i.e., kobs=kon [F−] + koff, where the slope and intercept correspond to kon and koff, respectively. Protein concentration was determined with an extinction coefficient of ɛ409=157 mm−1 cm−1 for WT Mb,18 ɛ413=135 mm−1 cm−1 for L29E Mb, and ɛ415=135 mm−1 cm−1 for L29E/F43H Mb.
Peroxidase reaction kinetics: Kinetic determinants for the reactions of WT Mb, L29E Mb, and L29E/F43H Mb with H2O2 were performed with a SF-61DX2 Hi-Tech KinetAsyst™ dual mixing stopped-flow spectrophotometer, similar to that for the fluoride binding studies. Typically, one syringe contains 10 μm of protein (in 100 mm KH2PO4 buffer, pH 7.0), and the second syringe contains H2O2 with concentrations ranging from 0.25 to 2 mm. The reaction was started by mixing equal volumes of solutions from the both syringes. 200 time-dependent spectra were collected over 5 s from 350 to 700 nm at 25 °C. The apparent rate constant, k1, was obtained by fitting the plot of the observed rate constants, kobs, versus the concentration of H2O2 to a linear regression model.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC) (No. 31370812 to Y.-W. Lin and No. 31270869 to X. Tan). The authors gratefully thank Prof. Stephen G. Sligar and Dr. Yi Lu at the University of Illinois at Urbana-Champaign, USA, for providing the Mb gene. X-ray diffraction data were collected at the Shanghai Synchrotron Radiation Facility (SSRF), China.
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
References
- 1a.Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1b.Coelho PS, Brustad EM, Kannan A, Arnold FH. Science. 2013;339:307–310. doi: 10.1126/science.1231434. [DOI] [PubMed] [Google Scholar]
- 1c.Spiro TG, Soldatova AV, Balakrishnan G. Coord. Chem. Rev. 2013;257:511–527. doi: 10.1016/j.ccr.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1d.Lin Y-W, Sawyer EB, Wang J. Chem. Asian J. 2013;8:2534–2544. doi: 10.1002/asia.201300291. [DOI] [PubMed] [Google Scholar]
- 1e.Lin Y-W, Wang J. J. Inorg. Biochem. 2013;129:162–171. doi: 10.1016/j.jinorgbio.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 1f.Zhong F, Lisi GP, Collins DP, Dawson JH, Pletneva EV. Proc. Natl. Acad. Sci. USA. 2014;111:306–315. doi: 10.1073/pnas.1317173111. , E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1g.Poulos TL. Chem. Rev. 2014;114:3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Wang Z-H, Lin Y-W, Rosell FI, Ni F-Y, Lu H-J, Yang P-Y, Tan X-S, Li X-Y, Huang Z-X, Mauk AG. ChemBioChem. 2007;8:607–609. doi: 10.1002/cbic.200600547. [DOI] [PubMed] [Google Scholar]
- 2b.Wang Z, Matsuo T, Nagao S, Hirota S. Org. Biomol. Chem. 2011;9:4766–4769. doi: 10.1039/c1ob05552f. [DOI] [PubMed] [Google Scholar]
- 3.Urayama P, Phillips GN, Jr, Gruner SM. Structure. 2002;10:51–60. doi: 10.1016/s0969-2126(01)00699-2. . [DOI] [PubMed] [Google Scholar]
- 4a.Burmester T, Ebner B, Weich B, Hankeln T. Mol. Biol. Evol. 2002;19:416–421. doi: 10.1093/oxfordjournals.molbev.a004096. [DOI] [PubMed] [Google Scholar]
- 4b.Sugimoto H, Kaino M, Sawai H, Kawada N, Yoshizato K, Shiro Y. J. Mol. Biol. 2004;339:873–885. doi: 10.1016/j.jmb.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 5a.Burmester T, Weich B, Reinhardt S, Hankeln T. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 5b.Guimarães RG, Hamdane D, Lechauve C, Marden MC, Golinelli-Pimpaneau B. Acta. Cryst. Sect. D. 2014;70:1005–1014. doi: 10.1107/S1399004714000078. [DOI] [PubMed] [Google Scholar]
- 6.Hargrove MS, Brucker EA, Stec B, Sarath G, Arredondo-Peter R, Klucas RV, Olson JS, Phillips GN., Jr Structure Fold. Des. 2000;8:1005–1014. doi: 10.1016/s0969-2126(00)00194-5. . [DOI] [PubMed] [Google Scholar]
- 7a.Kiger L, Uzan J, Dewilde S, Burmester T, Hankeln T, Moens L, Hamdane D, Baudin-Creuza V, Marden M. IUBMB Life. 2004;56:709–719. doi: 10.1080/15216540500037711. [DOI] [PubMed] [Google Scholar]
- 7b.Sturms R, DiSpirito AA, Hargrove MS. Biochemistry. 2011;50:3873–3878. doi: 10.1021/bi2004312. [DOI] [PubMed] [Google Scholar]
- 8a.Ozaki S, Matsui T, Roach MP, Watanabe Y. Coord. Chem. Rev. 2000;198:39–59. [Google Scholar]
- 8b.Köhler V, Ward TR. ChemBioChem. 2010;11:1049–1051. doi: 10.1002/cbic.201000093. [DOI] [PubMed] [Google Scholar]
- 8c.Ueno T. Angew. Chem. Int. Ed. 2010;49:3868–3869. doi: 10.1002/anie.201000337. [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2010;122 [Google Scholar]
- 8d.Oohora K, Hayashi T. Curr. Opin. Chem. Biol. 2014;19:154–161. doi: 10.1016/j.cbpa.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 8e.Lin Y-W, Wang J, Lu Y. Sci. China Chem. 2014;57:346–355. [Google Scholar]
- 9.Dou Y, Admiraal SJ, Ikeda-Saito M, Krzywda S, Wilkinson AJ, Li T, Olson JS, Prince RC, Pickering IJ, George GN. J. Biol. Chem. 1995;270:15993–16001. doi: 10.1074/jbc.270.27.15993. [DOI] [PubMed] [Google Scholar]
- 10.Santoni E, Jakopitsch C, Obinger C, Smulevich G. Biochemistry. 2004;43:5792–5802. doi: 10.1021/bi035835b. [DOI] [PubMed] [Google Scholar]
- 11.Redaelli C, Monzani E, Santagostini L, Casella L, Sanangelantoni AM, Pierattelli R, Banci L. ChemBioChem. 2002;3:226–233. doi: 10.1002/1439-7633(20020301)3:2/3<226::AID-CBIC226>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 12a.Lin Y-W, Yeung N, Gao Y-G, Miner KD, Lei L, Robinson H, Lu Y. J. Am. Chem. Soc. 2010;132:9970–9972. doi: 10.1021/ja103516n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b.Lin Y-W. Proteins Struct. Funct. Bioinf. 2011;79:679–684. doi: 10.1002/prot.22928. [DOI] [PubMed] [Google Scholar]
- 13a.Aime S, Fasano M, Paoletti S, Cutruzzolà F, Desideri A, Bolognesi M, Rizzi M, Ascenzi P. Biophys. J. 1996;70:482–488. doi: 10.1016/S0006-3495(96)79593-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13b.Neri F, Kok D, Miller MA, Smulevich G. Biochemistry. 1997;36:8947–8953. doi: 10.1021/bi970248+. [DOI] [PubMed] [Google Scholar]
- 13c.Merryweather J, Summers F, Vitello LB, Erman JE. Arch. Biochem. Biophys. 1998;358:359–368. doi: 10.1006/abbi.1998.0872. [DOI] [PubMed] [Google Scholar]
- 13d.Nicoletti FP, Droghetti E, Boechi L, Bonamore A, Sciamanna N, Estrin DA, Feis A, Boffi A, Smulevich G. J. Am. Chem. Soc. 2011;133:20970–20980. doi: 10.1021/ja209312k. [DOI] [PubMed] [Google Scholar]
- 13e.Droghetti E, Nicoletti FP, Bonamore A, Sciamanna N, Boffi A, Feis A, Smulevich G. J. Inorg. Biochem. 2011;105:1338–1343. doi: 10.1016/j.jinorgbio.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 13f.Kosowicz JG, Boon EM. J. Inorg. Biochem. 2013;126:91–95. doi: 10.1016/j.jinorgbio.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Matsui T, Ozaki S, Liong E, Phillips GN, Jr, Watanabe Y. J. Biol. Chem. 1999;274:2838–2844. doi: 10.1074/jbc.274.5.2838. . [DOI] [PubMed] [Google Scholar]
- 15.Sundaramoorthy M, Termer J, Poulos TL. Structure. 1995;3:1367–1377. doi: 10.1016/s0969-2126(01)00274-x. [DOI] [PubMed] [Google Scholar]
- 16.Finzel BC, Poulos TL, Kraut J. J. Biol. Chem. 1984;259:13027–13036. [PubMed] [Google Scholar]
- 17.Gajhede M, Schuller DJ, Henriksen A, Smithm AT, Poulos TL. Nat. Struct. Biol. 1997;4:1032–1038. doi: 10.1038/nsb1297-1032. [DOI] [PubMed] [Google Scholar]
- 18.Springer A, Sligar SG. Proc. Natl. Acad. Sci. USA. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Acta Crystallogr. Sect. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Crystallogr. Sect. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. Sect. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary


