Abstract
Background and Purpose
Moyamoya disease (MMD) is a rare, genetically heterogeneous cerebrovascular disease resulting from occlusion of the distal internal carotid arteries. A variant in the Ring Finger 213 gene (RNF213), altering arginine at position 4810 (p.R4810K), is associated with MMD in Asian populations. However, there is a lack of data on the role of RNF213 in MMD patients of additional ethnicities and diasporic Asian populations. We investigate the contribution of RNF213 alterations to MMD in an ethnically diverse population based in the United States (U.S).
Methods
We initially sequenced RNF213 exons 43, 44, 45 (encoding the eponymous RING finger domain), and exon 60 (encoding p.R4810K), in 86 ethnically diverse patients with MMD. Comprehensive exome sequencing data from 24 additional MMD patients was then analyzed to globally identify RNF213 variants. Segregation of variants with MMD and other vascular diseases was assessed in families.
Results
RNF213 p.R4810K was identified in 56% (9/16) of MMD patients of Asian descent, and not in 94 patients of non-Asian descent. 3.6% (4/110) of patients had variants in the exons encoding the RING finger domain. Seven additional variants were identified in 29% (7/24) of MMD patients who underwent exome sequencing. Segregation analysis supported an association with MMD for two variants, and a lack of association with disease for one variant.
Conclusions
These results confirm that alterations in RNF213 predispose patients of diverse ethnicities to MMD, and that the p.R4810K variant predisposes individuals of Asian descent in the U.S. to MMD.
Keywords: moyamoya disease, stroke, RNF213, genetic, rare genetic variants
INTRODUCTION
Moyamoya disease (MMD) is a vasculopathy characterized by progressive stenosis of the internal carotid arteries and its proximal branches accompanied by the development of a compensatory collateral vessel network.1 Affected individuals often present with strokes, transient ischemic attacks (TIA), and intracerebral hemorrhage.1 There are two incidence peaks for MMD, one in children around 5 years of age and another in adults in their mid-40s.1, 2 Patients with MMD present with a variety of comorbidities, including headaches, migraines, seizures, and cognitive impairment. Medical treatments to reverse or inhibit the progression of the arterial occlusion are not currently available. Therefore, neurosurgical intervention in the form of direct and indirect revascularization procedures to reduce the risk for strokes has remained the mainstay treatment for these patients.1 Epidemiological data have shown a higher incidence and prevalence of MMD in Japan.2, 3
MMD occurs in two or more members of a family in approximately 9–15% of cases, which supports a genetic basis to disease predisposition.4 Pedigree analysis of these families with MMD suggests an autosomal dominant inheritance pattern with reduced penetrance.5 A strong association between a RNF213 variant (p.R4810K) and MMD has been demonstrated in Japanese, Korean, and Chinese patients, but not in individuals of European descent.6 The RNF213 p.R4810K variant has been shown to segregate with disease in Asian families and predominantly demonstrates an autosomal dominant inheritance pattern with reduced penetrance.7 In addition to the p.R4810K variant found in East Asian patients, other RNF213 variants have been identified in both East Asian and Europeans (MMD patients from Germany and Czechoslovakia). Interestingly, the frequency of RNF213 variants in Europeans is lower than in Asian patients and RNF213 p.R4810K was not identified in Europeans.7
Since MMD occurs in diverse ethnic populations, we sought to examine the prevalence of RNF213 genetic variants in a multi-ethnic cohort of MMD patients from the U.S. In this study, we identified the RNF213 p.R4810K variant in MMD patients of Asian descent and confirm that RNF213 rare variants are associated with MMD in patients of non-Asian descent. Interestingly, RNF213 rare variants in non-Asian families also segregate with early onset occlusive diseases, such as coronary artery disease (CAD), but additional studies are needed to confirm that RNF213 variants also predispose to the additional vascular diseases.
MATERIALS AND METHODS
Study population and sample collection
This study was approved by the University of Texas Health Science Center at Houston Institutional Review Board and informed consent was obtained from study participants. A total of 110 families who had at least one family member diagnosed with MMD were recruited and/or referred to an ongoing research study at UTHealth between 2007 and 2013. Demographic data, vascular disease presentation, radiologic findings, and surgical and clinical histories were abstracted from the patient medical records when available, or obtained by patient self-report. Three generation family histories were obtained via interviews conducted by a genetic counselor or medical student. Consenting relatives of the affected probands with MMD were recruited into the study if available. In addition to MMD, diagnoses of other vascular diseases such as stroke, myocardial infarction, stenosis of other arteries, arterial aneurysms and dissections, and congenital defects affecting the heart or other vascular structures (e.g., aortic coarctation) were recorded for probands and their family members. Medical records documenting disease and risk factor status were obtained when available.
Diagnosis of MMD was based on magnetic resonance (MR), computed tomography (CT), or diagnostic angiogram findings demonstrating stenosis or occlusion of the terminal portion of the internal carotid artery with the formation of collateral vessels compensating for the arterial occlusion. Patients diagnosed with both unilateral and bilateral MMD were included in this study. Exclusion of other causes of arterial occlusion such as atherosclerosis was completed via medical record and imaging review. Premature coronary artery disease (CAD) and stroke were defined as onset of disease at the age of 55 years or younger in men, and 60 years or younger in women. Individuals of all ethnicities diagnosed with MMD at any age were included in this study. Syndromic cases of MMD (Moyamoya syndrome) and those with another established genetic cause for their MMD (e.g., ACTA2 mutations) were excluded.
Blood or buccal cells were collected for DNA extraction. Mutation status of individuals was determined by DNA sequencing or inferred based on their location in the pedigree.
DNA sequencing
Based on Ensembl, RNF213 has 5 splice variants and the longest isoform is NM_001256071, which includes 67 coding exons encoding 5207 amino acids8. Bidirectional Sanger sequencing of exons 43–45 along with exon 60 was performed to identify rare RNF213 functional variants, and to confirm the rare variants identified by exome sequencing. PCR and sequencing primers were designed 60–120 base pairs from the intron-exon boundaries. Polymerase chain reaction (PCR) was performed using HotStar Taq™ DNA polymerase (Qiagen Inc., CA). PCR products were treated with EXOSAP-IT (Affymetrix, Inc., OH) to digest the primers and subsequently sequenced using the BigDye™ chemistry (Applied Biosystems, CA). The sequencing product was purified using BigDye XTerminator (Applied Biosystems, CA) and then loaded on an ABI3730xl sequencing instrument using the Rapid36 run module. Sequencing results were analyzed using Mutation Surveyor software (SoftGenetics, PA).9
Exome sequencing
One microgram of barcoded shotgun library was hybridized for capture of probes targeting 64 Mb of coding exons (Roche/NimbleGen SeqCap EZ Cap v2) according to the manufacturer’s protocol, and custom blockers complimentary to the full length of the flanking adaptor and barcodes were added. Enriched libraries were amplified via PCR before sequencing (BioRad iProof). Pooled, barcoded libraries were sequenced via paired-end 50 bp reads with an 8 bp barcode read on Illumina HiSeq sequencers. Read data from a flow-cell lane were treated independently for alignment and quality-control purposes in instances where the merging of data from multiple lanes was required. Variant detection and genotyping were performed with the UnifiedGenotyper tool from GATK (v.1.529). Variant data for each sample were formatted as “raw” calls that contained individual genotype data for one or multiple samples and were flagged with the filtration walker (GATK) for marking sites that were of lower quality and potential false positives.
Exome analysis was performed using the Variant Association Tools platform10, with prioritization based on segregation of rare, damaging variants with disease in families. Additional analyses were also performed using SNP & Variation Suite v8.0.1 (Golden Helix, Bozeman, MT). Heterozygous variants which potentially altered amino acids and were observed at a minor allele frequency (MAF) less than 0.03% in the NHLBI Exome Sequencing Project were considered candidate mutations. The MAF for each variant was also checked in 1000 Genomes (1000genomes.org). For each variant, conservation scores at nucleotide residues were derived from the UCSC Genome Bioinformatics website (genome.ucsc.edu), and additional bioinformatics analyses were performed using CADD, MutationAssessor, MutationTaster, SIFT, PolyPhen2, and PROVEAN11–16.
Statistics
The segregation of the RNF213 p.D4013N variant in family MMD096 was assessed by two-point linkage analysis and was performed using the MLINK program of the FASTLINK package.17, 18 All individuals with MMD were designated as affected. Additional linkage analyses were performed designating individuals with MMD, premature onset CAD, stroke, or subarachnoid hemorrhage (SAH) as affected. All linkage analyses were done using affected individuals only, an autosomal dominant mode of inheritance, and a disease allele frequency of 0.0001. For each model, simulation analyses were performed using the MSIM of the SLINK package to obtain the expected maximum LOD score (EMLOD) given the pedigree structure, affection status, and availability of genotype data19.
RESULTS
Demographic and clinical description of MMD patients
In this study, DNA from 110 probands with MMD underwent targeted sequencing of RNF213 or whole exome sequencing (Table 1). All affected individuals who were sequenced had diagnoses of MMD of unknown genetic etiology. Of the 110 total probands, the 24 affected individuals who underwent exome sequencing were more likely to be early-onset cases with unaffected parents, familial MMD probands, or probands with MMD and other comorbid vascular diseases. The median age of diagnosis of the MMD probands was 28 years, with a range of 9 months to 59 years and the average age of diagnosis was 26.7 years. Approximately 75.5% of the MMD cohort was female (83/110). Of the patients whose medical records documented the laterality of their MMD, 73.6% had bilateral involvement (78/106). The majority of the MMD families were of European descent (74.5%; 82/110), but Hispanic, African American, and Korean patients each made up 5.5% of the cohort. The remainder of the cohort was composed of individuals from a variety of other Asian ethnicities.
Table 1.
Demographic characteristics
| Variable | Number (Range or percentage) |
|---|---|
| Median Age (IQR) | 28 (9 – 40) |
| Sex | |
| Male | 27 (24.5%) |
| Female | 83 (75.5%) |
| Ethnicity | |
| European American | 82 (74.5%) |
| Hispanic | 6 (5.5%) |
| African American | 6 (5.5%) |
| Korean | 6 (5.5%) |
| Japanese | 2 (1.8%) |
| Filipino | 2 (1.8%) |
| Indian | 1 (0.9%) |
| Bangladeshi | 1 (0.9%) |
| Chinese | 1 (0.9%) |
| Pakistani | 1 (0.9%) |
| Vietnamese | 1 (0.9%) |
| Japanese/Filipino | 1 (0.9%) |
| Laterality | |
| Bilateral | 78 (70.9%) |
| Unilateral | 28 (25.5%) |
| Unknown | 4 (3.6%) |
| n=110 | |
Identification of RNF213 variants in patients with MMD
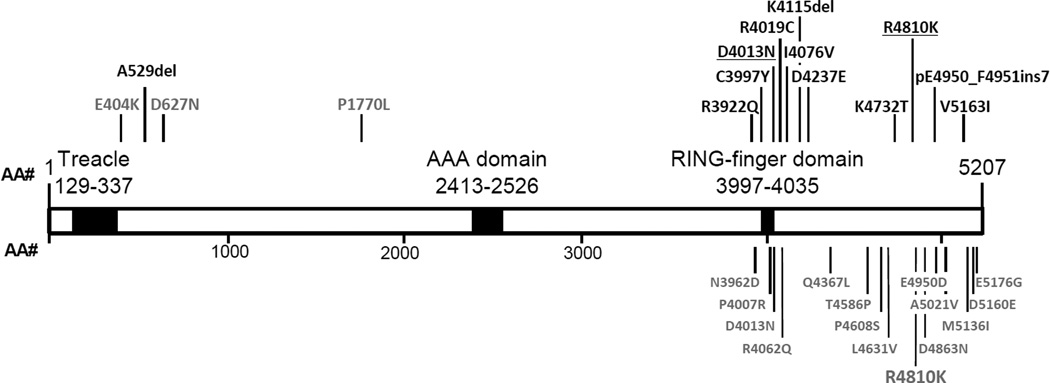
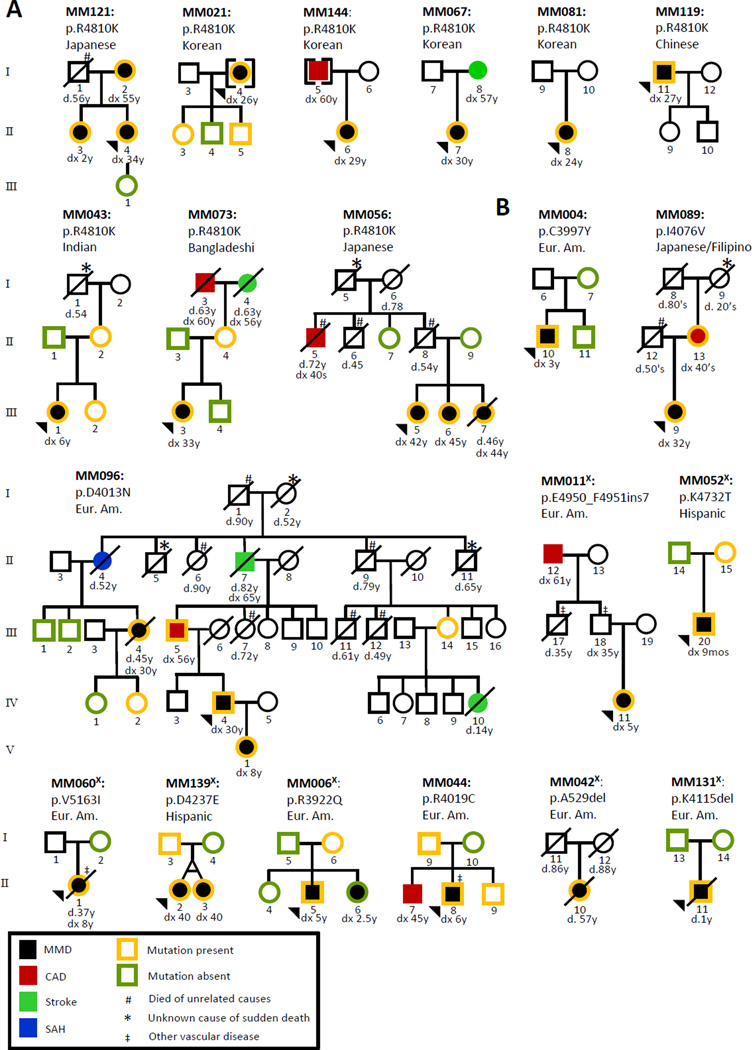
The RNF213 variant previously associated with MMD in Asian patients, p.R4810K, is located in exon 60 and the four possibly disease-causing variants (p.N3962, p.D4013N, p.R4062, and p.P4608S) identified in European MMD patients are located in exons 43–45, which encodes the RING finger domain (one of three functional domains identified in RNF213).7 Therefore, exons 43, 44, 45 and 60, along with the flanking introns, were initially sequenced in 86 MMD probands to determine if the RNF213 p.R4810K Asian founder mutation and rare variants in the RING finger domain were present in this U.S. based cohort. RNF213 p.R4810K was identified in 56% (9/16) of the unrelated MMD families of Asian descent, but was not identified in any European American or Hispanic families. RNF213 p.R4810K segregated with MMD in 2 of the 9 families with this variant (MM121, MM056; Figure 1, Figure 2A, Table 2) but was not fully penetrant, as previously reported. This variant was also identified in novel groups not previously reported. These included patients of Bangladeshi, Indian, and Filipino origin (Table 3). Four rare RNF213 variants located in and around the RING finger domain were identified via Sanger sequencing in this cohort (p.C3997Y, p.I4076V, p.D4013N, p.R4019C; Table 2, Figure 1, Figure 2). These rare variants were novel based on an Ensembl search or had a minor allele frequency (MAF) less than 0.03% in the Exome Variant Server database (EVS; evs.gs.washington.edu/EVS/).
Figure 1. RNF213 Alterations in MMD patients.
The 12 alterations identified in MMD patients in this study are shown in black above the schematic clustering at the C-terminus of RNF213 protein, with one alteration at the N-terminus. Three alterations identified in probands with familial TAAD are shown in grey above the schematic at the N-terminus of RNF213 protein. Alterations reported by other groups are shown in gray below the protein.6, 7 Known protein domains are indicated in black.
Figure 2. Segregation of RNF213 alterations with MMD and other vascular diseases in MMD families.
A. Pedigrees of 9 families of various Asian ethnicities with the RNF213 p.R4810K variant. The variant segregates with MMD in 2 families (MM121 and MM056). Reduced penetrance for MMD in individuals with the RNF213 p.R4810K is also demonstrated. B. Pedigrees of 11 families with other RNF213 rare variants. The p.D4013N alteration segregates with MMD, premature CAD, stroke and presentation with subarachnoid hemorrhage (SAH) in a large European American family (MM096). The p.R3922Q variant does not segregate with MMD in MM006. Disease and mutation status are indicated in the figure key. Circles represent females, squares represent males, and arrowheads indicate the proband in the family. A diagonal line through a circle or square indicates that the individual is deceased, with their age of death shown below. Age at onset of the vascular disease (dx) is shown below each individual when applicable; each disease is coded and correlates with the figure key. Ethnicities are indicated above each pedigree. The superscript X (X) indicates families in which RNF213 variants were identified through exome sequencing.
Table 2.
RNF213 rare variants identified and predicted impact of amino acid substitutions
| Aa Alt | A529delx | R3922Qx | C3997Y | D4013N | R4019C | I4076V | K4115delx | D4237Ex | K4732Tx | R4810K | E4950_F4951ins7x | V5163Ix |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs ID | 397514563 | 139265462 | 148776624 | 112735431 | 201733659 | |||||||
| position (chr17:) | 78268634 | 78338247 | 78341778 | 78341825 | 78341843 | 78346494 | 78343583 | 78357601 | 78343372 | 78358945 | 78360605 | 78367161 |
| PhastConsa | 0.992 | 1 | 1 | 0 | 0.843 | 0 | 0.228 | 0 | 0.008 | |||
| GERPb | 3.22 | 4.54 | 4.67 | −6.62 | 4.32 | −3.21 | −1.11 | 2.04 | 2.47 | |||
| MutationTaster | Pd | Pd | Dc | Pd | Pd | Pd | Dc | Pd | Pd | Pd | Dc | Pd |
| Polyphen2_HVAR | Benign | Pro De | Benign | Pos Df | Pos Df | Benign | Benign | Benign | Benign | |||
| SIFT | Damaging | Tolerated | Damaging | Tolerated | Damaging | Damaging | Damaging | Damaging | Damaging | |||
| MutationAssessor | Low | High | Low | Medium | Medium | Low | Neutral | Medium | Medium | |||
| Cscore_PHREDg | 10.59 | 14.8 | 16.19 | 19.28 | 9.963 | 5.689 | 8.108 | 6.746 | 1.25 | |||
| MAF_EVS-EAh | 0 | 0 | 0 | 0 | 0.00093 | 0 | 0 | 0.000581 | 0 | 0 | 0 | 0.000116 |
| MAF_EVS-AAh | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1kG_AA&EURi | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1kG_ASN_AFi | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | phase_1 PTj=0.011 |
0 | 0 |
Conservation;
Conservation;
Disease-causing;
Polymorphism;
Probably damaging;
Possibly damaging;
see reference11;
Minor allele frequencies in European American and African American cohorts in the NHLBI Exome Sequencing Project;
Minor allele frequencies in 1000 Genome project;
Japanese cohort in phase 1 of 1000 Genomes project;
Indicates families in which RNF213 variants were identified through exome sequencing
Table 3.
Clinical Characteristics of MMD patients with RNF213 Rare Variants
| Family Number |
Individual ID Number |
RNF213 Alteration |
Ethnicity | Age at Diagnosisa |
Sex (M/F) |
Lateralityb | Surgical Procedurec |
|---|---|---|---|---|---|---|---|
| MM121 | II-4 | p.R4810K | Japanese | 35 | F | Bilateral | Y; 35 |
| II-3 | p.R4810K | Japanese | 2 | F | Unilateral | N | |
| II-2 | p.R4810K | Japanese | 55 | F | Unilateral | N | |
| MM021 | I-4 | p.R4810K | Korean | 26 | F | Bilateral | Y; 26 |
| MM144 | II-6 | p.R4810K | Korean | 29 | F | Unilateral | Y; 30 |
| MM067 | II-7 | p.R4810K | Korean | 30 | F | Bilateral | Y; 31 |
| MM081 | II-8 | p.R4810K | Korean | 24 | F | Unilateral | Y; 25 |
| MM119 | I-11 | p.R4810K | Chinese | 27 | M | Bilateral | Y; 30 |
| MM043 | III-1 | p.R4810K | Indian | 6 | F | Bilateral | Y; 6 |
| MM073 | III-3 | p.R4810K | Bangladeshi | 33 | F | Bilateral | Y; 34 |
| MM056 | III-5 | p.R4810K | Japanese | 42 | F | Bilateral | Y; 44 |
| III-6 | p.R4810K | Japanese | 45 | F | Unilateral | Y; 45 | |
| III-7 | p.R4810K | Japanese | 44 | F | Unknown | Y; 45 | |
| MM004 | II-10 | p. R4019C | European American | 3 | M | Bilateral | Y; 3 |
| MM089 | III-9 | p. I4076V | Japanese/Filipino | 32 | F | Bilateral | Y; 33 |
| MM096 | IV-4 | p.D4013N | European American | 30 | M | Bilateral | Y; unk |
| V-1 | p.D4013N | European American | 8 | M | Bilateral | Y; 8 | |
| III-4 | p.D4013N | European American | 30 | F | Unknown | Y; 45 | |
| MM011x | IV-11 | p.E4950_F4951ins7 | European American | 5 | F | Bilateral | Y; 5 |
| MM052x | II-1 | p.K4732T | Hispanic | 9 mos | M | Bilateral | Y; 10 mos |
| MM060x | II-2 | p.V5163I | European American | 8 | F | Bilateral | Y; 36 |
| MM139x | II-3 | p.D4237E | Hispanic | 40 | F | Bilateral | Y; 41 |
| II-4 | p.D4237E | Hispanic | 40 | F | Bilateral | Y; 40 | |
| MM006x | II-6 | p.R3922Q | European American | 5 | M | Bilateral | Y; 5 |
| II-7 | p.R3922Q | European American | 2.5 | F | Bilateral | Y; 2.5 | |
| MM044 | II-9 | p.R4019C | European American | 6 | M | Bilateral | Y; 7 |
| MM042X | II-10 | p.A529del | European American | 50 | F | Bilateral | Y;51 |
| MM131X | II-11 | p.K4115del | European American | 10 mos | M | Bilateral | Y;1 |
Age of MMD onset shown in years unless otherwise indicated;
Laterality of MMD involvement;
Denotes whether a revascularization procedure has been performed for each patient and at what age (in years);
Indicates families in which RNF213 variants were identified through exome sequencing.
To determine whether other RNF213 rare variants were present in MMD patients apart from exons that were sequenced, exome sequencing was performed on 36 individuals from 24 unrelated families, including 24 individuals with MMD and 12 unaffected parents of childhood onset MMD patients. Gene variants identified via exome sequencing were filtered, and only rare variants that changed the amino acid sequence with a minor allele frequency (MAF) less than 0.03% in Exome Variant Server database (evs.gs.washington.edu/EVS/) were pursued for further investigation. The MAF threshold was set at 0.03% because we sought to identify RNF213 rare variants that conferred a significant risk for disease and expected to identify variants that are uncommon in the general population, since MMD is a rare condition. Seven RNF213 rare variants were identified via exome analysis in 7 unrelated families, including a 21 bp in-frame insertion starting at amino acid 4951, p.R3922Q, p.D4237E, p.K4732T, p.V5163I, and two 3 bp deletions, p.A529del and p.K4115del (Table 2, Table 3). All variants were confirmed by Sanger sequencing. Rare variants in the EVS database were spread evenly throughout RNF213 gene, while rare variants identified in our cohort were all located in exons 42 through 68, which encodes the C-terminus of RNF213, with a single exception, the p.A529del variant (Figure 1). We have also performed exome sequencing analysis on 86 unrelated probands with thoracic aortic aneurysms and dissections (TAAD) and only identified three rare RNF213 variants (Figure 1; unpublished data). All three rare variants found in the TAAD patients were located in the exons encoding the N-terminus of RNF213 protein. Furthermore, rare RNF213 variants were found at a significantly higher frequency in the MMD cohort compared with our TAAD cohort (p-value < 0.05).
RNF213 rare variants were identified in 8 out of 82 European Americans and in 2 out of 6 Hispanic families. Only one of these rare variants had been previously identified in a MMD patient7, and eight of these variants are novel and not present in exome databases (Table 2). Six of the eight missense variants are predicted to be possibly damaging or damaging by 2 out of 4 functional prediction programs. A family of Asian descent (Japanese/Filipino) was found to have a RNF213 variant different than the p.R4810K founder mutation. A novel 3 bp deletion (p.K4115del) was confirmed to be de novo in a child with severe and early onset MMD (MM131; Figure 2B). Segregation of the RNF213 rare variant with MMD was confirmed in 3 out of 20 families and decreased penetrance for MMD was noted in 5 families (Figure 2B). Monozygotic twins with the p.D4237E alteration were both diagnosed with MMD at 40 years (MM139; Figure 2B). One twin presented to medical attention with transient ischemic attacks and imaging revealed several ischemic infarcts and bilateral MMD. This diagnosis prompted cerebrovascular screening in the other twin, which revealed bilateral MMD; both underwent revascularization procedures. Only one variant, p.R3922Q variant, was found to be discordant with the disease status in a family (MM006; Figure 2B).
RNF213 variants and other vascular diseases
Two of the MMD probands with RNF213 rare variants presented with comorbid vascular diseases (Figure 2B). The proband in MM060 was diagnosed with coarctation of the aorta at 6 years old, thoracic aortic disease at 35 years old, and had a history of multiple intracranial aneurysms (IA). In MM044, the proband also had unilateral renal artery stenosis.
While MMD was the primary disease identified in the RNF213 alteration carriers, other vascular diseases occurred in members of half the families, including premature CAD and stroke, subarachnoid hemorrhage (SAH), aortic coarctation, thoracic aortic aneurysm (TAA), and stenosis of other arteries. Premature CAD and stroke were seen in 4 of the 9 Asian families (44.5%) who carry the p.R4810K mutation (MM144, MM067, MM073, MM056); segregation analysis was not possible in these families due to the unavailability of familial samples (Figure 2A).
Additional and diverse vascular diseases were also present in families with other RNF213 variants. The most informative family for assessing vascular diseases in family members with a RNF213 variant was MM096, who carried a previously reported RNF213 variant for MMD, p.D4013N.7 This family had three family members who presented with MMD, but individuals with the variant also presented with early onset CAD and stroke, and SAH (Figure 2B). Additionally, a 14 year-old girl who was at risk for inheriting the p.D4013N variant died suddenly of a stroke without MMD being previously diagnosed. For two point linkage analysis using only family members affected with MMD, the observed LOD is equal to EMLOD at 1.20. For analysis using all family members affected with all vascular diseases, the observed LOD score was 1.81, which is close to the EMLOD of 2.08. Therefore, the observed LOD score obtained using only affected individuals in family MM096 only is equal to or close to EMLOD, indicating that the RNF213 variant is likely to be disease-causing.
Approximately 45% (5/11) of the families with the non-Asian founder variants had other vascular diseases (MM089, MM096, MM011, MM060, MM044; Figure 2B). The mother of the proband in MM089 also has the novel p.I4076V variant and was diagnosed with CAD in her 40’s. Although a sample for segregation analysis in MM011 was not available, the proband’s father was diagnosed with unilateral carotid artery stenosis and had a paternal uncle who died from complications secondary to pulmonary artery stenosis at 35 years of age.
DISCUSSION
The results of this study indicate that the Asian founder mutation in RNF213, p.R4810K, is also present in Asian American MMD patients and segregates with disease in these families in an autosomal dominant manner with reduced penetrance. It is notable that the RNF213 p.R4810K variant was not identified in MMD patients of European, Hispanic, or African American descent. However, other rare variants in RNF213 were identified in European and Hispanic American populations. Of the 24 MMD probands who underwent exome sequencing, 22 were of European American descent and 2 were Hispanic, and RNF213 variants were identified in 5 out of the 22 European American families (23%) and in both of the Hispanic American probands. Thus, the data presented here confirms that the Asian founder mutation, p.R4810K, is a major predisposing allele in Asian Americans, encompassing individuals from Japan, China, Korea, Philippines, India, and Bangladesh. In addition, these data provide novel evidence that novel RNF213 variants are present in a substantial proportion of MMD in patients of European and Hispanic descent, and strong genetic data indicated that two of these variants are disease causing. A novel de novo p.K4115del was identified in an affected individual with severe, early-onset MMD in family MM131, and segregation of a previously identified RNF213 variant, p.D4013N, with disease was confirmed in family MM096.
Although limited segregation could be done in the families with novel RNF213 variants, there is evidence suggesting that many of these variants are disease causing. First, all but one of these variants are located in the C-terminus of the RNF213 protein, which is where the RNF213 p.R4810K founder variant is located, and also where other variants have been identified in MMD patients.6, 7 The variants are either not present, or present at extremely low frequencies in the NHLBI Exome Database (EVS database). The only variant with evidence that it may not be disease-causing is p.R3922Q, which does not segregate with disease in the family. It is interesting to note that all of the RNF213 variants identified in MMD patients to date are predicted to produce a mutant protein, i.e., no frameshift or nonsense mutations predicted to lead to degradation of the message have been identified.
In comparison to our data (Table 2), the EVS database reports 397 variants in RNF213. 284 of these (71.5%) have a MAF less than 0.03%, including 11 stop-gain, 4 frame-shift, 3 splice site, and 266 missense variants. PolyPhen-2 analysis suggests that 125 of these missense variants are benign, 55 are possibly damaging, 81 are probably damaging, and 5 have unknown effects. 185 of the 397 rare variants (46.6%) are located in exons 2–41 and 99 (24.9%) are located in exons 42–68. In addition, one frame-shift variant, c.2735del1, has a MAF of 0.59% in ESP database but this variant is likely to be a false positive because all of the 37 individuals who carry this variant are homozygous for the deletion. In the 1000 Genomes database, there are 141 missense variants and one stop-gain variant (MAF 0.05%) in RNF213 and the average overall MAF for these variants is 3.76%. Ninety-three missense variants have MAF less than 1%. Among all missense variants, 98 are predicted to be benign, 15 possibly damaging, and 24 probably damaging based on PolyPhen-2 analysis.
Comprehensive family history of vascular diseases, including early onset and unusual vascular diseases, was collected for all patients and family members included in these studies. In two families with distinct RNF213 rare variants, MM096 and MM089, family members with early onset CAD also carried RNF213 variants. We have previously reported that mutations in ACTA2, which encodes the smooth muscle specific isoform of α-actin, cause a predisposition for both a MMD-like cerebrovascular disease and early onset CAD in individuals with little to no cardiovascular risk factors.20–22 Further studies are needed to assess whether or not RNF213 alterations contribute to CAD and other vascular diseases.
MMD exhibits significant genetic heterogeneity and occurs as a clinical manifestation in several well-known genetic conditions. These include Down syndrome, sickle cell disease, Alagille syndrome, neurofibromatosis type 1, individuals with heterozygous ACTA2 mutation (including patients with Mutisystemic Smooth Muscle Dysfunction Syndrome), Majewski osteodysplastic primordial dwarfism type II (MOPD II), and Turner syndrome.20, 21, 23–25 None of the patients with RNF213 variants in our cohort were diagnosed with any of these conditions. In many of these conditions, patients harboring these gene mutations are also predisposed to other vascular diseases. Occlusive lesions in the large arteries show some pathologic features that are similar to atherosclerotic lesions, specifically increased proliferation of smooth muscle-like cells in the lumen or intimal layer. However, MMD occlusive lesions do not have evidence of other features of atherosclerotic lesions, such as cholesterol deposition and inflammatory cells.26 ACTA2 mutations in human smooth muscle cells (SMCs) and Acta2 (Acta2−/−) deficiency in mouse SMCs have both been shown to increase rates of proliferation of these cells.20, 27 The Acta2−/− proliferation could be blocked both in vitro and in vivo using imatinib, which blocks signaling through tyrosine kinase receptors like the PDGF receptors. Although mouse models deficient in RNF213 have been made, these models have not provided further information on the connection between RNF213 alterations and MMD.
MMD is a progressive disease in the majority of patients, including in individuals with the RNF213 p.R4810K variant.1, 28–30 Prevention of strokes and the resulting comorbidities depends on the early identification of at risk individuals predisposed to MMD. Early diagnosis allows for timely surgical intervention to reduce the risk of stroke and possibly decrease cognitive deficits. For example, screening for MMD in patients with unilateral MMD and in high risk populations, such as those with neurofibromatosis 1 or Down syndrome, has been shown to decrease the prevalence of strokes in these patients.31–33 A meta-analysis of 5 case-control studies demonstrated a strong association between the p.R4810K mutation and MMD (OR 157.53; 95% CI 85.37 – 290.70, P<0.00001). Accordingly, these data and the results presented here suggest that diagnostic screening for the RNF213 p.R4810K variant should be pursued in MMD patients of Asian descent, and family members screened for the variant if the MMD index case is positive. Baseline cerebrovascular screening for MMD is indicated in family members with the RNF213 p.R4810K variant. Three dimensional time-of-flight MRA imaging for individuals with the RNF213 p.R4810K would be reasonable given the sensitivity for detecting disease and lack of radiation exposure. The frequency of cerebrovascular imaging in asymptomatic p.R4810K carriers should be tailored individually until more data is made available on outcomes. The data presented here also suggest that screening for rare variants in the C-terminus domain of RNF213 (exons 42–68) should be considered for all MMD patients. Variants previously determined to cause MMD or those that segregate with disease in families could be used to identify other family members at risk for MMD. Ultimately, prevention of future strokes and the resulting comorbidities depends on the early identification of at risk individuals predisposed to MMD.
Acknowledgments
The authors are grateful to patients and their families for their participation in this study.
Sources of Funding
This work was supported by RO1 HL62594 (D.M.M) from the National Institutes of Health and funds from the Vivian L. Smith Foundation (D.M.M) and NIH funding to the University of Washington Center for Mendelian Genomics (1U54HG006493) and to the University of Texas Health Science Center at Houston (UL1 RR024148).
Footnotes
Disclosures
None.
References
- 1.Scott RM, Smith ER. Moyamoya disease and moyamoya syndrome. N Engl J Med. 2009;360:1226–1237. doi: 10.1056/NEJMra0804622. [DOI] [PubMed] [Google Scholar]
- 2.Kleinloog R, Regli L, Rinkel GJ, Klijn CJ. Regional differences in incidence and patient characteristics of moyamoya disease: a systematic review. J Neurol Neurosurg Psychiatry. 2012;83:531–536. doi: 10.1136/jnnp-2011-301387. [DOI] [PubMed] [Google Scholar]
- 3.Wakai K, Tamakoshi A, Ikezaki K, Fukui M, Kawamura T, Aoki R, et al. Epidemiological features of moyamoya disease in Japan: findings from a nationwide survey. Clin Neurol Neurosurg. 1997;99:S1–S5. doi: 10.1016/s0303-8467(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 4.Kuriyama S, Kusaka Y, Fujimura M, Wakai K, Tamakoshi A, Hashimoto S, et al. Prevalence and clinicoepidemiological features of moyamoya disease in Japan: findings from a nationwide epidemiological survey. Stroke. 2008;39:42–47. doi: 10.1161/STROKEAHA.107.490714. [DOI] [PubMed] [Google Scholar]
- 5.Mineharu Y, Takenaka K, Yamakawa H, Inoue K, Ikeda H, Kikuta KI, et al. Inheritance pattern of familial moyamoya disease: autosomal dominant mode and genomic imprinting. J Neurol Neurosurg Psychiatry. 2006;77:1025–1029. doi: 10.1136/jnnp.2006.096040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamada F, Aoki Y, Narisawa A, Abe Y, Komatsuzaki S, Kikuchi A, et al. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet. 2011;56:34–40. doi: 10.1038/jhg.2010.132. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Morito D, Takashima S, Mineharu Y, Kobayashi H, Hitomi T, et al. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE. 2011;6:e22542. doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo DC, Regalado E, Casteel DE, Santos-Cortez RL, Gong L, Kim JJ, et al. Recurrent Gain-of-Function Mutation in PRKG1 Causes Thoracic Aortic Aneurysms and Acute Aortic Dissections. Am J Hum Genet. 2013;93:398–404. doi: 10.1016/j.ajhg.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang GT, Peng B, Leal SM. Variant association tools for quality control and analysis of large-scale sequence and genotyping array data. Am J Hum Genet. 2014;94:770–783. doi: 10.1016/j.ajhg.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 14.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cottingham RW, Jr, Idury RM, Schaffer AA. Faster sequential genetic linkage computations. Am J Hum Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 18.Fishelson M, Geiger D. Exact genetic linkage computations for general pedigrees. Bioinformatics. 2002;18:S189–S198. doi: 10.1093/bioinformatics/18.suppl_1.s189. [DOI] [PubMed] [Google Scholar]
- 19.Ott J. Computer-simulation methods in human linkage analysis. Proc Natl Acad Sci U S A. 1989;86:4175–4178. doi: 10.1073/pnas.86.11.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo DC, Papke CL, Tran-Fadulu V, Regalado ES, Avidan N, Johnson RJ, et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and moyamoya disease, along with thoracic aortic disease. Am J Hum Genet. 2009;84:617–627. doi: 10.1016/j.ajhg.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milewicz DM, Ostergaard JR, la-Kokko LM, Khan N, Grange DK, Mendoza-Londono R, et al. De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am J Med Genet A. 2010;152A:2437–2443. doi: 10.1002/ajmg.a.33657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munot P, Saunders DE, Milewicz DM, Regalado ES, Ostergaard JR, Braun KP, et al. A novel distinctive cerebrovascular phenotype is associated with heterozygous Arg179 ACTA2 mutations. Brain. 2012;135:2506–2514. doi: 10.1093/brain/aws172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manjila S, Miller BR, Rao-Frisch A, Otvos B, Mitchell A, Bambakidis NC, et al. Moyamoya Disease Associated with Asymptomatic Mosaic Turner Syndrome: A Rare Cause of Hemorrhagic Stroke. J Stroke Cerebrovasc Dis. 2014;23:1242–1244. doi: 10.1016/j.jstrokecerebrovasdis.2013.07.038. [DOI] [PubMed] [Google Scholar]
- 24.Achrol AS, Guzman R, Lee M, Steinberg GK. Pathophysiology and genetic factors in moyamoya disease. Neurosurg Focus. 2009;26:E4. doi: 10.3171/2009.1.FOCUS08302. [DOI] [PubMed] [Google Scholar]
- 25.Kamath BM, Spinner NB, Emerick KM, Chudley AE, Booth C, Piccoli DA, et al. Vascular anomalies in Alagille syndrome: a significant cause of morbidity and mortality. Circulation. 2004;109:1354–1358. doi: 10.1161/01.CIR.0000121361.01862.A4. [DOI] [PubMed] [Google Scholar]
- 26.Reid AJ, Bhattacharjee MB, Regalado ES, Milewicz AL, El-Hakam LM, Dauser RC, et al. Diffuse and uncontrolled vascular smooth muscle cell proliferation in rapidly progressing pediatric moyamoya disease. J Neurosurg Pediatr. 2010;6:244–249. doi: 10.3171/2010.5.PEDS09505. [DOI] [PubMed] [Google Scholar]
- 27.Papke CL, Cao J, Kwartler CS, Villamizar C, Byanova KL, Lim SM, et al. Smooth muscle hyperplasia due to loss of smooth muscle alpha-actin is driven by activation of focal adhesion kinase, altered p53 localization and increased levels of platelet-derived growth factor receptor-beta. Hum Mol Genet. 2013;22:3123–3137. doi: 10.1093/hmg/ddt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imaizumi T, Hayashi K, Saito K, Osawa M, Fukuyama Y. Long-term outcomes of pediatric moyamoya disease monitored to adulthood. Pediatr Neurol. 1998;18:321–325. doi: 10.1016/s0887-8994(97)00209-9. [DOI] [PubMed] [Google Scholar]
- 29.Kronenburg A, Braun KP, van der ZA, Klijn CJ. Recent advances in moyamoya disease: pathophysiology and treatment. Curr Neurol Neurosci Rep. 2014;14:423. doi: 10.1007/s11910-013-0423-7. [DOI] [PubMed] [Google Scholar]
- 30.Mineharu Y, Takagi Y, Takahashi JC, Hashikata H, Liu W, Hitomi T, et al. Rapid progression of unilateral moyamoya disease in a patient with a family history and an RNF213 risk variant. Cerebrovasc Dis. 2013;36:155–157. doi: 10.1159/000352065. [DOI] [PubMed] [Google Scholar]
- 31.Jea A, Smith ER, Robertson R, Scott RM. Moyamoya syndrome associated with Down syndrome: outcome after surgical revascularization. Pediatrics. 2005;116:e694–e701. doi: 10.1542/peds.2005-0568. [DOI] [PubMed] [Google Scholar]
- 32.Smith ER, Scott RM. Progression of disease in unilateral moyamoya syndrome. Neurosurg Focus. 2008;24:E17. doi: 10.3171/FOC/2008/24/2/E17. [DOI] [PubMed] [Google Scholar]
- 33.Rosser TL, Vezina G, Packer RJ. Cerebrovascular abnormalities in a population of children with neurofibromatosis type 1. Neurology. 2005;64:553–555. doi: 10.1212/01.WNL.0000150544.00016.69. [DOI] [PubMed] [Google Scholar]