Abstract
Recent studies have demonstrated that allelic variation in daily expression profiles of the GIGANTEA gene may account for the variance in plant growth in different accessions. Studying natural variation in daily transcriptional patterns of circadian-clock regulated genes provides new insights into plant adaptive strategies to different geographical regions.
Keywords: natural variation, quantitative trait loci, gene expression profiles, circadian clock, luciferase bioluminescence imaging
Theoretical mechanism of photoperiodic regulation and its potential advantage to local adaptation
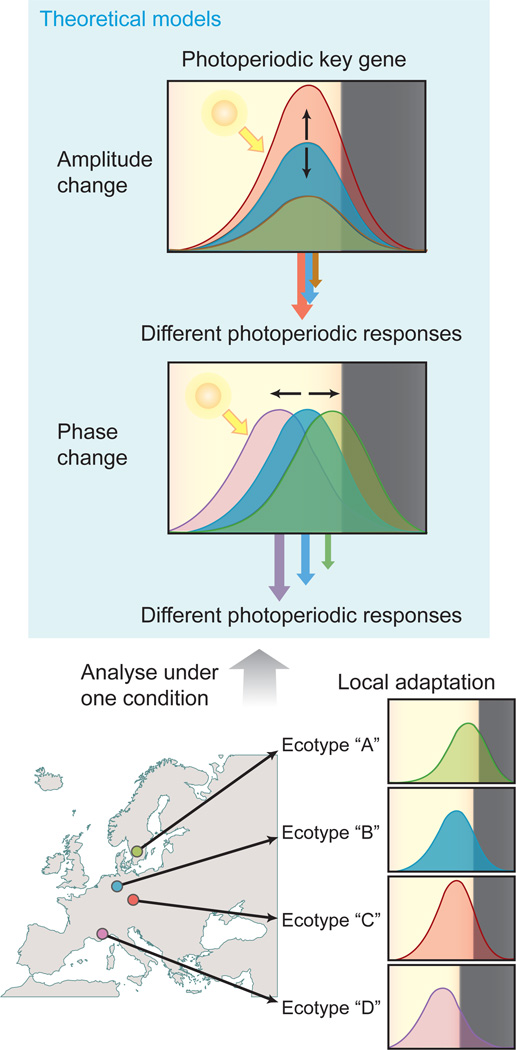
The circadian clock regulates various physiological and developmental events throughout the life cycle of plants, and enables them to anticipate upcoming daily changes and adjust the timing of responses to occur under the most appropriate conditions. Mutants defective in the clock have reduced fitness under normal daily light-dark cycles [1]. The circadian clock also controls seasonal responses, especially photoperiodic responses, such as flowering time and stem growth. Correctly timing these responses is crucial, as a failure in the proper flowering response jeopardizes successful fertilization and hinders the subsequent gamete maturation. The mechanisms underlying plant photoperiodic responses can be explained by the external coincidence model [2]. In this model, the circadian clock sets the timing of expression of a key regulator to the late afternoon. Photoperiodic responses are induced only when the expression of the key regulator coincides with the presence of external stimuli, such as light. Many plant species, including Arabidopsis (Arabidopsis thaliana), grow in wide geographic locations with latitudinal differences, with a wide range of day length changes throughout the year. How have plants genetically evolved the proper photoperiodic response for each location? Based on the external coincidence model, we can deduce that either changing the amplitude or phase of the peak expression of the key regulator will alter photoperiodic response (Figure 1). Studies using various ecotypes that are adapted to different local environments have provided evidence that plants have altered these parameters to optimize their seasonal responses.
Figure 1.
Models of how natural variation in circadian rhythms alter photoperiodic responses. Based on the external coincidence model, changing either the amplitude of expression of a hypothetical key regulator (the model shown at the top) or changing the phase of its expression (the model shown in the middle) will alter the degree of photoperiodic responses. Black arrows show the directions of the expression changes. The size of the different colored arrows located at the bottom of the panels indicates the strength of the response caused by the changes in the expression patterns of the key regulator and light stimuli. The color of the arrows matches the color of each hypothetical expression pattern of the key gene. At the bottom, a conceptual diagram depicts that analyzing the relationship between the detailed expression profiles of clock/photoperiod genes and photoperiodic responses under a certain condition among ecotypes may provide us with information regarding plant adaptive strategies to the local environment. Plants may change the phase and/or amplitude of the key regulator expression to adapt to the environment of their geographical location (indicated by circles). The effects of the natural variation can be analyzed and assessed if these ecotypes were grown under one growth condition.
Natural variation in expression patterns of clock-associated components affects photoperiodic responses
To examine the natural variation of circadian rhythms, Michael et al. analyzed the rhythms of circadian leaf movement in 150 Arabidopsis accessions [3]. These accessions showed variation in period length, phase, and amplitude. Among these variations, period length had a significant correlation with the length of the longest day in locations where the accessions were from. The accessions that originated from higher latitudes (where plants experience longer day lengths) tended to have longer circadian periods. Quantitative trait loci (QTL) analysis revealed that multiple loci interactively determine the period length, amplitude and phase of leaf movement rhythms. Several PSEUDO RESPONSE REGULATOR (PRR) genes, which encode components of the circadian oscillator, were located at some of the QTL for circadian periodicity, suggesting that polymorphism in the PRR genes may have some advantage for adaptation to local environments.
A study using natural accessions of Capsella bursa-pastoris (a close relative to Arabidopsis) also indicates that altering circadian timing may have an adaptive advantage. The flowering time of various C. bursa-pastoris ecotypes showed a strong correlation with the latitudes of their original habitats [4]. Gene expression analysis between two ecotypes from lower and higher latitudes, which showed early- and late-flowering phenotypes in long days, revealed that the difference in the expression of the genes involved in the circadian clock and GA-mediated flowering regulation may be the cause of flowering time difference. The timing of core clock gene expression in the ecotype from the lower latitude was earlier than in the one from the higher latitude, indicating that the phase advance of clock gene expression may contribute to earlier flowering.
Similarly, natural variation of photoperiodic flowering response in the tree species Populus trichocarpa may also be caused by the phase shift of clock-regulated gene expression [5]. The peak expression of the poplar CONSTANS homolog PtCO2 appeared earlier in the day in the southern populations compared to the northern populations, and PtCO2 expression under light correlated with the expression of the poplar homolog of FLOWERING LOCUS T, PtFT1. Although a recent work showed that PtCO2 does not control PtFT1 expression [6], the original observation indicated that the phase shift of circadian timing in those populations may induce PtFT1 differently.
These studies suggest that allelic difference in the expression patterns of clock and photoperiodic regulator genes may account for adaptive responses (such as growth and flowering time) to local environments. However, the mechanisms that connect the variation in circadian gene expression with the photoperiodic responses are still not well understood. Also, due to the difficulty of fine-scale large gene expression profiling, the number of accessions in which the daily expression patterns of circadian-regulated genes were analyzed has been limited. To more accurately examine the effects of variation of rhythmic expression on photoperiodic responses, measuring precise temporal gene expression profiles among large populations is required. A recent work circumvented this technical issue by employing quantitative live luciferase bioluminescence imaging assay [7].
de Montaigu et al. analyzed allelic variation in detailed transcriptional profiles of the evening clock gene GIGANTEA (GI) among 77 Arabidopsis accessions using the GI promoter-fused luciferase reporter under different day-length conditions [7]. The phases of GI peaks within these accessions varied more in long days than in short days. The GI phase variations observed in long days in approximately half of the accessions correlated with circadian period-length variation (i.e. later phases of the GI peak were found in plants that have longer periods). These variations imply that the period lengths of internal clocks contribute in some accessions to the adjusting of the peak phases of GI (and potentially other evening expressed genes), but the individual responses to afternoon light conditions in other accessions also set the phases regardless of the free running periods of their clocks.
QTL mapping using two ecotypes, which possess early and late GI peaks, indicated that PHYTOCHROME B (PHYB) was one of the loci that control the timing of GI. Gene expression analysis in phyB-9 mutants confirmed that PHYB regulates the phase and amplitude of GI in long days. The study also found that the change in GI peak timing and amplitude correlates with hypocotyl length and the expression level of PHYTOCHROME INTERACTING FACTOR 4 (PIF4), which is a major regulator of hypocotyl elongation [8]. Although the question of whether or not the GI pattern difference within the two ecotypes is a major cause of the changes in PIF4 expression still needs further investigation, these results implied that natural variation can adjust photoperiodic hypocotyl growth, potentially through the regulation of phase and amplitude of GI profiles in long days, and that the change may alter the expression pattern of PIF4 and subsequently hypocotyl growth rates.
Concluding remarks and future perspectives
To understand how the circadian clock increases plant fitness to local environments, it is important to investigate how natural variation in the clock modulates photoperiodic responses. Utilizing the promoter-luciferase reporter was an excellent approach for analyzing the allelic variation of gene expression rhythm quantitatively among many ecotypes. As the cost of sequencing continues to decrease, detailed time course transcriptome analysis of large accessions will be increasingly practical for the identification of novel correlations between diurnal gene expression profiles and physiological responses. In addition, because posttranscriptional regulation also plays an important role in the regulation of photoperiodic response, applying the translational clock-related gene luciferase fusion protein reporters to monitor protein accumulation patterns may further facilitate our understanding of how changes in daily protein expression profiles modulate photoperiodic output. One challenge for these approaches will be providing further mechanistic evidence that the relationships between expression pattern variation and response changes are not merely correlations. As these changes are often quantitative, mathematical modeling could help us estimate the contribution of each variation in these responses. These approaches will broaden our understanding of the evolution of the clock system during the course of plant adaptation to natural environments.
Acknowledgements
We thank H. Kinmonth-Schultz for critical reading of the manuscript. This work was supported by funding from the National Institutes of Health (GM079712) to T.I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- 2.Song YH, et al. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol. 2014 Dec 12; doi: 10.1146/annurev-arplant-043014-115555. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michael TP, et al. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- 4.Slotte T, et al. Differential expression of genes important for adaptation in Capsella bursa-pastoris (Brassicaceae) Plant Physiol. 2007;145:160–173. doi: 10.1104/pp.107.102632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böhlenius H, et al. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- 6.Hsu CY, et al. Overexpression of CONSTANS homologs CO1 and CO2 fails to alter normal reproductive onset and fall bud set in woody perennial poplar. PLoS One. 2012;7:e45448. doi: 10.1371/journal.pone.0045448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Montaigu A, et al. Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc Natl Acad Sci U S A. 2015;112:905–910. doi: 10.1073/pnas.1422242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]