Abstract
Pituitary adenylate-cyclase-activating polypeptide (PACAP) has widespread physiological/pathophysiological actions and there is increased interest for its use therapeutically, especially in the CNS (neuroprotection). Unfortunately, no selective PACAP-analogs exist for PACAP-preferring PAC1-receptors, primarily because of its high sequence identity to VIP and particularly, because of the inability of structure-function studies to separate the pharmacophore of PAC1-R from VPAC1-R, which has high affinity for PACAP and VIP. The present study attempted to develop PAC1-R-selective agonists primarily by making conformationally-restricted PACAP -analogs in positions important for receptor-selectivity/affinity. Forty-six PACAP-related-analogs were synthesized with substitutions in positions 1–4, 14–17, 20–22 ,28,34,38 and receptor-selectivity determined in PAC1-R,VPAC1-R,VPAC2-R-transfected or native cells from binding or cAMP-generation experiments. Fifteen PACAP-analogs had 6–78-fold higher affinities for PAC1-R than VPAC1-R and 13 were agonists. Although binding-affinities correlated significantly with agonist potency, the degree of receptor-spareness varied markedly for the different PACAP-analogs, resulting in selective potencies for activating the PAC1 receptor over the VPAC1 receptor from 0- to-103-fold. In addition, a number of PACAP-analogs were identified that had high selectivity for PAC1-R over VPAC2-R as well as PACAP-analogs that could prove more useful therapeutically because of substitutions known to extend their half-lives (substitutions at potential sites of proteolysis and attachment of long-chain fatty acids). This study provides for the first time a separation of the pharmacophores for PAC1-R and VPAC1-R, resulting in PACAP-related analogs that are PAC1-R-preferring. Some of these analogs, or their modifications, could prove useful as therapeutic agents for various diseases.
Keywords: PACAP, vasoactive intestinal peptide, neuroprotection, stroke, traumatic brain injury, structure-function study
1. Introduction
Pituitary adenylate cyclase-activating polypeptide, first isolated from hypothalamus, occurs in two forms: either with 38-amino acid residues (PACAP38) or the other, containing only the first 27 residues (PACAP27)[35]. PACAP has a high sequence identity to vasoactive intestinal polypeptide (VIP) with PACAP27 sharing 68% sequence identity with VIP[54,55]. Their actions are mediated by three Class II (secretin-type) G-protein-coupled receptors: PAC1 receptor (PAC1-R), VPAC1 receptor (VPAC1-R) and VPAC2 receptor (VPAC2-R)[20]. PACAP38/27 have high affinities for all three receptors and VIP only has high affinities for the latter two receptors[20,55]. These receptors all signal through cAMP with PAC1-R also activating phospholipase C in many tissues [12,20,55].
PACAP and its receptors have widespread distributions in both the central nervous system (CNS) and peripheral tissues with wide-ranging activities in different tissues, both physiologically and pathophysiological [12,20,43,55]. PACAP has numerous functions in the CNS (release of oxytocin/vasopressin, circadian rhythm, behavioral changes, neurotrophic-development, glial cell activity, pituitary cell function), the cardiovascular system (vasorelaxant effects on vascular tone, direct cardiac effects), immune system (monocyte differentiation, regulatory of inflammatory process), the urogenital and respiratory systems (smooth muscle activity, mucus secretion, urinary bladder function), gastrointestinal tract (alters secretion [salivary, pancreatic, gastric], proliferation) and endocrine glands (adrenal, thyroid, islets), as well as the gonads (regulates gonadal activity)[12,20,54,55]. In pathologic processes, PACAP has been shown to have a number of beneficial effects, especially related to its neuroprotective and neurotrophic actions manifested by its ability to stimulate the migration, proliferation, differentiation, and survival of neural cells[20,54]; its ability to exert potent immuno-modulatory actions that are primarily anti-inflammatory[57]; its ability to enhance insulin release and ameliorate islet injury[1] and its ability to ameliorate renal failure/injury[3,31]. Primarily due to these actions, PACAP has been shown to have beneficial effects in models of stroke (focal cerebral ischemia), traumatic brain/spinal injury, various CNS/neurological diseases (Parkinson’s disease, Huntington’s chorea, Alzheimer’s disease, schizophrenia)[14,33,46,47,57], diabetes[1], kidney failure due to various diseases (myeloma, diabetic nephropathy, contrast-induced nephropathy)[3,30,31] and to prevent the toxic effects of numerous neurotoxins (glutamate, lipopolysaccharide, ethanol, oxidative stress, β-amyloid)[7,20,37,43,54]. Furthermore, PAC1-R is frequently overexpressed by various human tumors and PACAP has been shown to have both stimulatory and inhibitory effects on the growth of various tumors leading to the proposal that development of PAC1-R selective agonists or antagonists could give rise to useful tools for the treatment of cancers [36,48,55].
Most of the beneficial effects of PACAP are mediated via PAC1-R and, therefore, there is considerable interest in identifying PAC1-R selective agonists for possible therapeutic effects [32]. At present, no suitable VIP/PACAP-analog exists and the only selective PAC1 agonist, maxadilan, is a 61-amino-acid peptide with no sequence similarity to PACAP[29] and is rarely used. The development of PAC1 selective agonists has been difficult because PACAP38, which is the predominant form in both the CNS and peripheral tissues, is rapidly degraded [4,54]; has high sequence identity to VIP, and interacts with high affinities with the other VIP-PACAP family of receptors (VPAC1-R and VPAC2-R)[20,54]. To identify VIP/PACAP-analogs with selectivity for PAC1-R over VPAC1-R has proven particularly difficult, and none have been described.
The purpose of this study was to attempt to identify, using information from previous structure-function and conformations studies of PACAP/VIP, VIP/PACAP-analogs with selectivity for PAC1-R over VPAC1-R. To accomplish this goal, 46 PACAP-analogs (primarily PACAP38 analogs) were synthesized with substitutions that were conformationally-restricting to attempt to identify analogs with preferential binding/activation of PAC1-R. Using this approach, we identified for the first time PACAP-analogs that preferentially bound to PAC1-R and/or activated PAC1-R over VPAC1-R, and also identified strategies that could be used to reduce interactions with VPAC2-R.
2. Material and Methods
2.1. Materials
NIH 3T3 cells, PANC-1 cells and Sup-T1 cells were from American Type Culture Collection (ATCC), Rockville, MD; Dulbecco’s minimum essential medium (DMEM), phosphate-buffered saline (PBS), G418 sulfate, fetal bovine serum (FBS), RPMI 1640 medium, penicillin, streptomycin and sodium pyruvate from Gibco Life Technology (Grand Island, NY); bacitracin, soybean trypsin inhibitor, 3-isobutyl-1-methylxanthine (IBMX), formic acid, ammonium formate, disodium tetraborate, and alumina were obtained from Sigma-Aldrich (St. Louis, MO); iodine-125 (100 mCi/ml) and [2,8-3H]adenine were from Perkin Elmer Life Sciences (Boston, MA); 1,2,4,6-tetrachloro-3α-6α-diphenylglycouril (Iodo-Gen) from Pierce Chemical Co. (Rockford, IL); AG 1-X8 resin from BIO-RAD (Richmond CA). Standard protected amino acids and other synthetic reagents were obtained from Bachem Bioscience Inc. (King of Prussia, PA).
2.2. Cell culture
VPAC1 stably transfected into PANC-1 cells(VPAC1-R /PANC-1 cells), VPAC2 stably-transfected into PANC-1 cells(VPAC2-R /PANC-1 cells) [23,25,42] and PAC1-R stably-transfected into NIH 3T3 cells(PAC1-R cells) [15,42,45] were used because they are well characterized and behave similar to wild type receptors. PAC1-R cells were a gift from Dr. J.R. Pisegna, UCLA. VPAC1, VPAC2 PANC-1 cells and PAC-1R cells were grown in DMEM media supplemented with 10% FBS, 100 U/ml of penicillin, 100 mg/ml of streptomycin and 300 mg/l of G418 sulfate . T47D breast cancer cells naturally containing VPAC1-R (T47D cells) [23] were grown in DMEM media supplemented with 10% FBS, 100 U/ml of penicillin, 100 mg/ml of streptomycin and 1 mM sodium pyruvate. Sup-T1 lymphoblastoma cells (Sup-T1 cells) naturally expressing VPAC2-R [23] were grown in RPMI supplemented with 10% FBS and 100 U/ml of penicillin, 100 mg/ml of streptomycin. All cells were incubated at 37°C in a 5% CO2 atmosphere.
2.3. Preparation of peptides
PACAP-analogs were synthesized with solid-phase methods as described previously [23–25]. Briefly, solid-phase syntheses of peptide amides were carried out using Boc chemistry on methylbenzhydrylamine resin (Advanced ChemTech, Louiville, KY) followed by HF-cleavage of free peptides amides. The crude peptides were purified on 92.5 × 50 cm columns of Vydac C18 silica (10 µm), which was eluted with linear gradients of acetonitrile in 0.1% (v/v) trifluoroacetic acid. Homogeneity of the peptides was assessed by analytical reverse-phase high-pressure liquid chromatography (HPLC) and purity was usually 97% or higher. Amino acid analysis (only amino acids with primary amino acid groups were quantitated) gave the expected amino acid ratios. Peptide molecular masses were obtained by matrix-assisted laser desorption mass spectrometry (Finegan Lasermat) and all corresponded well with calculated values.
2.4. Preparation of 125I-PACAP27, 125I-VIP and 125I-Ro 25–1553
These radioligands, with specific activities of 2200 Ci/mmol were prepared as previously described [23–25,27]. Briefly, 0.8 µg of Iodo-Gen solution (0.01 µg/ µl in chloroform) was added to a 5-ml plastic test tube, dried under nitrogen, and washed with 100 µl of a 0.5 M potassium phosphate solution (pH 7.4). Eight µg of peptide in 4 µl of water and 2 mCi (20 µl) of Na125I were added and incubated at room temperature for 6 minutes. The incubation was stopped by the addition of 100 µl of water. The radiolabeled peptides were separated using a SEP-Pak cartridge (Waters Associates, Milford, Mass) and further purified by reverse-phase HPLC as previously described [23–25,27]. The fractions with the highest radioactivity and binding were neutralized with 0.2 M Tris buffer (pH 9.5) and stored with 0.5% BSA (w/v) at −20°C.
2.5. Binding studies
Binding to VPAC1-R, VPAC2-R, and PAC1-R containing cells was assayed using a variation of previous described methods [23–25,27]. Specifically, for VPAC1-R/PANC-1 cells (0.2 × 106 cells/ml) and for T47D cells [23–25] (1.2 × 106 cells/ml), 50 pM 125I-VIP was used. For VPAC2-R /PANC-1 cells (0.1 × 106 cells/ml) and PAC1-R /3T3 cells (0.2 × 106 cells/ml) 50 pM 125I-PACAP27 was used [23–25]and for Sup-T1 cells (2.5 × 106 cells/ml), 75 pM 125I-Ro 25–1553 was used [24,25]. Binding was performed by incubation in standard incubation solution containing 24.5 mM HEPES (pH 7.4), 98 mM NaCl, 6 mM KCl, 2.5 mM Na H2PO4, 5 mM sodium pyruvate, 5 mM sodium fumarate, 5 mM Mg Cl2, 0.01% (w/v) soybean trypsin inhibitor, 1% (w/v) bovine serum albumin, and 0.05% (w/v) bacitracin for 60 minutes at room temperature for PAC1-R /3T3 cells, VPAC1-R /PANC-1 cells, VPAC2-R /PANC-1 cells and T47D cells. To assess VPAC2-R affinities in Sup-T1 cells, binding was performed at 37°C with 125I-Ro 25–1553, because 125I-VIP and 125I-PACAP27 were rapidly degraded by these cells even with protease inhibitors present [24,25]. The separation of bound from free radioactivity was obtained by centrifugation (10,000 rpm for 1 min) of cells through 2% (w/v) BSA in a standard incubation solution. The tubes were washed twice with 2% (w/v) BSA in a standard incubation solution, and radioactivity was counted. Nonsaturable binding was determined using 1 µM unlabeled peptide and for 125I-VIP, 125I-PACAP27 and 125I-Ro 25–1553 was less than 10% of total binding. For all peptides, the IC50 (concentration that gave half-maximal inhibition of that seen with a saturating concentration of 1 µM PACAP38) was calculated using the curve-fitting program KaleidaGraph (Synergy Software, Reading, PA). To determinate relative affinities of the different PACAP-analogs for PAC1-R, VPAC1-R and VPAC-R, their IC50 from binding studies were expressed as a ratio for VPAC1-R /PANC-1 or VPAC2-R /PANC-1 cells compared to the IC50 for PAC1-R /3T3. To compare the effects of substitutions in PACAP38 or PACAP27, the affinities were compared and expressed as a ratio of the substituted analogs to native PACAP38 or PACAP27 depending on whether the substituted analogs were PACAP38- or PACAP27-substituted.
2.6. cAMP Assay
PAC1-R /3T3 cells, VPAC1-R /PANC-1 cells, VPAC2-R /PANC-1 cells and T47D cells were plated in 24-well plates (5.0 × 104 cells/ml) and incubated for 24 hours at 37°C with growth media containing 10% FBS (v/v). The media was then replaced with DMEM containing 2% FBS (v/v) and 2 Ci/ml [2,8-3H]adenine. Cells were incubated for an additional 48 hours at 37°C. The medium was removed and cells were washed by incubating at 37°C for 15 minutes with 1 ml of DMEM. The cells were then incubated in 500 µl of DMEM containing 1% (w/v) BSA and 0.5 mM IBMX in DMEM with a protease inhibitor cocktail (1 ml of Sigma P8340 in 1000 ml of DMEM) and incubated with or without peptides at various concentrations for 1 hour at 37°C. Reactions were terminated by the addition of 50 µL of 2.2 N HCl. Samples were stored at −20°C until analyzed. [3H]cAMP was isolated by a modification of the ion-exchange chromatographic method described by Alvarez and Daniel [2] using a single column of acidic aluminum oxide with dilute HCl and ammonium acetate. To make the columns, dry acidic alumina (0.75 g) was measured into a 5-ml glass pipet fitted with a glass ball to hold the alumina. The columns were placed on a Plexiglass rack designed to hold the columns and to fit over a box of 100 scintillation counting vials. The samples were loaded onto the dry alumina in the columns and washed with 4 ml of 0.005 N HCl followed by 1 ml of 0.1 M ammonium acetate. The cAMP was then eluted into fresh scintillation counting vials with 4 ml of 0.1 M ammonium acetate. Each eluate was mixed with scintillation cocktail and assayed for radioactivity in a liquid scintillation counter. For all peptides, the EC50 was calculated, which was the concentration of the peptide that gave half-maximal stimulation of a maximally effective concentration of PACAP38 (1 µM) or the peptide tested. The EC50 was calculated using the curve-fitting program KaleidaGraph 4.0.
2.7. Measurement of [3H]Inositol Phosphates
[3H]Inositol phosphates (IP) were determined in the different cells as described previously [6,50]. In brief, PAC1-R cells (4.0 × 104 cells/ml) were subcultured into 24-well plates in regular propagation media and then were incubated for 24 hours at 37°C in a 5% CO2 atmosphere. The cells were then incubated with 3 mCi/ml of myo-[2-3H]inositol in growth media supplemented with 2% FBS for an additional 24 hours. After the incubation, the 24-well plates were washed by incubating for 30 minutes at 37°C with 1 ml/well of PBS (pH 7.0) containing 20 mM lithium chloride. The wash buffer was aspirated and replaced with 500 ml of IP assay buffer containing 135 mM sodium chloride, 20 mM HEPES (pH 7.4), 2 mM calcium chloride, 1.2 mM magnesium sulfate, 1 mM EGTA, 20 mM lithium chloride, 11.1 mM glucose, and 0.05% BSA (w/v), and was incubated without (control) or with different concentrations of the peptides studied. After 60 minutes of incubation at 37°C, the experiments were terminated by the addition of 1 ml of ice-cold 1% (v/v) hydrochloric acid in methanol. The total [3H]IP was isolated by anion-exchange chromatography as described previously [6,50]. Samples were loaded onto Dowex AG1-X8 anion-exchange resin columns, washed with 5 ml of distilled water to remove free [3H]IP and then washed with 2 ml of 5 mM disodium tetraborate/60 mM sodium formate solution to remove [3H]glycerophosphorylinositol. Finally, 2 ml of 1 mM ammonium formate/100 mM formic acid solution were added to the columns to elute the total [3H]IP. Each eluate was mixed with scintillation cocktail and measured for radioactivity in a scintillation counter.
2.7. Statistical Analysis
The results are the mean and S.E.M. from at least three separate experiments. IC50 and EC50 were calculated using the curve-fitting program KaleidaGraph 4.0. Statistical comparisons were made using the Student’s t-test.
3. Results
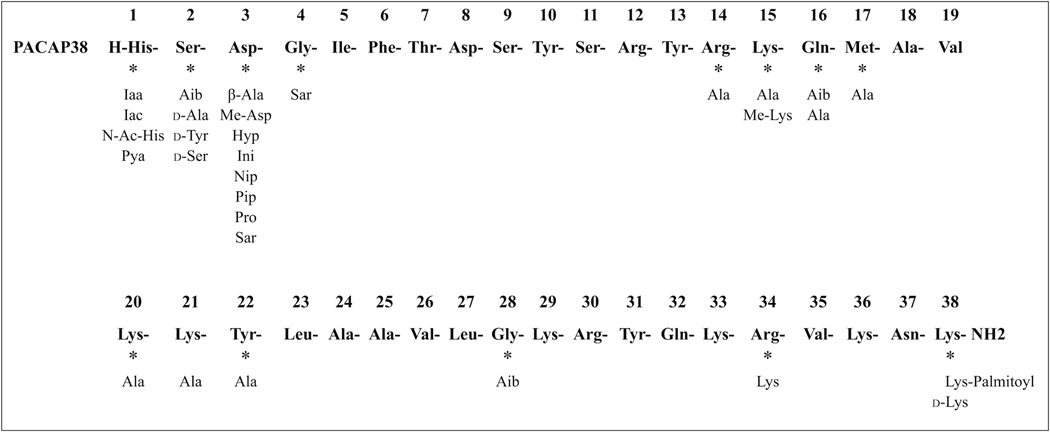
Forty-six different PACAP38-related peptide analogs were synthesized with 24 having one or two amino acid substitutions and 22 having more than two substitutions (Tables 1–4, Fig. 1). Substitutions were made in positions 1, 2, 3, 4, 14, 15, 16, 17, 20, 21, 22, 28, 34, and 38 of PACAP38, PACAP27 or VIP primarily based on previous studies of the importance of amino acids in these positions for affinity/activity/proteolysis of PACAP, VIP, secretin, and GHRH-related peptides (Fig. 1) [7–10,13,18,20,22,23,25,40,58].
TABLE 1.
Affinities of various single or double substituted PACAP-related analogs for PAC1, VPAC1-R and VPAC2 receptors.
| Peptide Number | Peptide structure | IC50 (nM) |
||||
|---|---|---|---|---|---|---|
| PAC1-R |
VPAC1-R |
VPAC2-R |
||||
| PAC1-R | VPAC1- R/PANC-1 |
T47D | VPAC2- R/PANC-1 |
Sup-T1 | ||
| PACAP27 | 2.82 ± 0.17 | 0.23 ± 0.09 | 0.20 ± 0.07 | 4.79 ± 0.20 | 24.5 ± 1.0 | |
| PACAP38 | 0.28 ± 0.02 | 1.35 ± 0.04 | 1.29 ± 0.05 | 1.00 ± 0.11 | 6.03 ± 0.30 | |
| VIP | >1,000 | 0.14 ± 0.01 | 0.20 ± 0.02 | 6.03 ± 0.30 | 16.6 ± 1.0 | |
| P42 | [Iaa1PACAP38 | 0.28 ± 0.02 | 2.10 ± 0.09 | 2.45 ± 0.12 | 1.00 ± 0.07 | 3.16 ± 0.21 |
| P44 | [Iac1]PACAP38 | 1.63 ± 0.10 | 5.01 ± 0.13 | 3.80 ± 0.11 | 9.40 ± 0.71 | 32.4 ± 1.3 |
| P43 | [Iaa1,d-Ser2]PACAP38 | 2.19 ± 0.16 | 23.4 ± 0.9 | 12.9 ± 0.4 | 21.9 ± 0.9 | 25.7 ± 1.4 |
| P27 | [N-Ac-His1,Pip3]PACAP38 | 46.8 ± 3.3 | 316 ± 9 | 288 ± 16 | 37.2 ± 1.9 | 87 ± 2 |
| P45 | [Iaa1,Ala22]PACAP38 | 1.74 ± 0.73 | 16.3 ± 0.4 | 9.12 ± 0.57 | 676 ± 34 | >1,000 |
| P46 | [Iac1,Ala22]PACAP38 | 7.41 ± 0.47 | 53.7 ± 2.7 | 37.2 ± 3.0 | >1,000 | >1,000 |
| P47 | [Pya1,Ala22]PACAP38 | 83.2 ± 3.8 | 186 ± 8 | 162 ± 8 | 490 ± 28 | 692 ± 17 |
| P15 | [Aib2]PACAP38 | 4.68 ± 0.29 | 7.08 ± 0.35 | 4.37 ± 0.22 | 2.04 ± 0.11 | 3.16 ± 0.15 |
| P4 | [d -Ser 2 ]PACAP27 | 54.9 ± 2.4 | 8.51 ± 0.47 | 4.68 ± 0.20 | 31.6 ± 1.6 | 72.4 ± 6.5 |
| P16 | [d -Ser 2 ]PACAP38 | 7.94 ± 0.50 | 417 ± 19 | 170 ± 8 | 14.8 ± 0.9 | 63.1 ± 1.6 |
| P14 | [Aib2,Lys(Palmitoyl)38]PACAP38 | 6.31 ± 0.22 | 14.5 ± 1.1 | 3.16 ± 0.72 | 55 ± 4 | 13.2 ± 0.2 |
| P13 | [d-Ser2,Lys(Palmitoyl)38]PACAP38 | 7.94 ± 0.48 | 11.5 ± 0.9 | 3.90 ± 0.29 | 15.8 ± 0.1 | 13.2 ± 0.5 |
| P26 | [Pip3]PACAP38 | 48.9 ± 2.6 | 575 ± 43 | 468 ± 56 | 74.1 ± 2.5 | 257 ± 4 |
| P5 | [Pro3]PACAP27 | >1,000 | 467 ± 28 | 149 ± 12 | 316 ± 22 | >1,000 |
| P18 | [Pro3]PACAP38 | 79.4 ± 2.4 | 302 ± 9 | 151 ± 9 | 83.2 ± 2.5 | 229 ± 13 |
| P6 | [Sar3]PACAP27 | >1,000 | 251 ± 9 | 83.2 ± 2.9 | 631 ± 42 | >1,000 |
| P19 | [Sar3]PACAP38 | 32.4 ± 0.8 | 288 ± 13 | 263 ± 8 | 91.2 ± 5.4 | 214 ± 13 |
| P25 | [Pro3,Lys34]PACAP38 | 32 ± 2 | 44.7 ± 2.7 | 64.6 ± 4.3 | 38 ± 2 | 107 ± 4 |
| P22 | [Sar4]PACAP27 | 69 ± 4 | 675 ± 37 | 575 ± 27 | 30.2 ± 0.7 | 302 ± 15 |
| P2 | [Me-Lys15]PACAP27 | >1,000 | 398 ± 19 | 339 ± 25 | 327 ± 25 | >5000 |
| P17 | [Ala22]PACAP38 | 3.72 ± 0.12 | 17.4 ± 0.4 | 20.4 ± 1.3 | >1,000 | >1,000 |
| P24 | [Lys34]PACAP38 | 1.38 ± 0.06 | 0.96 ± 0.05 | 1.10 ± 0.10 | 2.42 ± 0.18 | 4.20 ± 0.16 |
| P12 | [Lys(palmitoyl)38]PACAP38 | 6.31 ± 0.26 | 8.13 ± 0.46 | 4.40 ± 0.20 | 21.4 ± 0.5 | 5.89 ± 0.40 |
| P50 | [Pro3]VIP | >1,000 | 457 ± 16 | 692 ± 28 | >1,000 | >1,000 |
The indicated cell type was incubated with 50 pM 125I-labeled peptide and various concentrations of the unlabeled PACAP27 and PACAP38 analog as described under Materials and Methods. The IC50 was the concentration causing half-maximal inhibition of the saturable binding, calculated using the curve-fitting program KaleidaGraph. In each experiment each value was determined in duplicate, and values given are means and S.E.M. from at least three separate experiments. Abbreviations: VIP, vasoactive intestinal peptide; PAC1-R, PAC1-receptor; VPAC1-R, VPAC1-receptor; VPAC2-R, VPAC2-receptor; PAC1-R stably transfected into NIH 3T3 cells; VPAC1-R and VPAC2-R stably transfected into PANC1 cells; T47D breast cancer cells and Sup-T1 lymphoblastoma cells containing native VPAC1-Rand VPAC2-R respectively, others-see Fig. 1 legend.
TABLE 4.
The potency of various multi-substituted PACAP-related analogs for PAC1-R, VPAC1-R and VPAC2-R.
| Peptide Number | Peptide structure | [3H]cAMP |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| PAC1-R |
VPAC1-R |
VPAC2-R |
|||||||
| PAC1-R |
VPAC1-R/PANC-1 |
T47D |
VPAC2-R/PANC-1 |
||||||
| % max | EC50 (nM) | % max | EC50 (nM) | % max | EC50 (nM) | % max | EC50 (nM) | ||
| PACAP27 | Ag | 0.03 ± 0.00 | Ag | 0.20 ± 0.01 | Ag | 0.79 ± 0.07 | Ag | 1.00 ± 0.04 | |
| PACAP38 | Ag | 0.03 ± 0.03 | Ag | 0.54 ± 0.05 | Ag | 3.24 ± 0.19 | Ag | 1.38 ± 0.10 | |
| VIP | No Act | 174 ± 10 | Ag | 0.18 ± 0.01 | Ag | 1.00 ± 0.07 | Ag | 1.50 ± 0.07 | |
| P48 | [Iaa1 ,d-Ser2,Ala22]PACAP38 | Ag | 2.19 ± 0.17 | Ag | 83 ± 9 | Ag | 501 ± 27 | Ag (1 µM, 43) | >1,000 |
| P49 | [Iaa1,d-Tyr2,Ala22]PACAP38 | Ag | 98 ± 2 | Ag | 389 ± 33 | Ag (1 µM, 54) | 832 ± 63 | Ag | 501 ± 36 |
| P51 | [Iaa1,d-Ala2,Ala22]PACAP38 | Ag | 9.12 ± 0.67 | Ag | 20.4 ± 1.5 | Ag | 7.10 ± 0.47 | Ag | 323 ± 20 |
| P29 | [Iac1,Ala16,17,d-Lys38]PACAP38 | Ag | 0.05 ± 0.00 | Ag | 1.45 ± 0.08 | Ag | 7.08 ± 0.44 | Ag | 2.53 ± 0.07 |
| P28 | [N-Ac-His1,Ala16,17,d-Lys38]PACAP38 | Ag | 0.09 ± 0.01 | Ag | 0.95 ± 0.12 | Ag | 7.08 ± 0.71 | Ag | 3.02 ± 0.10 |
| P8 | [N-Ac-His1,Pip3,Aib16,Ala17]PACAP27 | No Act | >1,000 | pAg (24) | >1,000 | No Act | >1,000 | Ag (1 µM, 13) | >1,000 |
| P39 | [Pip3,Ala14,17,Aib16,28,Lys34,d-Lys38]PACAP38 | Ag (1 µM, 14) | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P38 | [Pip3,Ala15,17,Aib16,28,Lys34,d-Lys38]PACAP38 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P7 | [Pip3,Aib16,Ala17]PACAP27 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P35 | [Pip3,Aib16,28,Ala17,Lys34,d-Lys38]PACAP38 | Ag (1 µM, 81) | 407 ± 20 | Ag (1 µM, 48) | >1,000 | No Act | >1,000 | Ag (1 µM, 33) | >1,000 |
| P41 | [Pip3,Aib16,28,Ala17,20,Lys34,d-Lys38]PACAP38 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P40 | [Pip3,Aib16,28,Ala17,21,Lys34,d-Lys38]PACAP38 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P21 | [β-Ala3,Ala16,17]PACAP38 | Ag (1 µM, 61) | 602 ± 37 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P20 | [Hyp3,Ala16,17]PACAP38 | Ag | 139 ± 10 | pAg (25) | >1,000 | No Act | >1,000 | Ag (1 µM, 35) | >1,000 |
| P31 | [Hyp3,Aib16Ala17,Lys34]PACAP38 | Ag | 1.51 ± 0.10 | Ag (1 µM, 80) | 155 ± 13 | Ag (1 µM, 55) | >1,000 | Ag | 155 ± 11 |
| P33 | [Me-Asp3,Aib16,28,Lys34,d-Lys38]PACAP38 | Ag | 1.00 ± 0.06 | Ag | 16.2 ± 1.3 | Ag | 47.9 ± 2.6 | Ag | 23.4 ± 0.9 |
| P37 | [Ini3,Aib16,28,Ala17,Lys34,d-Lys38]PACAP38 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P36 | [Nip3,Aib16,28,Ala17,Lys34,d-Lys38]PACAP38 | Ag (1 µM, 39) | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P32 | [Ala15,17,Aib16,28,Lys34,d-Lys38]PACAP38 | Ag | 1.38 ± 0.07 | Ag | 1.95 ± 0.16 | Ag | 30.2 ± 2.9 | Ag | 7.41 ± 0.39 |
| P23 | [Ala16,17,d-Lys38]PACAP38 | Ag | 0.35 ± 0.02 | Ag | 3.80 ± 0.33 | Ag | 12.9 ± 1.0 | Ag | 5.37 ± 0.36 |
| P30 | [Aib16,Ala17,Lys34]PACAP38 | Ag | 0.25 ± 0.02 | Ag | 1.38 ± 0.21 | Ag | 3.24 ± 0.24 | Ag | 3.80 ± 0.18 |
| P34 | [Aib16,28,Ala17,Lys34,d-Lys38]PACAP38 | Ag | 0.19 ± 0.01 | Ag | 1.58 ± 0.08 | Ag | 30.2 ± 1.9 | Ag | 2.63 ± 0.15 |
cAMP generation was determined in the indicated cell type loaded with [3H]adenine for 48 hours as described under Materials and Methods. Results are expressed as a percentage of the maximal stimulation of cAMP accumulation caused by 10 µM PACAP38, which was calculated using the curve-fitting program KaleidaGraph. The EC50 was calculated as the concentration causing half-maximal stimulation with each analog. In each experiment, each value was determined in duplicate, and values given are and means S.E.M. from at least three separate experiments. Abbreviations: see Fig. 1, Table 1 and Table 2 legends.
Figure 1.
Positions of amino acid substitutions in PACAP38. The positions of substitutions made either alone or in combination. Abbreviations used: Ac, acetyl; Aib, α-amino-isobutyric acid (α-methylalanine); Ala, alanine; Arg, arginine; Asn, asparagine; Asp, aspartic acid; Gln, glutamine; Gly, glycine; His, histidine; Hyp, hydroxyproline; Iaa, 4-imidazole acetic acid; Iac, 4-imidazole acrylic acid; Ini, isonipecotic acid (piperidine-4-carboxylic acid); Ile, isoleucine; Leu, leucine; Lys, lysine; Me, methyl; Met, methionine; Nip, nipecotic acid (piperidine-3-carboxylic acid); PACAP, pituitary adenylate cyclase-activating polypeptide; Phe, phenylalanine; Pip, pipecolic acid (piperidine-2-carboxylic acid); Pro, proline; Pya, 3-pyridyl acetic acid; Sar, sarcosine (N-methylglycine); Ser, serine; Thr, threonine; Tyr, tyrosine; Val, valine.
3.1. Binding Affinity of PACAP38, PACAP27 and VIP (Tables 1 and 2, Fig. 2)
TABLE 2.
Affinities of various multi-substituted PACAP-related analogs for PAC1-R, VPAC1-R and VPAC2-R.
| Peptide Number | Peptide structure | IC50 (nM) |
||||
|---|---|---|---|---|---|---|
| PAC1-R |
VPAC1-R |
VPAC2-R |
||||
| PAC1-R | VPAC1-R/ PANC-1 |
T47D | VPAC2-R/ PANC-1 |
Sup-T1 | ||
| PACAP27 | 2.82 ± 0.17 | 0.23 ± 0.09 | 0.20 ± 0.07 | 4.79 ± 0.20 | 24.5 ± 1.0 | |
| PACAP38 | 0.28 ± 0.02 | 1.35 ± 0.04 | 1.29 ± 0.05 | 1.00 ± 0.11 | 6.03 ± 0.30 | |
| VIP | >1,000 | 0.14 ± 0.01 | 0.20 ± 0.02 | 6.03 ± 0.30 | 16.6 ± 1.0 | |
| P48 | [Iaa1,d-Ser2,Ala22]PACAP38 | 32.4 ± 1.3 | 426 ± 19 | 200 ± 9 | 85.1 ± 6.3 | 295 ± 16 |
| P49 | [Iaa1,d-Tyr2,Ala22]PACAP38 | 123 ± 8 | 251 ± 18 | 371 ± 22 | 55 ± 4 | 200 ± 16 |
| P51 | [Iaa1,d-Ala2,Ala22]PACAP38 | 58.9 ± 3.6 | 105 ± 7 | 55.2 ± 2.2 | 42 ± 3 | 162 ± 10 |
| P29 | [Iac1,Ala16,17,d-Lys38]PACAP38 | 0.30 ± 0.02 | 23.4 ± 1.9 | 15.5 ± 1.0 | 2.75 ± 0.11 | 5.13 ± 0.26 |
| P28 | [N-Ac-His1,Ala16,17,d-Lys38]PACAP38 | 1.45 ± 0.03 | 17.4 ± 1.0 | 16.2 ± 0.6 | 1.82 ± 0.16 | 0.32 ± 0.02 |
| P8 | [N-Ac-His1,Pip3,Aib16,Ala17]PACAP27 | >1,000 | 436 ± 28 | 139 ± 5 | 525 ± 11 | >1,000 |
| P39 | [Pip3,Ala14,17,Aib16,28,Lys34,d-Lys38]PACAP38 | >1,000 | >1,000 | >1,000 | 847 ± 17 | >1,000 |
| P38 | [Pip3,Ala15,17,Aib16,28,Lys34,d-Lys38]PACAP38 | >1,000 | >1,000 | >1,000 | 708 ± 39 | >1,000 |
| P7 | [Pip3,Aib16,Ala17]PACAP27 | >1,000 | 363 ± 19 | 239 ± 5 | >1,000 | >1,000 |
| P35 | [Pip3,Aib16,28,Ala17,Lys34,d-Lys38]PACAP38 | 631 ± 38 | 832 ± 40 | >1,000 | 398 ± 18 | 660 ± 20 |
| P41 | [Pip3,Aib16,28,Ala17,20,Lys34,d-Lys38]PACAP38 | >1,000 | >1,000 | >1,000 | >1,000 | >1,000 |
| P40 | [Pip3,Aib16,28,Ala17,21,Lys34,d-Lys38]PACAP38 | >1,000 | >1,000 | >1,000 | >1,000 | >1,000 |
| P21 | [β-Ala3,Ala16,17]PACAP38 | 100 ± 5 | 794 ± 39 | 646 ± 14 | 263 ± 14 | 323 ± 14 |
| P20 | [Hyp3,Ala16,17]PACAP38 | 114 ± 3 | 707 ± 39 | 427 ± 22 | 100 ± 8 | 380 ± 15 |
| P31 | [Hyp3,Aib16,Ala17,Lys34]PACAP38 | 51.3 ± 2.4 | 363 ± 21 | 478 ± 31 | 92 ± 3 | 100 ± 4 |
| P33 | [Me-Asp3,Aib16,28,Lys34,d-Lys38]PACAP38 | 36.3 ± 3.4 | 42.7 ± 2.4 | 53.7 ± 3.5 | 50 ± 1 | 30.2 ± 1.3 |
| P37 | [Ini3,Aib16,28,Ala17,Lys34,d-Lys38]PACAP38 | 851 ± 34 | >1,000 | >1,000 | 100 ± 4 | 794 ± 63 |
| P36 | [Nip3,Aib16,28,Ala17,Lys34,d-Lys38]PACAP38 | 933 ± 65 | >1,000 | >1,000 | 436 ± 15 | >1,000 |
| P32 | [Ala15,17,Aib16,28,Lys34,d-Lys38]PACAP38 | 100 ± 6 | 87.1 ± 2.4 | 692 ± 26 | 84 ± 3 | 42.7 ± 1.1 |
| P23 | [Ala16,17,d-Lys38]PACAP38 | 1.15 ± 0.05 | 0.16 ± 0.01 | 0.21 ± 0.01 | 3.16 ± 0.21 | 5.62 ± 0.28 |
| P30 | [Aib16,Ala17,d-Lys34]PACAP38 | 3.39 ± 0.14 | 3.39 ± 0.13 | 3.16 ± 0.14 | 3.63 ± 0.16 | 2.95 ± 0.07 |
| P34 | [Aib16,28,Ala17,Lys34,d-Lys38]PACAP38 | 6.30 ± 0.31 | 5.37 ± 0.35 | 11.7 ± 0.5 | 6.31 ± 0.20 | 7.76 ± 0.34 |
The indicated cell type was incubated with 50 pM 125I-labeled peptide and various concentrations of the unlabeled PACAP27 and PACAP38 analog as described under Materials and Methods. The IC50 was the concentration causing half-maximal inhibition of the saturable binding, calculated using the curve-fitting program KaleidaGraph. In each experiment each value was determined in duplicate, and values given are means and S.E.M. from at least three separate experiments. Abbreviations: see Fig. 1 and Table 1 legends.
Figure 2.
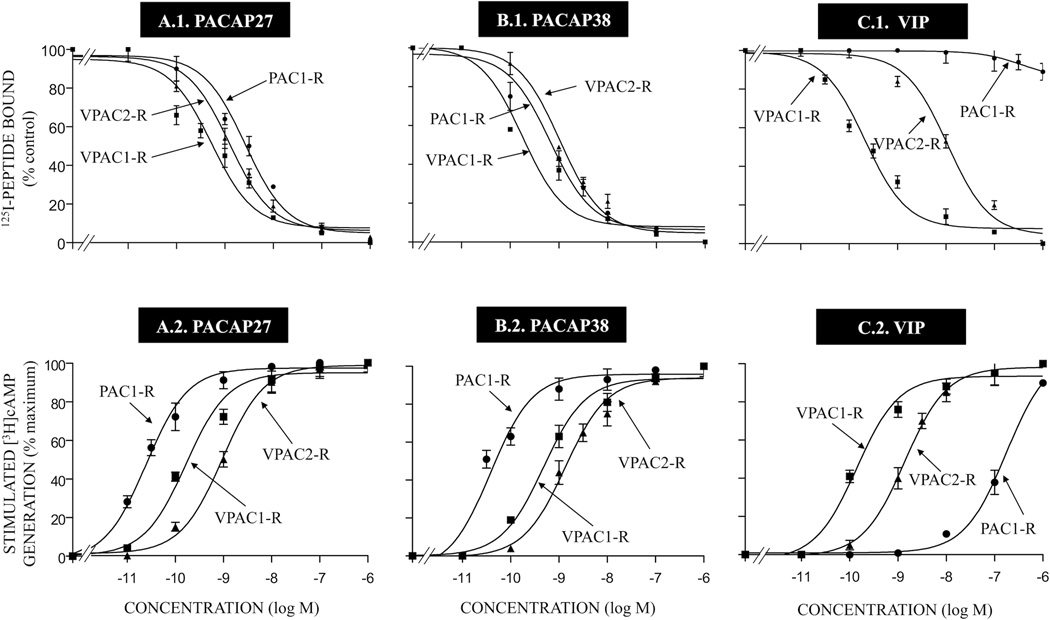
Comparison of the ability of PACAP27, PACAP38 and VIP to inhibit binding of 125I-labeled peptide [upper panels] and their ability to stimulate [3H]cAMP accumulation [lower panels] in cells containing PAC1, VPAC1, VPAC2 receptors. Receptor binding was performed with 50 pM 125I-ligand as described in Materials and Methods for PAC1-R cells, VPAC1-R/PANC-1 cells and VPAC2-R/PANC-1 cells. Results are expressed as the percentage of saturable binding without unlabeled peptide added (percent control). Stimulation of cAMP accumulation [lower panels] was performed as described in Materials and Methods using 2 µCi/ml [3H]adenine. Values are expressed as percentage of maximal stimulated [3H]cAMP accumulation by 1 µM PACAP38. Control and 1 µM PACAP38 (as maximum) stimulated values for PAC1-R cells were 5328 ± 828 and 21717 ± 8759 dpm, respectively; for VPAC1-R/PANC-1 cells, 2020 ± 364 and 10084 ± 5213, respectively and for VPAC2-R/PANC-1 cells, 4444 ± 814 and 28951 ± 5213 dpm, respectively. The results are the mean and S.E.M. from at least three separate experiments. Abbreviations: see legends for Fig. 1 and Table 1.
To initially investigate the abilities of the naturally occurring peptides PACAP38, PACAP27 and VIP to interact with PAC1-R, VPAC1-R and VPAC2-R, we determined their affinities for each receptor subtype by performing binding studies (Fig. 2). Binding studies were performed using both transfected cells (PAC1 in NIH 3T3, VPAC1 in PANC-1 and VPAC2 in PANC-1) and two cells lines possessing native VPAC1-R (T47D cells) [23] or native VPAC2-R (Sup T-1) [23] (Tables 1 and 2). PACAP38 had the highest affinity for PAC1-R cells (IC50: 0.28 nM; Table 1, Fig. 2A.1), but also had high affinity for VPAC1-R/PANC-1 cells (IC50: 1.35 nM; Table 1, Fig. 2A.1) and VPAC2-R/PANC-1 cells (IC50: 1.00 nM, Table 1, Fig. 2A.1). PACAP27 had a 10-fold lower affinity than PACAP38 for PAC1-R cells (IC50: 2.82 nM, Table 1, Fig. 2B.1), but had a 6-fold higher affinity than PACAP38 for VPAC1-R/PANC-1 cells (IC50: 0.23 nM; Table 1, Fig. 2B.1) and also had a high affinity for VPAC2-R/PANC-1 cells (IC50: 4.8 nM; Table 1, Fig. 2B.1). In contrast, VIP had a very low affinity for the PAC1 receptor (IC50: >1,000 nM; Table 1, Fig. 2C.1), but had a high affinity for VPAC1-R/PANC-1 cells and VPAC2-R/PANC-1 cells (IC50: 0.14–6 nM, Table 1, Fig. 2C.1). These results agree with previous studies that have reported [11,13,16,41,61] that PACAP38 and PACAP27 bind with high affinity to all three PACAP-VIP receptors (PAC1-R, VPAC1-R and VPAC2-R).
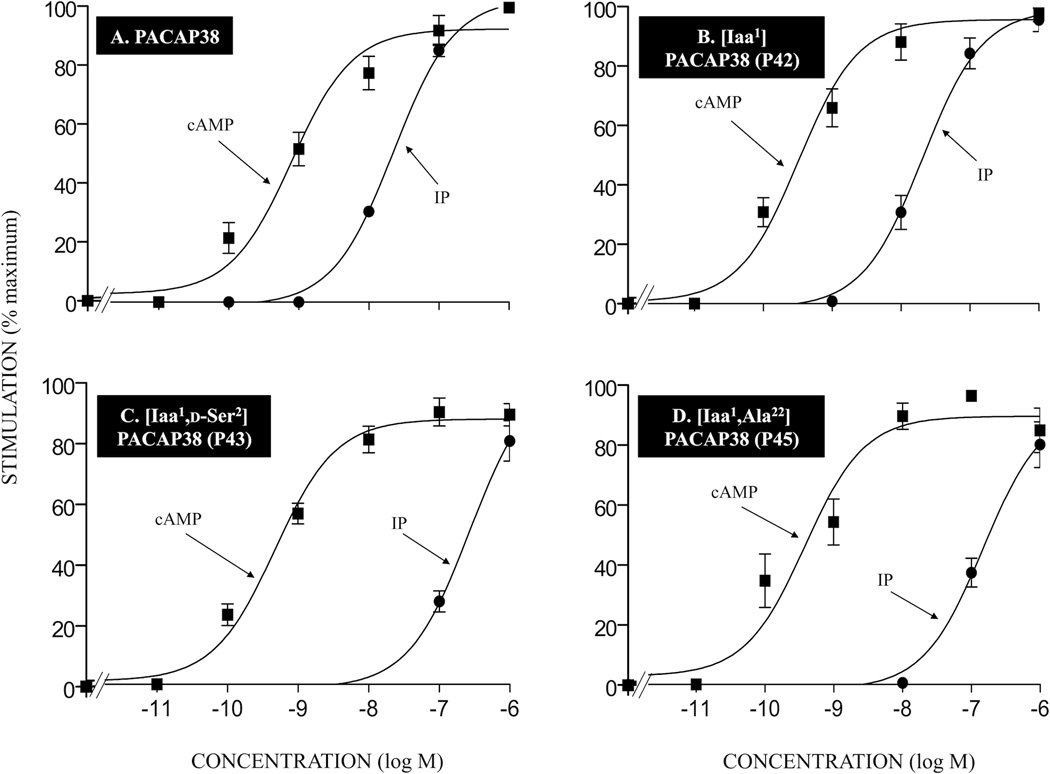
3.2.1. Affinities of single- or double-substituted PACAP-analogs compared to PACAP38 (or PACAP27) for PAC1-R (Table 1, Fig. 3)
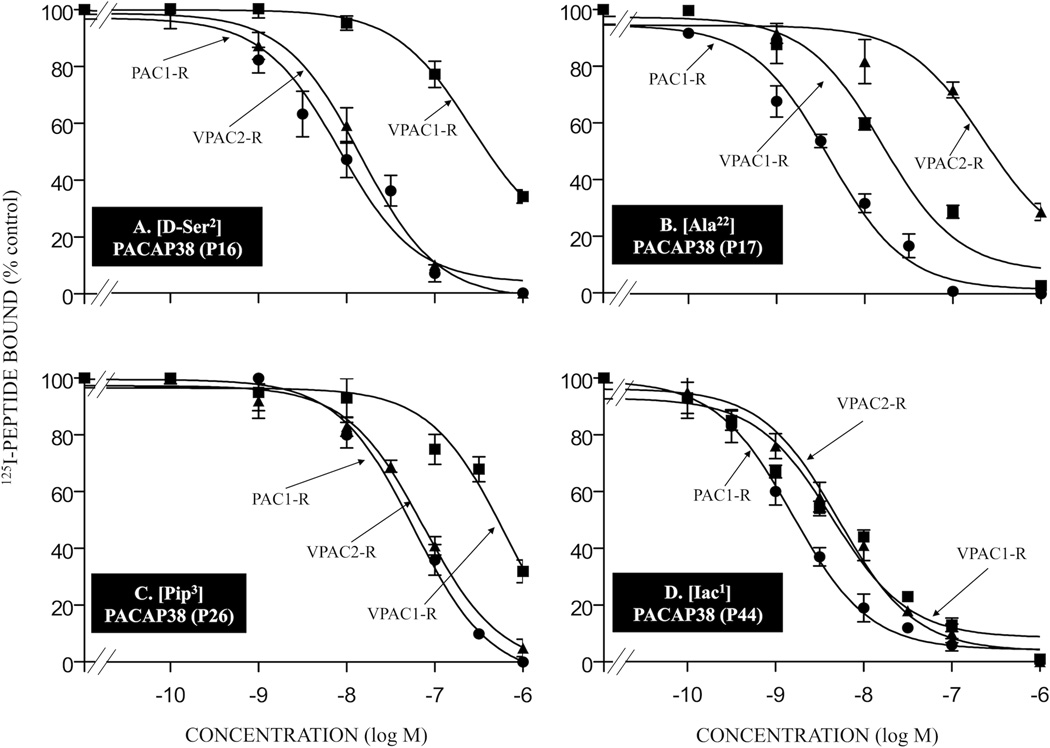
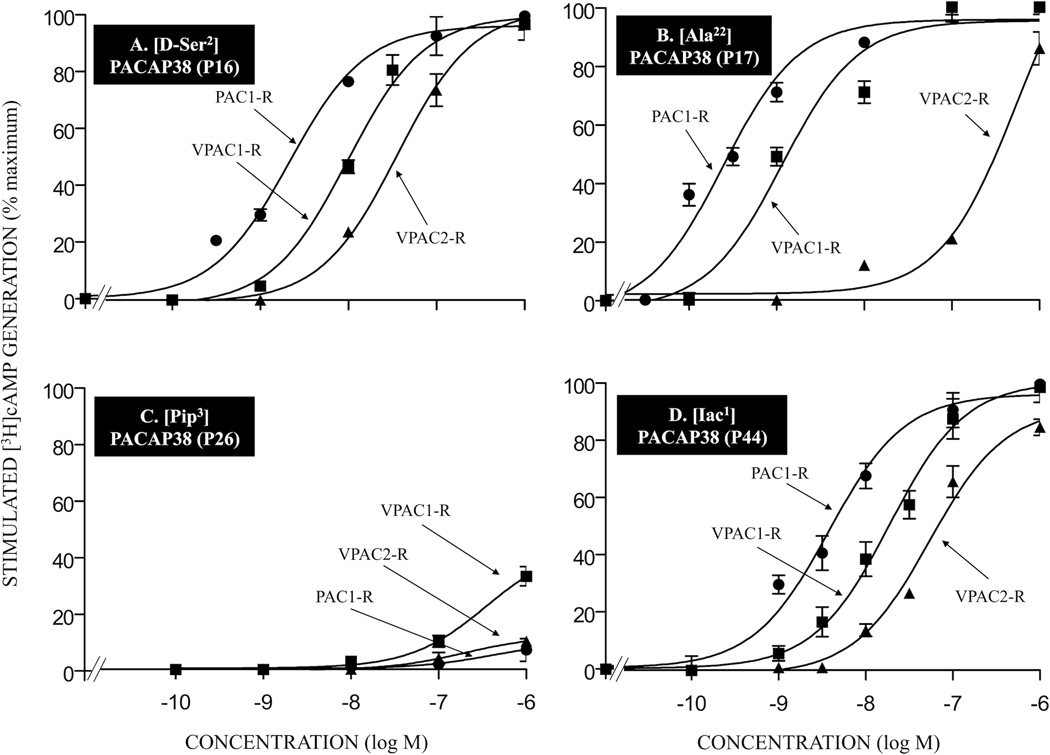
Figure 3.
Affinities of various single amino acid substituted PACAP-related analogs for PAC1-R, VPAC1-R and VPAC2-R. Briefly, cells containing PAC1-R, VPAC1-R and VPAC2-R were incubated for 60 min at room temperature with 50 pM 125I-ligand, with or without the indicated concentrations of unlabeled single amino acid substituted PACAP38 analogs. The results for the PACAP-analogs P16, P17, P26, and P44 are shown in Table 1. The experimental conditions and expression of the results are the same as in the legend for Fig. 2. Abbreviations: see legend for Table 1.
Only one of the 18 single- or double-substituted PACAP-analogs had an affinity equal to native PACAP38 for PAC1-R cells (analog P42; IC50: 0.28 nM; Table 1) and one VIP analog showed similar affinity to native VIP for PAC1-R cells (analog P50; IC50: >1,000 nM, Table 1). Four PACAP38 analogs showed a 5–10-fold decrease in affinity compared to PACAP38 for PAC1-R cells (analogs P24, P43, P44, and P45; IC50: 1.4–2.2 nM; Table 1, Fig. 3D, 4B and 4C). Seven PACAP38 analogs had a 11–30-fold decrease in affinity for PAC1-R cells (analogs P12, P13, P14, P15, P16, P17, and P41; IC50: 3.7–7.8 nM; Table 1, Fig. 3A and 3B) and two PACAP27 analogs showed a 15–25-fold lower affinity for PAC1-R cells (analogs P4 and P22; IC50: 55–69 nM, Table 1). Six PACAP38 analogs showed a 100–300-fold decrease in affinity for PAC1-R cells (analogs P18, P19, P25, P26, P27, and P47; IC50: 32–83 nM; Table 1, Fig. 3C). The remaining three PACAP27 analogs had >1,000-fold decrease in affinity compared with PACAP27 in PAC1-R cells (analogs P2, P5 and P6; IC50: >1,000 nM; Table 1).
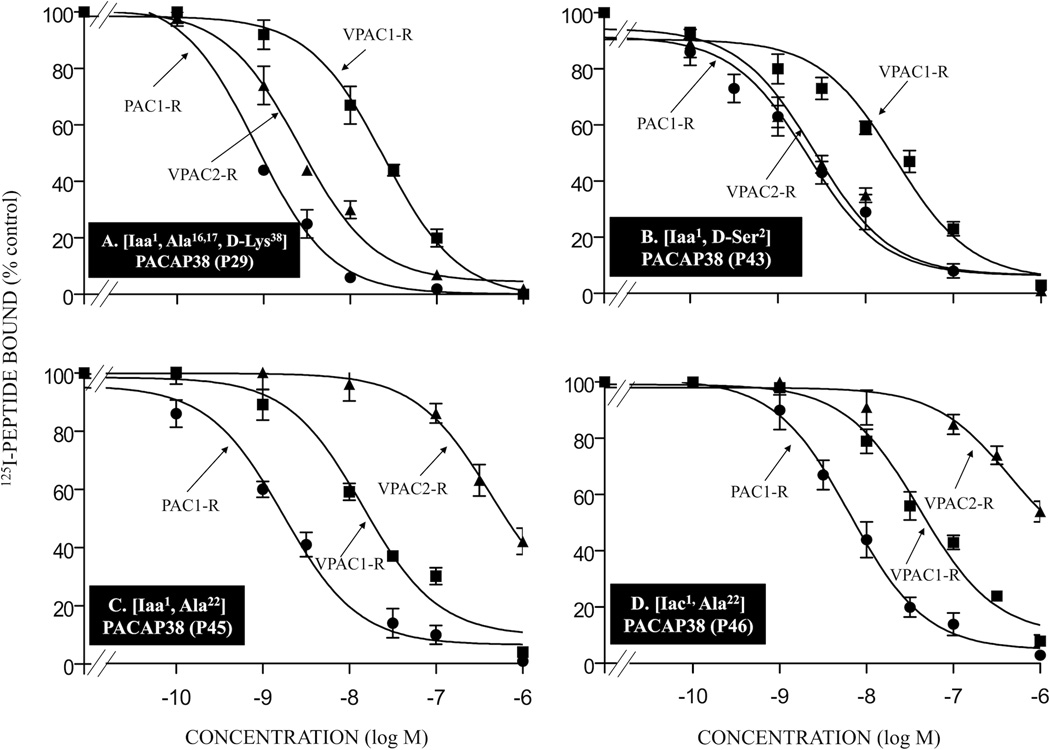
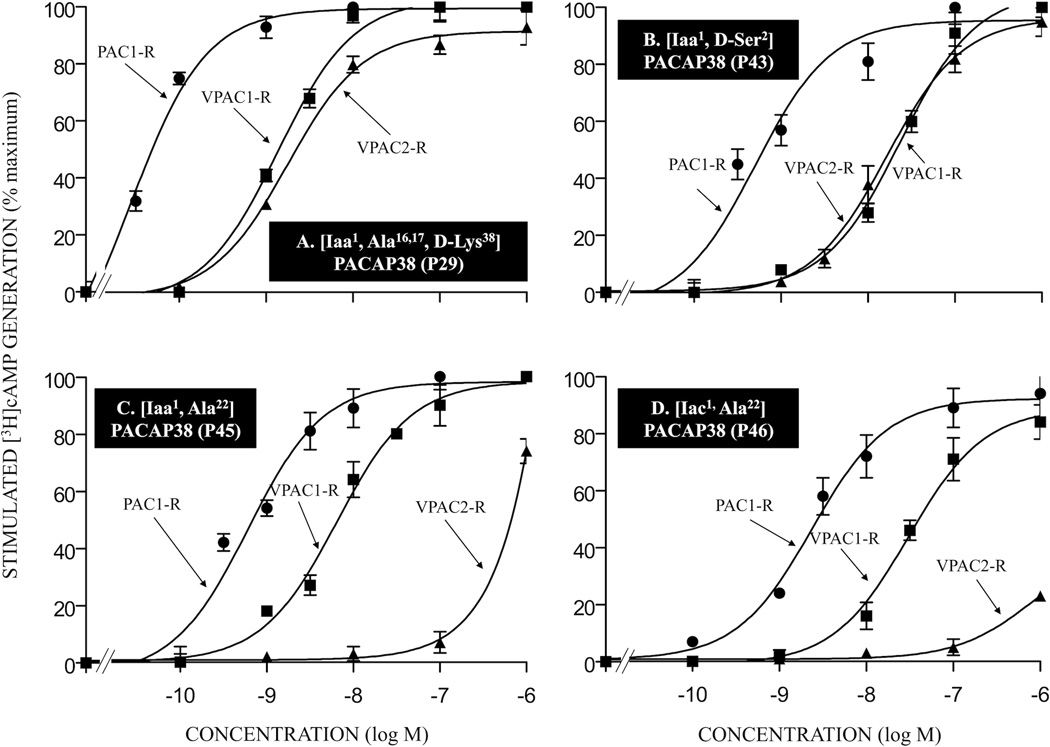
Figure 4.
Affinities of various double- or multi-substituted PACAP-related analogs for PAC1-R, VPAC1-R and VPAC2-R. The experimental conditions were similar to those in the legend for Fig. 2 and Fig. 3. The values are the mean and S.E.M. from at least three separate experiments. The results for PACAP-analogs P29, P43, P45, and P46 are shown in Tables 1 and 2. Abbreviations: see legends for Fig. 1 and Table 1.
3.2.2. Affinities of multiple-substituted PACAP-analogs compared to PACAP38 (or PACAP27) related peptides for PAC1-R cells (Table 2, Fig. 3)
To try to improve the possible PAC1 selectivity and maintain the affinity for PAC1, multi-substitutions were made in PACAP38, PACAP27 or VIP (Table 2). Only one of the 20 multi-substituted PACAP38 analogs or 2 substituted PACAP27 analogs had similar affinities to native PACAP38 or PACAP27 for PAC1-R cells (analog P29; IC50: 0.3 nM; Table 2, Fig. 4A). However, two PACAP38 analogs had a 4–5-fold decrease in affinity for PAC1-R cells compared to PACAP38 (analogs P23 and P28; IC50: 1 nM, Table 2). Two PACAP38 analogs showed a 10–30-fold lower affinity for PAC1-R cells compared to PACAP38 (analogs P30 and P34; Table 1). Four PACAP38 analogs showed a 100–300-fold lower affinity for PAC1-R cells than PACAP38 (analogs P31, P33, P48, and P51; IC50: 32–59 nM; Table 2) and the other four PACAP38 analogs showed a greater decrease (>350-fold) in affinity for PAC1-R cells compared to PACAP38 (analogs P20, P21, P32, and P49; IC50: 100–123 nM; Table 2). The seven remaining PACAP38 analogs (analogs P35, P36, P37, P38, P39, P40, and P41; IC50: 631 to >1,000 nM; Table 2) and the two PACAP27 analogs (P7 and P8; IC50: >1,000 nM; Table 2) had a >1,000-fold decrease in affinity compared with PACAP38 or PACAP27 in PAC1-R cells.
3.3.1. Affinities of single- or double-substituted PACAP-analogs compared to PACAP38 (or PACAP27) for VPAC1-R or VPAC2-R (Table 1, Fig. 3)
Only one PACAP38 analog had an affinity similar to PACAP38 for VPAC1-R/PANC-1 cells (analog P24, IC50: 0.96 nM, Table 1) and one PACAP38 analog had similar affinity to PACAP38 for VPAC2-R/PANC-1 cells (analog P42; IC50: 1 nM; Table 1). Five PACAP38 analogs showed 2–10-fold lower affinity than PACAP38 for VPAC1-R/PANC-1 cells (analogs P12, P13, P15, P42, and P44; IC50: 2–12 nM; Table 1, Fig. 3D). Three of these PACAP38 analogs showed a similar decrease for VPAC2-R/PANC-1 cells (analogs P15, P24 and P44; IC50: 2.0–9.4 nM; Table 1, Fig. 3D), whereas the other two PACAP38 analogs had a 16–21-fold decrease compared to PACAP38 (analogs P12 and P13, IC50: 15.8–31.6 nM, Table 1). Six PACAP38 analogs had a 11–40-fold lower affinity than PACAP38 for VPAC1-R/PANC-1 cells (analogs P14, P17, P25, P27, P43, and P45; IC50: 15–54 nM; Table 1, Fig. 3B and 4B) and one PACAP27 analog showed a 37-fold decrease in affinity compared to PACP27 for VPAC1-R/PANC-1 cells (analog P4; IC50: 8.5 nM; Table 1). Two of these PACAP38 analogs showed a similar decrease for VPAC2-R/PANC-1 cells (analogs P25 and P43; IC50: 22–38 nM, Table 1, Fig. 4B), whereas the other four PACAP38 analogs had a 55 to >1,000-fold lower affinity for VPAC2-R/PANC-1 cells (analogs P14, P15, P17, and P46; IC50: 55 to >1,000 nM; Table 1, Fig. 3B and 4D) and one PACAP27 analog had a 7-fold decrease in affinity for PACAP27 in VPAC2-R/PANC-1 cells (analog P4; IC50: 31.6 nM, Table 1). The remaining six PACAP38 and four PACAP27 analogs had a 100–500-fold lower affinity for VPAC1-R/PANC-1 cells than PACAP38 (analogs P16, P18, P19, P26, P27, and P47; IC50: 15–575 nM; Table 1, Fig. 3A and 3C) or PACAP27 (analogs P2, P5, P6, and P22; IC50: 251 to >675 nM; Table 1). These six PACAP38 analogs had 15–500-fold lower affinity than PACAP38 for VPAC2-R/PANC-1 cells (analogs P16, P18, P19, P26, P27, and P47; IC50: 14–500 nM; Table 1, Fig. 3A and 3C) and these four PACAP27 analogs had a 6–132-fold decrease in affinity compared to PACAP27 for VPAC2-R/PANC-1 cells (analogs P2, P5, P6, and P22; IC50: 31–631 nM; Table 1). One VIP analog had >500-fold decrease in affinity for VPAC1-R/PANC-1 cell and VPAC2-R/PANC-1 cell (analog P50; IC50: >500 nM; Table 1).
3.3.2. Affinities of multiple substituted PACAP-analogs compared to PACAP38 (or PACAP27) for VPAC1-R or VPAC2-R (Table 2, Fig. 4)
One PACAP38 analog had greater affinity than PACAP38 for VPAC1-R/PANC-1 cells (analog P23, IC50: 0.16 nM, Table 2). Four PACAP38 analogs had a 3–20-fold decrease in affinity compared to PACAP38 for VPAC1-R/PANC-1 cells (analogs P28, P29, P30, and P34; IC50: 3–23 nM; Table 2, Fig. 4A). However, these five analogs showed 2-to-6-fold lower affinity than PACAP38 for VPAC2-R/PANC-1 cells (analogs P23, P28, P29, P30, and P34; IC50: 2–6 nM; Table 2, Fig. 4A). Three PACAP38 analogs had a 30-to-100-fold lower affinity for VPAC1-R/PANC-1 cells than PACAP38 (analogs P32, P33 and P51; IC50: 43–105 nM; Table 2) and a 40–85-fold decrease in affinity for VAPC2/PANC-1 cells compared to PACAP38 (analog P33; IC50: 42–84 nM, Table 2). Six PACAP38 analogs had 180-to-620-fold low affinity for VPAC1-R/PANC-1 cells than PACAP38 (analogs P20, P21, P31, P35, P48, and P49; IC50: 251–832 nM; Table 2), and these analogs also showed a 50–400-fold decrease in affinity for VPAC2-R/PANC-1 cells compared to PACAP38 (analogs P20, P21, P31, P35, P48, and P49; IC50: 55 to >1,000 nM; Table 2). The six remaining analogs showed a >1,000-fold lower affinity for VPAC1-R/PANC-1 cells compared to PACAP38 (analogs P36, P37, P38, P39, P40, and P41; IC50: >1,000 nM; Table 2) or PACAP27 (analogs P7 and P8; IC50: 363–436 nM; Table 2) and 100- to >1,000-fold lower affinity for VPAC-2/PANC-1 cells compared to PACAP38 (analogs P36, P37, P38, P39, P40, and P41; IC50: 100-to->1,000 nM; Table 2) or PACAP27 (analogs P7 and P8; IC50: 525-to ->1,000 nM, Table 2).
In general, there was a close correlation between the binding affinities found for the different PACAP-analogs from studies on VPAC1/PANC1 cells and from studies of VPAC1 receptors occurring naturally in T47 breast cancer cells (r = 0.94, p < 0.0001, Tables 1,2). Similarly, despite the different radiolabeled ligands and experimental conditions (incubation at room temperature or 37°C), there was also a close correlation with binding of the various PACAP-analogs to VPAC2/PANC1 cells or the VPAC2 Sup TI cells natively expressing these receptors (r = 0.68, p < 0.0001), Tables 1,2) However, in the later case the absolute affinities determined on VPAC2 Sup T 1 cells was on the average 3.0 ± 0.4 lower than seen on VPAC2/PANC1 cells, likely due to the different ligands/binding conditions used (Tables 1 and 2) .
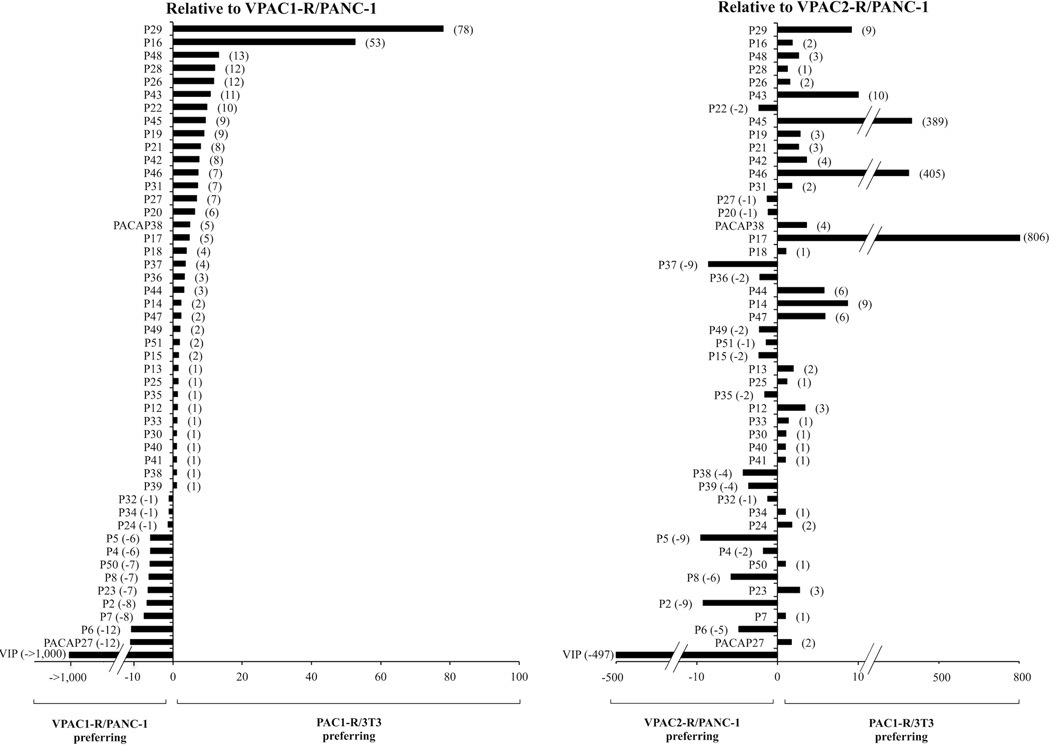
3.4. Relative affinities of single, double or multi-substituted PACAP-analogs for PAC1-R compared to VPAC1-R/PANC-1 or VPAC2-R/PANC-1 cells (Fig. 5)
Figure 5.
Relative affinities of PACAP-related analogs for VPAC1-R/PANC-1 or VPAC2-R/PANC-1 cells compared to affinity for PAC1-R cells. The results are expressed as the ratio of the affinity (IC50) for VPAC1-R/PANC-1(Left panel) or VPAC2-R /PANC-1(Right panel) cells divided by the affinity (IC50) for PAC1-R cells. The results are ordered by selectivity for PAC1-R cells compared to VPAC1-R PANC-1.
These results are summarized in Fig. 5 where the relative affinities are expressed as a ratio of the affinity for VPAC1-R/PANC-1 or VPAC2-R/PANC-1 cells (IC50) compared to PAC1-R . In terms of the relative affinities for PAC1-R, VPAC1-R and VPAC2-R, two analogs had a 53–78-fold higher affinity for PAC1-R cells over VPAC1-R/PANC-1 (analogs P16 and P29; Tables 1 and 2, Fig. 3A, 4A and 5). These two analogs showed a 2–9-fold greater affinity for PAC1-R cells than for VPAC2-R/PANC-1 cells (analogs P16 and P29; Tables 1 and 2, Fig. 3A, 4A and 5). Four analogs had an 11–13-fold higher affinity for PAC1-R cells than VPAC1-R/PANC-1 cells (analogs P26, P28, P43, and P48; Tables 1 and 2, Fig. 3C, 4B and 5). Three of these four analogs showed a 2–10-fold greater affinity for PAC1-R cells than for VPAC2-R/PANC-1 cells (analogs P26, P43 and P48; Tables 1 and 2, Fig. 3C, 4B and 5) and one had same affinity for PAC1-R cells and VPAC2-R/PANC-1 cells (analog P28; Table 2, Fig. 5). Nine analogs showed a 6–10-fold selectivity for PAC1-R cells over VPAC1-R/PANC-1 cells (analogs P19, P20, P21, P22, P27, P31, P42, P45, and P46; Tables 1 and 2, Fig. 4C, 4D and 5). Six of these analogs had greater affinity (2–405-fold) for PAC1-R cells over VPAC2-R/PANC-1 cells (analogs P19, P21, P31, P42, P45, and P46; Tables 1 and 2, Fig. 4C, 4D and 5), whereas two analogs had same affinity for PAC1-R cells and VPAC2-R/PANC-1 cells (analogs P20 and P27; Tables 1 and 2, Fig. 5) and one analog had a 2-fold decrease in affinity for PAC1-R cells compared to VPAC2-R/PANC-1 cells (analog P22; Table 1, Fig. 5). Ten analogs and PACAP38 showed a 2–5-fold selectivity for PAC1-R cells over VPAC1-R/PANC-1 cells (analogs P14, P15, P17, P18, P36, P37, P44, P47, P49, and P51; Tables 1 and 2, Fig. 3B, 3D and 5). One of these analogs, compared to VPAC2-R/PANC-1 cells, had a >800-fold selectivity for PAC1-R cells (analog P17; Table 1, Fig. 3B and 5), whereas three analogs and PACAP38 had a 4–9-fold greater affinity for PAC1-R cells (analogs P14, P44 and P47; Table 1, Fig. 3B and 5), two analogs had similar affinities for PAC1-R cells and VPAC2-R/PANC-1 cells (analog P18 and P51, Tables 1 and 2) and four analogs showed a 2–9-fold lower affinity for PAC1-R cells than for VPAC2-R/PANC-1 cells (analog P15, P36, P37, P49, and P51; Tables 1 and 2, Fig. 5). Thirteen analogs had similar affinities for VPAC1-R/PANC-1 cells and PAC1-R cells (analogs P12, P13, P24, P25, P30, P32, P33, P34, P35, P38, P39, P40, and P41; Tables 1 and 2, Fig. 5). Similarly, seven of these three analogs (analogs P25, P30, P32, P33, P34, P40, and P41; Table 1, Fig. 5) had similar affinities for PAC1-R cells and VPAC2-R/PANC-1 cells, whereas three showed a slight selectivity (2–3-fold) for PAC1-R cells over VPAC2-R/PANC-1 cells (analogs P12, P13 and P24; Table 1, Fig. 5) and the other three analogs showed the opposite effect with a 2–4-fold higher affinity for VPAC2-R/PANC-1 cells than for PAC1-R cells (analogs P35, P38 and P39; Table 2, Fig. 5). Seven analogs had a 6–10-fold lower affinity for PAC1-R cells than VPAC1-R/PANC-1 cells (analogs P2, P4, P5, P7, P8, P23, and P50; Tables 1 and 2, Fig. 5). However, one of these analogs showed a higher affinity (2-fold) for PAC1-R cells over VPAC2-R/PANC-1 cells (analog P23; Table 2, Fig. 5), two analogs had similar affinity for PAC1-R cells and VPAC2-R/PANC-1 cells (analog P7 and P50; Tables 1 and 2, Fig. 5) and four analogs showed a 2–9-fold decrease in affinity for PAC1-R cells than for VPAC2-R/PANC-1 cells (analogs P2, P4, P5, and P8; Tables 1 and 2, Fig. 5). One analog and PACAP27 showed 12-fold lower affinity for PAC1-R cells compared to VPAC1-R/PANC-1 cells (analog P6; Table 1, Fig. 5). This analog had a 5-fold decrease in affinity for PAC1-R cells than for VPAC1-R/PANC-1 cells (analog P6; Table 1, Fig. 5), whereas PACAP27 showed a 2-fold greater affinity for PAC1-R cells over VPAC2-R/PANC-1 cells. VIP had a >1,000-fold decrease in affinity for PAC1-R cells than VPAC1-R/PANC-1 cells and >400-fold decrease for VPAC2-R/PANC-1 cells.
3.5. Potency of PACAP38, PACAP27 and VIP (Tables 3 and 4, Fig. 2)
TABLE 3.
The potency of various single or double substituted PACAP-related analogs for PAC1-R, VPAC1-R and VPAC2-R
| Peptide Number |
Peptide structure | [3H]cAMP |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| PAC1-R |
VPAC1-R |
VPAC2-R |
|||||||
| PAC1-R |
VPAC1-R/PANC-1 |
T47D |
VPAC2-R/PANC-1 |
||||||
| % max | EC50 (nM) | % max | EC50 (nM) | % max | EC50 (nM) | % max | EC50 (nM) | ||
| PACAP27 | Ag | 0.03 ± 0.01 | Ag | 0.20 ± 0.01 | Ag | 0.79 ± 0.07 | Ag | 1.00 ± 0.04 | |
| PACAP38 | Ag | 0.03 ± 0.01 | Ag | 0.54 ± 0.05 | Ag | 3.24 ± 0.19 | Ag | 1.38 ± 0.10 | |
| VIP | No Act | 174 ± 10 | Ag | 0.18 ± 0.01 | Ag | 1.00 ± 0.07 | Ag | 1.51 ± 0.07 | |
| P42 | [Iaa1]PACAP38 | Ag | 0.40 ± 0.01 | Ag | 2.63 ± 0.37 | Ag | 5.75 ± 0.73 | Ag | 5.13 ± 0.42 |
| P44 | [Iac1]PACAP38 | Ag | 13.8 ± 0.7 | Ag (1 µM, 85) | 2.63 ± 1.50 | Ag | 224 ± 16 | Ag | 66 ± 4 |
| P43 | [Iaa1,d-Ser2]PACAP38 | Ag | 0.50 ± 0.01 | Ag | 2.63 ± 1.80 | Ag | 56.2 ± 3.9 | Ag | 20.1 ± 1.1 |
| P27 | [N-Ac-His1,Pip3]PACAP38 | No Act | >1,000 | Ag (1 µM, 72) | 182 ± 9 | Ag (1 µM, 18) | >1,000 | Ag (1 µM, 18) | >1,000 |
| P45 | [Iaa1,Ala22]PACAP38 | Ag | 0.60 ± 0.05 | Ag (1 µM, 74) | 2.63 ± 0.46 | Ag | 118 ± 10 | Ag | 550 ± 17 |
| P46 | [Iac1,Ala22]PACAP38 | Ag | 2.46 ± 0.22 | Ag | 2.63 ± 2.70 | Ag (1 µM, 23) | 69 ± 6 | Ag (1 µM, 23) | >1,000 |
| P47 | [Pya1,Ala22]PACAP38 | Ag | 10.00 ± 0.54 | Ag | 2.63 ± 1.30 | Ag | 199 ± 16 | Ag | 126 ± 6 |
| P15 | [Aib2]PACAP38 | Ag | 0.46 ± 0.02 | Ag | 2.19 ± 2.24 | Ag | 16.6 ± 0.9 | Ag | 11.7 ± 0.7 |
| P4 | [d-Ser2]PACAP27 | Ag | 0.47 ± 0.03 | Ag | 1.58 ± 0.19 | pAg (90) | 3.39 ± 0.29 | Ag | 7.41 ± 0.32 |
| P16 | [d-Ser2]PACAP38 | Ag | 2.57 ± 0.07 | Ag | 11 ± 1 | Ag | 19.9 ± 1.8 | Ag | 33.1 ± 1.9 |
| P14 | [Aib2,Lys(palmitoyl)38]PACAP38 | Ag | 0.45 ± 0.02 | pAg (80) | 9.12 ± 0.56 | pAg (62) | 6.68 ± 0.28 | Ag | 3.31 ± 0.20 |
| P13 | [d-Ser2,Lys(palmitoyl) 38]PACAP38 | Ag | 0.15 ± 0.01 | Ag | 4.07 ± 0.27 | pAg (78) | 5.25 ± 0.25 | Ag | 3.47 ± 0.20 |
| P26 | [Pip3]PACAP38 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P5 | [Pro3]PACAP27 | Ag (1 µM, 55) | 776 ± 67 | Ag (1 µM, 23) | >1,000 | Ag (1 µM, 16) | >1,000 | No Act | >1,000 |
| P18 | [Pro3]PACAP38 | Ag | 129 ± 6 | Ag (1 µM, 40) | >1,000 | No Act | >1,000 | Ag (1 µM, 26) | >1,000 |
| P6 | [Sar3]PACAP27 | Ag (1 µM, 63) | 602 ± 25 | Ag | 22.9 ± 0.8 | Ag (1 µM, 69) | 776 ± 49 | No Act | >1,000 |
| P19 | [Sar3]PACAP38 | Ag | 96 ± 6 | Ag | 182 ± 23 | Ag (1 µM, 53) | 891 ± 49 | Ag (1 µM, 35) | >1,000 |
| P25 | [Pro3,Lys34]PACAP38 | Ag | 19.4 ± 1.2 | Ag | 204 ± 9 | Ag (1 µM, 60) | 7.24 ± 0.42 | Ag | 252 ± 13 |
| P22 | [Sar4]PACAP27 | Ag (1 µM, 50) | >1,000 | No Act | >1,000 | No Act | >1,000 | No Act | >1,000 |
| P2 | [Me-Lys15]PACAP27 | Ag | 190 ± 12 | Ag (1 µM, 68) | 692 ± 50 | No Act | >1,000 | Ag (1 µM, 19) | >1,000 |
| P17 | [Ala22]PACAP38 | Ag | 0.32 ± 0.11 | Ag | 1.07 ± 0.06 | Ag | 41.7 ± 3.9 | Ag | 282 ± 12 |
| P24 | [Lys34]PACAP38 | Ag | 0.29 ± 0.01 | Ag | 2.88 ± 0.16 | pAg (85) | 5.50 ± 0.35 | Ag | 2.57 ± 0.17 |
| P12 | [Lys(palmitoyl)38]PACAP38 | Ag | 0.21 ± 0.02 | Ag | 5.01 ± 0.35 | Ag | 5.45 ± 0.42 | Ag | 5.37 ± 0.25 |
| P50 | [Pro3]VIP | No Act | >1,000 | Ag (1 µM, 68) | 324 ± 32 | Ag (1 µM, 68) | 468 ± 22 | Ag (1 µM, 52) | 933 ± 21 |
cAMP generation was determined in the indicated cell type loaded with [3H]adenine for 48 hours as described under Materials and Methods. Results are expressed as a percentage of the maximal stimulation of cAMP accumulation caused by 10 µM PACAP38, which was calculated using the curve-fitting program KaleidaGraph. The EC50 was calculated as the concentration causing half-maximal stimulation with each analog. In each experiment, each value was determined in duplicate, and values given are means and S.E.M. from at least three separate experiments. Abbreviations: Ag, Full Agonist = PACAP38; Ag (1 µM, % maximum), % agonist activity at max concentration of 1 µM, still increasing; No Act, No agonist activity at 1 µM; pAg (% maximum), partial Agonist ; others-see Fig. 1 and Table 1 legends.
To investigate the abilities of the natural PACAP-related peptides (PACAP27, PACAP38 and VIP) as well as the substituted PACAP-analogs, to activate each of the three PACAP-VIP-related receptors, we determined their abilities to stimulate adenylate cyclase, by assessing cAMP generation. The results are summarized in Tables 3 and 4 from the studies that were performed using both transfected cells (PAC1-R in BALB 3T3, VPAC1-R in PANC-1 and VPAC2-R in PANC-1) and one cell line possessing native VPAC1-R (T47D cells) [23] (Tables 3 and 4). cAMP studies were not performed in the Sup-T1 cell line containing native VPAC2-R [23], because of the low stimulation obtained made it difficult to determine dose-response curves.
Each of the natural peptides, PACAP27, PACAP38 and VIP, had similar efficacy in stimulating cAMP in PAC1-R cells, VPAC1-R/PANC-1 and VPAC2-R/PANC-1 cells (Tables 3 and 4, Fig. 2). Specifically, each stimulated a 4.1 ± 1.5-fold maximal increase in PAC1-R cells, 5.0 ± 0.7-fold increase in VPAC1-R/PANC-1 cells and 6.5 ± 2.0-fold increase in VPAC2-R/PANC-1 cells (Tables 3 and 4, Fig. 2). PACAP38 and PACAP27 were equipotent at stimulating PAC1-R cells (EC50: 0.03 nM), whereas VIP was >800-fold less potent. In contrast, with VPAC1-R/PANC-1 cells and VPAC2-R/PANC-1 cells, each of the three peptides was potent (EC50: 0.18–0.54 nM VPAC1-R/PANC-1 cells and 1–1.5 nM VPAC2-R/PANC-1 cells, Tables 3 and 4, Fig. 2).
3.6.1. Potencies of single-, double- or multi-substituted PACAP-analogs compared to PACAP38 (or PACAP27) for PAC1-R (Tables 3 and 4, Fig. 6, 7 and 8)
Figure 6.
The potency of various single substituted PACAP-analogs for PAC1-R, VPAC1-R and VPAC2-R. Briefly, stably transfected PAC1-R, VPAC1-R/PANC-1 or VPAC2-R/PANC-1 cells were preincubated for 48 hours with [3H]adenine, washed and incubated for 60 minutes at 37°C with or without the indicated concentrations of the various PACAP38 analogs. The experimental conditions and expression of results were as describe in the legend Fig. 2. Values are means and S.E.M. from at least three separate experiments. The results for PACAP-related analogs P16, P17, P26, and P44 are shown in Table 3. Abbreviations: see legends for Fig. 1 and Table 1.
Figure 7.
The potency of various double- or multi-substituted PACAP-analogs for PAC1-R, VPAC1-R and VPAC2-R. The experimental conditions were similar to those in the legends for Fig. 2 and Fig. 6. The values are the mean and S.E.M. from at least three separate experiments. The results for PACAP-analogs P29, P43, P45, and P46 are shown in Tables 2 and 3. Abbreviations: see legends for Fig. 1, Table 1 and Table 2.
Figure 8.
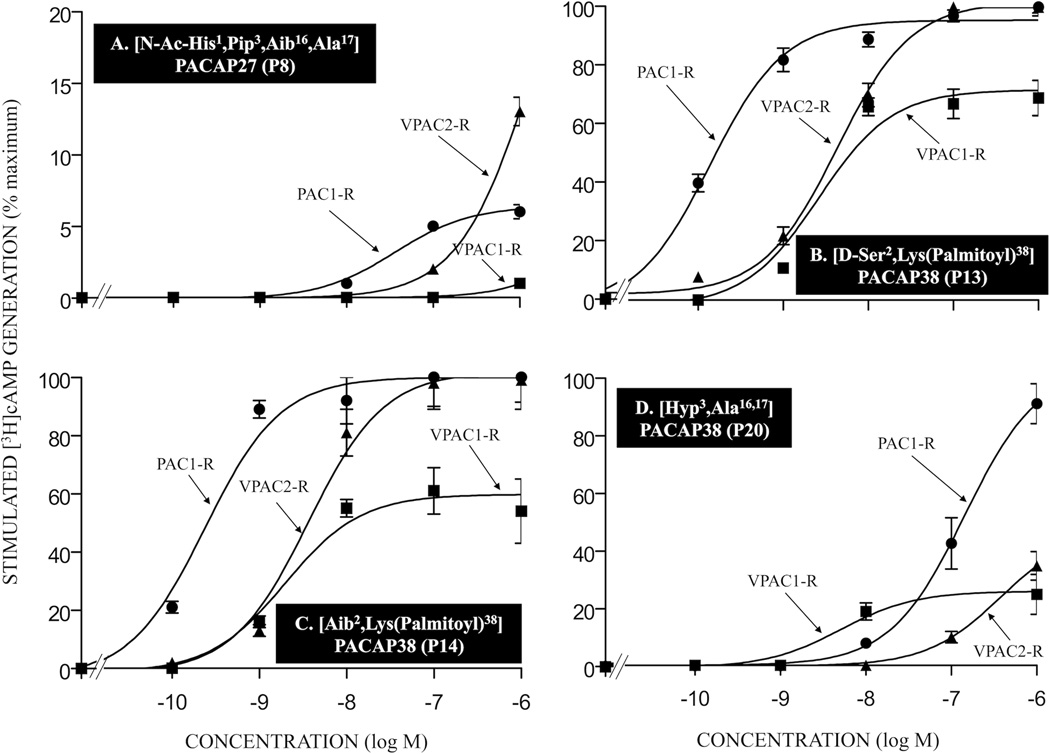
The potency of various double- or multi-substituted PACAP-analogs for PAC1-R, VPAC1-R and VPAC2-R. The experimental conditions were similar to those in the legend to Fig. 2 and Fig. 6. Values are the mean and S.E.M. from at least three separate experiments. The results for PACAP-analogs P8, P13, P14, and P20 are shown in Tables 1 and 2. Abbreviations: see legends for Fig. 1, Table 1 and Table 2.
To investigate the effect of the amino acid substitutions on efficacy, we determined the ability of each PACAP-analog to maximally stimulate adenylate cyclase by PAC1, VPAC1 and VPAC2 receptor activation with concentrations up to 1 µM (Tables 3 and 4, Fig. 5, 6, 7 and 8). In PAC1-R cells, twenty-eight of the thirty-eight PACAP38 analogs and two of the seven PACAP27 analogs were full agonists (Tables 3 and 4, Fig. 6 and 7). None of the ten remaining PACAP38 related peptides were partial agonists. Six of these ten PACAP38 analogs (P26, P27, P37, P38, P40, and P41; Table 3 and 4, Fig. 6C), two PACAP27 analogs (P7 and P8; Table 4) and one VIP analog (P50; Table 3) had no agonist activity at up to 1 µM. The remaining four PACAP38 analogs (P21, P35, P36 and P39; Tables 3 and 4) and three PACAP27 analogs (P5, P6 and P22; Table 3) had agonist activity reaching 14 to 81% of maximal stimulation, which was still increasing in magnitude at a 1 µM concentration, the maximal peptide concentration that could be studied. In VPAC1-R/PANC-1 cells, twenty-two of the thirty-eight PACAP38 analogs and two of the seven PACAP27 analogs were full agonists (Tables 3 and 4, Fig. 5, 6, 7 and 8). Two PACAP38 analogs (P14 and P20; Tables 3 and 4, Fig. 8C) and one PACAP27 analog (P8; Table 4, Fig. 8A) were partial agonists stimulating 24, 25 and 80% of maximum. Eight PACAP38 analogs (P21, P26, P36, P37, P38, P39, P40, and P41; Tables 3 and 4, Fig. 6C) and two PACAP27 analogs (P7 and P22; Tables 3 and 4) had no agonist activity at up to 1 µM. Six PACAP38 analogs (P18, P27, P31, P35, P44, and P45; Tables 3 and 4, Fig. 6D and 7C), two PACAP27 analogs (P2 and P5, Table 3) and one VIP analog (P50, Table 3), had agonist activity of 23 to 85% of maximum, which was still increasing in magnitude at 1 µM. In VPAC2-R/PANC-1 cells, twenty-three of the thirty-eight PACAP38 analogs and one of the seven PACAP27 analogs were full agonists (Tables 3 and 4, Fig. 5, 6, 7 and 8) and none were partial agonists. Eight PACAP38 analogs (P21, P26, P36, P37, P38, P39, P40, and P41; Tables 3 and 4, Fig. 6C) and four PACAP27 analogs (P5, P6, P7, and P22; Tables 3 and 4) had no agonist activity at up to 1 µM. Seven PACAP38 analogs (P18, P19, P20, P27, P35, P46, and P48; Tables 3 and 4, Fig. 6D), two PACAP27 analogs (P2 and P8; Tables 3 and 4, Fig. 8A) and one VIP analog (P50, Table 3) had agonist activity of 13 to 52% of maximal with stimulation still increasing in magnitude at 1 µM. For the five PACAP38 analogs and one PACAP27 analog with no activity at 1 µM at any of the three receptors, no antagonist was identified using either 1 nM or 10 nM PACAP38 as an agonist and 1 µM of the possible antagonist in any of the cells, likely due to their low affinities for the receptors.
3.6.2. Potencies of single-, double- or multi-substituted PACAP-analogs compared to PACAP38 (or PACAP27) for VPAC1-R orVPAC2-R (Table 3 and 4, Fig. 6, 7 and 8)
To assess the selectivity of the analogs for PAC1-R cells over VPAC1-R/PANC-1 or VPAC2-R/PANC-1 cells based on their potencies for activation of each receptor, we also determined full dose-response curves for each analog to stimulate generation of cAMP for each of the VPAC cells. Analog 31 had the greatest increase (103-fold) in overall selectivity for PAC1-R cells over both VPAC1-R/PANC-1 and VPAC2-R/PANC-1 cells (Table 4). Eight analogs had a 16–50-fold higher potency for PAC1-R cells compared to VPAC1-R/PANC-1 cells (analogs P12, P13, P14, P18, P20, P29, P33, and P48; Tables 3 and 4, Fig. 7A, 8B, 8C and 8D). One of these eight analogs had a >1,000-fold increase in potency for PAC1-R cells over VPAC2-R/PANC-1 cells (analog 48, Table 4), six had a 22–55-fold greater potency for PAC1-R cells than for VPAC2-R/PANC-1 cells (analogs P12, P13, P18, P20, P29, and P33; Tables 3 and 4, Fig. 7A, 8B and 8D) and one analog showed 7.4-fold increase in potency for PAC1-R cells over VPAC2-R/PANC-1 cells (analog P14; Table 3, Fig. 8C). Eight showed 6–15-fold increase in potency for PAC1-R cells than for VPAC1-R/PANC-1 cells (analogs P23, P24, P25, P28, P30, P34, P35, and P42; Tables 3 and 4). One of these eight analogs had a 34-fold increase in potency for PAC1-R cells over VPAC2-R/PANC-1 cells (analog P28, Table 4) and seven analogs showed 7-to-15-fold higher potency for PAC1-R cells than for VPAC2-R/PANC-1 cells (analogs P23, P24, P25, P30, P34, P35, and P42; Tables 3 and 4). Twelve analogs showed 2-to-5-fold increase in potency for PAC1-R cells than for VPAC1-R/PANC-1 cells (P2, P4, P5, P15, P16, P17, P19, P21, P43, P45, P49, and P51; Tables 3 and 4, Fig. 6A, 6B, 7B and 7C). Two of these analogs had >800-fold higher potency for PAC1-R cells compared to VPAC2-R/PANC-1 cells (analogs P17 and P45, Tables 3 and 4, Fig. 6B and 7C), seven analogs showed 12–140-fold increase in potency in PAC1-R cells than for VPAC2-R/PANC-1 cells (analogs P2, P4, P15, P16, P19, P43, and P51; Tables 3 and 4, Fig. 6A and 7B) and three analogs had 2–5-fold greater potency for PAC1-R cells than for VPAC2-R/PANC-1 cells (analogs P5, P21, and P49; Tables 3 and 4). Twelve analogs had equal potency for PAC1-R cells and VPAC1-R/PANC-1 cells (analogs P7, P8, P22, P26, P32, P36, P37, P38, P39, P40, and P41; Tables 3 and 4, Fig. 6C and 7D). One of these twelve analogs showed >1,000-fold higher potency for PAC1-R cells compared to VPAC2-R/PANC-1 cells (analog P46; Table 2, Fig. 7D); one had a 5-fold increase in potency for PAC1-R cells than for VPAC2-R/PANC-1 cells (analog P32, Table 4) and the ten remaining analogs had similar potency for PAC1-R cells and VPAC2-R/PANC-1 cells (analogs P7, P8, P22, P26, P36, P37, P38, P39, P40, and P41; Tables 3 and 4, Fig. 6C).
In general, there was a strong correlation between the potencies for stimulating cAMP generation for each of the single, double or multi-substituted PACAP analogs for VPAC1-R in VPAC1-R/PANC-1 cells and T47D cells naturally expressing VPAC1-R (r = 0.792, p < 0.0001). There was an overall significant correlation (r = 0.594, p < 0.0001) between the ability of each single, double or multi-substituted PACAP-related analog to stimulate cAMP accumulation in PAC1-R cells and its affinity from binding studies for PAC1 in BALB cells. In addition, significant correlations existed between the potencies of the different analogs to stimulate increases in cAMP accumulation and their receptor affinity in VPAC1-R/PANC-1 cells (r = 0.704, p < 0.0001) and VPAC2-R/PANC-1 cells (r = 0.324, p < 0.02). However, the relationship between affinity for receptor occupation and potencies of receptor activation (i.e. coupling ratio) varied markedly for the different receptors and different analogs. In PAC1-R cells, the average coupling ratio (binding affinity/potency for cAMP) for all PACAP-related analogs was 11.69 ± 3.22, which was twice as great as VPAC1-R/PANC-1 cells (6.59 ± 1.97) and about 10-fold higher than VPAC2-R/PANC-1 cells (1.68 ± 0.48), demonstrating marked differences in receptor sparseness. This was also seen for PACAP27 and PACAP38 because in PAC1-R cells, the coupling ratio (IC50-binding affinity/EC50-cAMP generation) was 94- and 17-fold, demonstrating that minimal receptor occupation causes maximal cAMP generation, whereas with VPAC1-R/PANC-1 cells their coupling ratios were 1.2 for PACAP27 and 2.5-fold for PACAP38 and with VPAC2-R/PANC-1 cells the ratios were was 4.8 (PACAP27) and 1.1 (PACAP38). In PAC1-R cells, there was a wide variation in the coupling ratio with the PACAP-analogs varying from 116- to-1-fold in thirty-five analogs with greater potencies than affinities, whereas eleven analogs had 1.2- to 64-fold greater affinities than potencies (analogs P18, P19, P20, P21, P22, P26, P27, P36, P37, and P44; Tables 1, 2, 3, and 4). In VPAC1-R/PANC-1 cells, thirty-one analogs had a coupling ratio varying from 71- to 1-fold with greater potencies than affinities, whereas fifteen analogs had a 1.2–65.2-fold greater affinity than potencies for stimulating cAMP generation (analogs P2, P5, P7, P8, P18, P20, P21, P22, P23, P24, P25, P26, P35, P42, and P49; Tables 1, 2, 3, and 4). In VPAC2-R/PANC-1 cells, the coupling ratio varied from 17- to 1-fold in the eighteen analogs with greater potency than affinity, whereas twenty-eight analogs had from 99- to 1-fold higher affinity than potency for stimulating cAMP generation (analogs P2, P5, P6, P8, P15, P16, P18, P19, P20, P21, P22, P23, P24, P25, P26, P27, P28, P31, P35, P36, P37, P38, P39, P42, P44, P49, and P51; Tables 1, 2, 3 and 4).
3.7. Potencies of PACAP38, PACAP27 and selected PACAP-analogs to activate phospholipase C (Fig. 9)
Figure 9.
Comparison of the ability of PACAP38 and various single- or double-substituted PACAP-analogs to stimulate [3H]IP generation and cAMP generation in PAC1-R cells. For stimulation of cAMP, the cells were incubated with each ligand at the indicated concentration for 60 minutes. cAMP generation was determined in the indicated cell type loaded with [3H]adenine for 48 hours as described under Materials and Methods. [3H]IP was determined after a 60-minute incubation at 37°C after loading the cells for 48 hours with 3 mCi/ml myo-[2-3H]inositol as described in Materials and Methods. The control and maximal stimulated [3H]IP values for PAC1-R was 4047 ± 261 and 145873 ± 9962, respectively. The control and maximal cAMP generation values for PAC1-R was 5328 ± 828 and 21717 ± 8759, respectively. The values are the mean and S.E.M. from at least three separate experiments. The results are shown for PACAP38 and PACAP-analogs P42, P43 and P45. Abbreviations: see legends for Fig. 1, Table 1 and Table 2.
Previous studies report that activation of PAC1-R[12,20,44,51], in contrast to VPAC1-R and VPAC2-R [54,55], is also coupled to activation of phospholipase C (PLC). To investigate the ability of selected PACAP-analogs to activate both adenylate cyclase and stimulated PLC activation, we determined their abilities to stimulate increases in cAMP or generation of inositol phosphate (IP) in the same cells under similar conditions. We determined the full dose-response curves for activation of each cell line for PACAP38 and the selective PACAP related analogs (P42, P43 and P45; Tables 1, 2, 3, and 4). In PAC1-R , PACAP38 stimulated the generation of cAMP and [3H]IP, and each of the three PACAP-analogs was fully efficacious (Fig. 9). PACAP38 caused a half-maximal increase in [3H]IP at 10 nM (EC50: 21.44 nM; Fig. 9A) in PAC1-R cells and was 19-fold less potent at stimulating PLC than generation of cAMP (EC50: 1.15 nM; Fig. 9A). The PACAP-analogs P42, P43 and P45 caused half-maximal increases in [3H]IP at 10, 100 and 100 nM and were 25, 377 and 106-fold less potent at stimulating [3H]IP than cAMP (IP EC50: 19.34, 256 and 140 nM; cAMP EC50: 0.78, 0.68 and 1.32; Fig. 9B, 9C and 9D).
4. Discussion
The purpose of this study was to develop a selective PAC1-R agonist based on a detailed structure-activity analysis of VIP/PACAP-analogs. This was undertaken because of the importance of this receptor in numerous physiological/pathophysiological conditions [20,43,54,55] and because despite many structure-function studies of PACAP/VIP-analogs[7–11,13,16,20,22,23,54,55], no selective PAC1-R agonist has been identified. The only selective agonist available to investigators to study the role of PAC1-R in these processes is maxadilan[53], which was isolated from salivary gland of sand flies[29]; however, it is a 61-amino-acid peptide with three disulfide bonds that is not structurally related to PACAP/VIP. Unfortunately, maxadilan is difficult to synthesize and is, therefore, rarely used[20].
The selectivity of ligands for PAC1-R is a particular problem because, as seen in this study and numerous others[20,54], the naturally occurring agonists, PACAP27 and PACAP38, both have high affinities for all three of the receptor subtypes mediating the action of VIP-PACAP (i.e., PAC1-R,VPAC1-R and VPAC2-R). Furthermore, VIP and PACAP27 have high sequence identity (68%)[34] and have similar secondary structures[26,38,58,59], likely contributing to the difficulty in identifying selective PAC1-R analogs. In our study, the strategy used was based on the reported secondary structure of PACAP/VIP[26,38,52,58,59], previous data from structure-function studies of VIP-PACAP[8,10,13,23–25,40,49] and structure-proteolysis studies[7] and was primarily aimed at trying to develop a selective PACAP38-analog with particular emphasis on separating PAC1-R and VPAC1-R selectivity (Table 5). This approach was taken for a number of reasons. First, numerous studies exploring the secondary structure of PACAP provide evidence that the initial 5–9 amino acids have a disordered structure, followed by an α-helix for amino acids 9–27, and finally by amino acids 28–38 at the COOH terminus, which can also adopt an α-helical conformation[26,38,52,58,59]. High-affinity receptor interactions and the ability to activate any of the VIP-PACAP receptors is dependent on the amino terminus[10,13,20,22,49]. Because the NH2-terminus can adopt many different conformations with certain ones having high affinity for the PAC1-R, our approach was to systematically explore the initial 5 residues of PACAP using substitutions that resulted in conformationally-restricted analogs. We also explored the results of selective substitutions in other sites that might conformationally-restrict the PACAP-analog, which could affect degradation or could extend its half-life in the circulation, such as the attachment of long-chain fatty-acids[7,21,39]. The latter approach was included because PACAP38 has a plasma half-life of <5–10 minutes[4,4,7,31], which restricts its therapeutic potential. PACAP38 was concentrated on for a number of reasons. PACAP38 is the main native peptide in both brain (90%) and gastrointestinal tract, many of PACAP’s potential therapeutic uses are related to CNS disease and PACAP38, but not PACAP27, is transported into the CNS by a saturable-transporter[4,14]. Furthermore, PACAP38 is more stable in the CNS than PACAP27[4,14]. PACAP38 has greater specificity because it does not interact with formyl receptors, as does PACAP27[28], furthermore, in some systems it is more potent than PACAP27 and can activate signaling cascades not activated by PACAP27[5,20]. Lastly, we concentrated on identifying analogs separating PAC1-R from VPAC1-R because both receptors are widely distributed, especially in the CNS[20,54,55] and up to the present time, no PACAP-selective VIP/PACAP-analogs have been identified, whereas a few studies have reported VIP/PACAP-analogs that have some selectivity(< 50-fold) for PAC1-R/VPAC1-R over VPAC2-R[13,16].
Table 5.
Summary of most important findings.
|
A number of our results support the conclusion that for the first time we were successful at identifying PACAP-analogs that had PAC1-R selectivity over VPAC1-R (Table 5). First, in binding-studies, [Iac1,Ala16,17,Lys38]PACAP38 (P29) and [D-Ser2]PACAP38 (P16) had 78-fold and 53-fold, respectively, greater affinity for PAC1-R than VPAC1-R. Second, 5 analogs ([Iaa1,D-Ser2]PACAP38(P43), [Pip3]PACAP38 (P26), [N-Ac-His1,Ala16,17,D-Lys38]PACAP38 (P28), [Iac1,D-Ser2,Ala22]PACAP38(P48), and [Sar4]PACAP38(P22) had a 10–15 fold greater affinity for PAC1-R than VPAC1-R. Third, activation of each of the 3 VIP/PACAP receptors results in the stimulation of adenylate- cyclase and generation of cAMP as one of the main signaling cascades[20,27,37,44,51,62]. Five of the 7 PAC1-R selective-analogs described above on binding studies activated adenylate-cyclase (i.e., the analogs [Sar4]PACAP38(P22) and [Pip3]PACAP38(P26) were inactive), were also selective at activating PAC1-R over VPAC1-R varying from 6- to-250-fold. Furthermore, 4 other PACAP-analogs ([Hyp3,Aib16,Ala17,Lys34]PACAP38 (P31), [Me-Asp3,Aib16,28,Lys34,D-Lys38]PACAP3(P33), [Aib2,Lys(palmitoyl)38]PACAP38 (P14), and [D-Ser2,Lys(palmitoyl)38]PACAP38(P13)) that varied from 1.5-to-7-fold selective in binding studies, showed greater selectivity in activating PAC1-R varying from 15-to-666-fold. This divergence between the degrees of selectivity of various PACAP-analogs for PAC1-R over VPAC1-R from binding studies and cell activation studies occurred because of the wide variation in the degree of receptor-spareness for the different analogs reflected in the difference between their abilities to occupy the receptors (binding-IC50) and activate the receptors (EC50-cAMP) (Table 5). Even though for both PAC1-R and VPAC1-R, the IC50 and the EC50’s for the different analogs significantly correlated(p<0.0001), the degree of receptor-spareness for the different analogs varied from 0- to 116-fold for the PAC1-R and from 0-to-71-fold for VPAC1-R. Furthermore, the degree of receptor-spareness of a given analog for the PAC1-R did not correlate with the degree of receptor-spareness with the VPAC1-R. The result of this was that some analogs, such as the latter 4 above (i.e., P13,P14,P31,P33) showed much greater selectivity based on functional assays than on receptor-binding assays. This result shows that potentially useful receptor selective analogs can be missed in structure-function studies that examine only receptor binding or biological activity.
Our comparative structure-function study of PAC1-R and VPAC1-R showed a number of important points, some of which have been reported in previous studies. First, we found, as reported by others[7,10,22,49], the effect of a given substitution in PACAP27/PACAP38 can be markedly different. For example, [D-Ser2]PACAP38 (P16) was 53-fold selective for PAC1-R over VPAC1-R,whereas the [D-Ser2]PACAP27 (P4) was not more selective for VPAC1-R,which is similar to its only 3-fold PAC1-selectivity reported in a previous study[13]. Similar discordant results were seen in the relative selectivity with [D-Pro3]PACAP38 (P18) and [D-Pro3]PACAP27 (P5). Second, our results show that degree of selectivity of an analog with multiple substitutions may not be predicted from the individual substitutions. This is best shown with [Aib16,17,D-Lys38]PACAP38 (P23) and [Iac1]PACAP38(P44), which had minimal to no selectivity for PAC1-R; however, the combination analog,[Iac1,Aib16,17,D-Lys38]PACAP38 (P29) had the greatest selectivity of any analog in the present study (i.e.,78-fold). Third, our results are in agreement with previous studies[7,10,13,22] that replacement of serine in position 2 by nonbulky groups or by substitutions that altered conformation (i.e., Aib2,D-Phe2) had only a minimal effect on PAC1-R affinity/potency, whereas it had a marked effect on decreasing potency/affinity for the VPAC1-R and thus, could have an important effect on selectivity. Fourth, our results show that the attachment of a fatty-acid on the COOH-terminus lysine [Lys(palmitoyl)38] (P12, P13 and P14) had only a minimal effect on PAC1-affinity and thus could be an important substitution for PAC1-R selective agonists in the future. One of the main problems with the potential therapeutic use of PACAP is its short half-life[4,31]; however, recent studies with other peptides (insulin, glucagon,glucagon-like peptide 1) show that e attachment of long-chain fatty acids greatly extends their half-lives[21,39,56].
Although this study was primarily aimed at identifying VIP/PACAP-analogs with selectivity for PAC1 over VPAC1, their selectivity for PAC1 over VPAC2 was also examined (Table 5). Whereas, a number of selective VPAC1-R agonists ([Ala11,22,28]VIP, [Ala2,8,9,16,19,24]VIP and [Lys15,Arg16,Leu27]VIP(1–7)/GHRH(8–27)) and selective VAC2 agonists (Ro25-1553 and Ro25-1392) have been described[11,17,19,19,24,40,60], a few agonists selective for PAC1-R/VPAC1-R over VPAC2-R have also been described[13,16]. There are also a number of structure-function studies comparing VPAC1-R and VPAC2-R that have provided important insights into VIP analogs that can discriminate these two closely related receptors[11,23–25,40,41,55]. In regard to this latter point, a number of studies have shown that presence of Thr7, Tyr10, Thr11, Tyr22, Leu27, and Asn28 (present in both VIP and PACAP27) are particularly important for VPAC2 affinity[18,23–25,40]. Of these VPAC2 selective amino acids, the replacement of the aromatic residue Tyr22 (present in both PACAP and VIP) by an aliphatic hydrophobic residue such as Ala, results in an analog that is 100-fold more selective for VPAC1-R[18]. Our results demonstrate that [Ala22]PACAP38 (P17) also retains high affinity for PAC1-R, with an affinity 5-fold greater than for VPAC1 and >800-fold greater than for VPAC2-R. While these results suggest that in the future the insertion of Ala22 into the analogs that we found were selective for PAC1-R over VPAC1-R would result in a generally selective PAC1-R-agonist, a note of caution is needed. The insertion of Ala22 into two other PAC1 analogs in this study also resulted in >400-fold selectivity over VPAC2-R(P45,P46). However, in four other cases (P47, P48, P49,P51), the analogs had equal, low selectivity(2–6-fold) for both VPAC1-R and VPAC2-R. These results demonstrate that the impact on VPAC2-R selectivity of an Ala22 insertion varies markedly depending on the nature of other substitutions in the PACAP-analog.
Although the results of the present study are promising for developing PAC1-selective VIP/PACAP-based selective-agonists, it is important to remember that, in contrast to VPAC1-R/VPAC2-R, the PAC1-R has numerous splice-variants and some of these differ in relative affinities/potencies for PACAP27/PACAP38, as well as their abilities to activate intracellular signaling cascades[20,44,51,54], and, therefore, it is likely these will also vary for different PACAP-analogs. Therefore, in different tissues depending on the distribution of these splice-variants, in future studies, it is very possible that the selectivity of PAC1-R selective-analogs will vary markedly.
Highlights.
46 PACAP analogues were synthesized with conformationally restricted substitutions.
Their pharmacology (affinity, potency) was determined for VPAC1R, VPAC2R and PAC1-R.
15 peptides had 6–78-fold higher affinity[up to 103-fold potency] for PAC1-R
A number of analogs also had marked selectivity for PAC1-R over VPAC2
This study describes for first time, peptide agonists selective for PAC1-R.
Acknowledgments
This work was supported in part by intramural funds from the NIDDK branch of the National Institutes of Health. This work was also supported in part by 1 U54 GM104940 from the National Institute of General Medical Sciences of the National Institutes of Health, which funds the Louisiana Clinical and Translational Science Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Glossary
- BSA
bovine serum albumin fraction V
- cAMP
cyclic adenosine 3’:5’-monophosphate
- CNS
central nervous system
- DMEM
Dulbecco’s minimum essential medium
- DTT
dithiothreitol
- EC50
concentration causing half-maximal stimulation
- FBS
fetal bovine serum
- GRP
gastrin-releasing peptide
- GHRH
growth hormone-releasing hormone
- IC50
half-maximal inhibitory concentration
- IBMX
3-isobutyl-1-methylxanthine
- IP
inositol phosphate
- PACAP
pituitary adenylate cyclase-activating polypeptide
- PAC1-R
PACAP preferring receptor
- PACAP27
27-amino-acid form of PACAP
- PACAP38
38-amino-acid form of PACAP
- PBS
phosphate-buffered saline
- PLC
phospholipase C
- Ro 25–1553
Ac-His-Ser-Asp-Ala-Val-Phe-Thr-Glu-Asn-Tyr-Thr-Lys-Leu-Arg-Lys-Gln-Nle-Ala-Ala-Lys-cyclo[Lys-Tyr-Leu-Asn-Asp]-Leu-Lys-Lys-Gly-Gly-Thr-NH2
- Sup T1 cells
human Sup-T1 lymphoblastoma cells naturally containing VPAC2-R
- T47D cells
human T47D breast cancer cells naturally containing VPAC1-R
- VIP
vasoactive intestinal peptide
- VPAC1-R
VIP/PACAP receptor, subtype 1 (VIP1 receptor)
- VPAC2-R
VIP/PACAP receptor, subtype 2 (VIP2 receptor)
- 3T3
mouse embryonic fibroblast cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ahren B. Role of pituitary adenylate cyclase-activating polypeptide in the pancreatic endocrine system. Ann N Y Acad Sci. 2008;1144:28–35. doi: 10.1196/annals.1418.003. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez R, Daniels DV. A separation method for the assay of adenylylcyclase, intracellular cyclic AMP, and cyclic-AMP phosphodiesterase using tritium-labeled substrates. Anal Biochem. 1992;203:76–82. doi: 10.1016/0003-2697(92)90045-9. [DOI] [PubMed] [Google Scholar]
- 3.Banki E, Degrell P, Kiss P, Kovacs K, Kemeny A, Csanaky K, et al. Effect of PACAP treatment on kidney morphology and cytokine expression in rat diabetic nephropathy. Peptides. 2013;42:125–130. doi: 10.1016/j.peptides.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Banks WA, Kastin AJ, Komaki G, Arimura A. Passage of pituitary adenylate cyclase activating polypeptide1-27 and pituitary adenylate cyclase activating polypeptide1-38 across the blood-brain barrier. J Pharmacol Exp Ther. 1993;267:690–696. [PubMed] [Google Scholar]
- 5.Baun M, Pedersen MH, Olesen J, Jansen-Olesen I. Dural mast cell degranulation is a putative mechanism for headache induced by PACAP-38. Cephalalgia. 2012;32:337–345. doi: 10.1177/0333102412439354. [DOI] [PubMed] [Google Scholar]
- 6.Benya RV, Fathi Z, Battey JF, Jensen RT. Serines and threonines in the gastrin-releasing peptide receptor carboxyl terminus mediate internalization. J Biol Chem. 1993;268:20285–20290. [PubMed] [Google Scholar]
- 7.Bourgault S, Vaudry D, Botia B, Couvineau A, Laburthe M, Vaudry H, et al. Novel stable PACAP analogs with potent activity towards the PAC1 receptor. Peptides. 2008;29:919–932. doi: 10.1016/j.peptides.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Bourgault S, Vaudry D, Dejda A, Doan ND, Vaudry H, Fournier A. Pituitary adenylate cyclase-activating polypeptide: focus on structure-activity relationships of a neuroprotective Peptide. Curr Med Chem. 2009;16:4462–4480. doi: 10.2174/092986709789712899. [DOI] [PubMed] [Google Scholar]
- 9.Bourgault S, Vaudry D, Guilhaudis L, Raoult E, Couvineau A, Laburthe M, et al. Biological and structural analysis of truncated analogs of PACAP27. J Mol Neurosci. 2008;36:260–269. doi: 10.1007/s12031-008-9081-7. [DOI] [PubMed] [Google Scholar]
- 10.Bourgault S, Vaudry D, Segalas-Milazzo I, Guilhaudis L, Couvineau A, Laburthe M, et al. Molecular and conformational determinants of pituitary adenylate cyclase-activating polypeptide (PACAP) for activation of the PAC1 receptor. J Med Chem. 2009;52:3308–3316. doi: 10.1021/jm900291j. [DOI] [PubMed] [Google Scholar]
- 11.Dickson L, Aramori I, McCulloch J, Sharkey J, Finlayson K. A systematic comparison of intracellular cyclic AMP and calcium signalling highlights complexities in human VPAC/PAC receptor pharmacology. Neuropharmacology. 2006;51:1086–1098. doi: 10.1016/j.neuropharm.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Dickson L, Finlayson K. VPAC and PAC receptors: From ligands to function. Pharmacol Ther. 2009;121:294–316. doi: 10.1016/j.pharmthera.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Doan ND, Bourgault S, Dejda A, Letourneau M, Detheux M, Vaudry D, et al. Design and in vitro characterization of PAC1/VPAC1-selective agonists with potent neuroprotective effects. Biochem Pharmacol. 2011;81:552–561. doi: 10.1016/j.bcp.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 14.Dogrukol-Ak D, Tore F, Tuncel N. Passage of VIP/PACAP/secretin family across the blood-brain barrier: therapeutic effects. Curr Pharm Des. 2004;10:1325–1340. doi: 10.2174/1381612043384934. [DOI] [PubMed] [Google Scholar]
- 15.Germano PM, Stalter J, Le SV, Wu M, Yamaguchi DJ, Scott D, et al. Characterization of the pharmacology, signal transduction and internalization of the fluorescent PACAP ligand, fluor-PACAP, on NIH/3T3 cells expressing PAC1. Peptides. 2001;22:861–866. doi: 10.1016/s0196-9781(01)00410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gourlet P, Vandermeers A, Vandermeers-Piret MC, Rathe J, De Neef P, Robberecht P. C-terminally shortened pituitary adenylate cyclase-activating peptides (PACAP) discriminate PACAP I, PACAP II-VIP1 and PACAP II-VIP2 recombinant receptors. Regul Pept. 1996;62:125–130. doi: 10.1016/0167-0115(96)00010-9. [DOI] [PubMed] [Google Scholar]
- 17.Gourlet P, Vandermeers A, Vertongen P, Rathe J, DeNeef P, Cnudde J, et al. Development of high affinity selective VIP1 receptor agonists. Peptides. 1997;18:1539–1545. doi: 10.1016/s0196-9781(97)00228-3. [DOI] [PubMed] [Google Scholar]
- 18.Gourlet P, Vandermeers-Piret MC, Rathe J, De Neef P, Cnudde J, Robberecht P, et al. Vasoactive intestinal peptide modification at position 22 allows discrimination between receptor subtypes. Eur J Pharmacol. 1998;348:95–99. doi: 10.1016/s0014-2999(98)00133-2. [DOI] [PubMed] [Google Scholar]
- 19.Gourlet P, Vertongen P, Vandermeers A, Vandermeers-Piret MC, Rathe J, DeNeef P, et al. The long-acting vasoactive intestinal polypeptide agonist RO 25–1553 is highly selective of the VIP2 receptor subclass. Peptides. 1997;18:403–408. doi: 10.1016/s0196-9781(96)00322-1. [DOI] [PubMed] [Google Scholar]
- 20.Harmar AJ, Fahrenkrug J, Gozes I, Laburthe M, May V, Pisegna JR, et al. Pharmacology and functions of receptors for vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide: IUPHAR review 1. Br J Pharmacol. 2012;166:4–17. doi: 10.1111/j.1476-5381.2012.01871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holz GG, Chepurny OG. Glucagon-like peptide-1 synthetic analogs: new therapeutic agents for use in the treatment of diabetes mellitus. Curr Med Chem. 2003;10:2471–2483. doi: 10.2174/0929867033456648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou X, Vandermeers A, Gourlet P, Vandermeers-Piret MC, Robberecht P. Structural requirements for the occupancy of rat brain PACAP receptors and adenylate cyclase activation. Neuropharmacology. 1994;33:1189–1195. doi: 10.1016/s0028-3908(05)80009-7. [DOI] [PubMed] [Google Scholar]
- 23.Igarashi H, Ito T, Hou W, Mantey SA, Pradhan TK, Ulrich CD, II, et al. Elucidation of vasoactive intestinal peptide pharmacophore for VPAC1 receptors in human, rat and guinea pig. JPET. 2002;301:37–50. doi: 10.1124/jpet.301.1.37. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi H, Ito T, Mantey SA, Pradhan TK, Hou W, Coy DH, et al. Development of simplified vasoactive intestinal peptide analogs with receptor selectivity and stability for human vasoactive intestinal peptide/pituitary adenylate cyclase-activating polypeptide receptors. J Pharmacol Exp Ther. 2005;315:370–381. doi: 10.1124/jpet.105.088823. [DOI] [PubMed] [Google Scholar]
- 25.Igarashi H, Ito T, Pradhan TK, Mantey SA, Hou W, Coy DH, et al. Elucidation of the vasoactive intestinal peptide pharmacophore for VPAC2 receptors in human and rat, and comparison to the phrmacophore for VPAC1 receptor. J Pharmacol Exp Ther. 2002;302:445–460. doi: 10.1124/jpet.102.038075. [DOI] [PubMed] [Google Scholar]
- 26.Inooka H, Ohtaki T, Kitahara O, Ikegami T, Endo S, Kitada C, et al. Conformation of a peptide ligand bound to its G-protein coupled receptor. Nat Struct Biol. 2001;8:161–165. doi: 10.1038/84159. [DOI] [PubMed] [Google Scholar]
- 27.Ito T, Hou W, Katsuno T, Igarashi H, Pradhan TK, Mantey SA, et al. Rat and guinea pig pancreatic acini possess both VIP1 and VIP2 receptors, which mediate enzyme secretion. Am J Physiol (Gastrointest Liver Physiol ) 2000;278:G64–G74. doi: 10.1152/ajpgi.2000.278.1.G64. [DOI] [PubMed] [Google Scholar]
- 28.Kim Y, Lee BD, Kim O, Bae YS, Lee T, Suh PG, et al. Pituitary adenylate cyclase-activating polypeptide 27 is a functional ligand for formyl peptide receptor-like 1. J Immunol. 2006;176:2969–2975. doi: 10.4049/jimmunol.176.5.2969. [DOI] [PubMed] [Google Scholar]
- 29.Lerner EA, Ribeiro JM, Nelson RJ, Lerner MR. Isolation of maxadilan, a potent vasodilatory peptide from the salivary glands of the sand fly Lutzomyia longipalpis. J Biol Chem. 1991;266:11234–11236. [PubMed] [Google Scholar]
- 30.Li M, Maderdrut JL, Lertora JJ, Arimura A, Batuman V. Renoprotection by pituitary adenylate cyclase-activating polypeptide in multiple myeloma and other kidney diseases. Regul Pept. 2008;145:24–32. doi: 10.1016/j.regpep.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Li M, Maderdrut JL, Lertora JJ, Batuman V. Intravenous infusion of pituitary adenylate cyclase-activating polypeptide (PACAP) in a patient with multiple myeloma and myeloma kidney: a case study. Peptides. 2007;28:1891–1895. doi: 10.1016/j.peptides.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Huang S, Peng SB. Overexpression of G protein-coupled receptors in cancer cells: involvement in tumor progression. Int J Oncol. 2005;27:1329–1339. [PubMed] [Google Scholar]
- 33.Missig G, Roman CW, Vizzard MA, Braas KM, Hammack SE, May V. Parabrachial nucleus (PBn) pituitary adenylate cyclase activating polypeptide (PACAP) signaling in the amygdala: Implication for the sensory and behavioral effects of pain. Neuropharmacology. 2014;86C:38–48. doi: 10.1016/j.neuropharm.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyata A, Arimura A, Dahl RR, Minamino N, Uehara A, Jiang L, et al. Isolation of a novel 38 residue-hypothalamic polypeptide which stimulates adenylate cyclase in pituitary cells. Biochem Biophys Res Commun. 1989;164:567–574. doi: 10.1016/0006-291x(89)91757-9. [DOI] [PubMed] [Google Scholar]
- 35.Miyata A, Jiang L, Dahl RD, Kitada C, Kubo K, Fujino M, et al. Isolation of a neuropeptide corresponding to the N-terminal 27 residues of the pituitary adenylate cyclase activating polypeptide with 38 residues (PACAP38) Biochem Biophys Res Commun. 1990;170:643–648. doi: 10.1016/0006-291x(90)92140-u. [DOI] [PubMed] [Google Scholar]
- 36.Moody TW, Hill JM, Jensen RT. VIP as a trophic factor in the CNS and cancer cells. Peptides. 2003;24:163–177. doi: 10.1016/s0196-9781(02)00290-5. [DOI] [PubMed] [Google Scholar]
- 37.Moody TW, Ito T, Osefo N, Jensen RT. VIP and PACAP: recent insights into their functions/roles in physiology and disease from molecular and genetic studies. Curr Opin Endocrinol Diabetes Obes. 2011;18:61–67. doi: 10.1097/MED.0b013e328342568a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musso GF, Patthi S, Ryskamp TC, Provow S, Kaiser ET, Velicelebi G. Development of helix-based vasoactive intestinal peptide analogues: identification of residues required for receptor interaction. Biochemistry (Mosc) 1988;27:8174–8181. doi: 10.1021/bi00421a028. [DOI] [PubMed] [Google Scholar]
- 39.Myers SR, Yakubu-Madus FE, Johnson WT, Baker JE, Cusick TS, Williams VK, et al. Acylation of human insulin with palmitic acid extends the time action of human insulin in diabetic dogs. Diabetes. 1997;46:637–642. doi: 10.2337/diab.46.4.637. [DOI] [PubMed] [Google Scholar]
- 40.Nicole P, Lins L, Rouyer-Fessard C, Drouot C, Fulcrand P, Thomas A, et al. Identification of key residues for interaction of vasoactive intestinal peptide with human VPAC1 and VPAC2 receptors and development of a highly selective VPAC1 receptor agonist. J Biol Chem. 2000;275:24003–24012. doi: 10.1074/jbc.M002325200. [DOI] [PubMed] [Google Scholar]
- 41.Onoue S, Misaka S, Yamada S. Structure-activity relationship of vasoactive intestinal peptide (VIP): potent agonists and potential clinical applications. Naunyn Schmiedebergs Arch Pharmacol. 2008;377:579–590. doi: 10.1007/s00210-007-0232-0. [DOI] [PubMed] [Google Scholar]
- 42.Pisegna JR, Moody TW, Wank SA. Differential signaling and immediate-early gene activation by four splice variants of the human pituitary adenylate cyclase-activating polypeptide receptor (hPACAP-R) Ann N Y Acad Sci. 1996;805:54–64. doi: 10.1111/j.1749-6632.1996.tb17473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pisegna JR, Oh DS. Pituitary adenylate cyclase-activating polypeptide: a novel peptide with protean implications. Curr Opin Endocrinol Diabetes Obes. 2007;14:58–62. doi: 10.1097/MED.0b013e328012d605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pisegna JR, Wank SA. Molecular cloning and functional expression of the pituitary adenylate cyclase activating polypeptide (PACAP) Type I receptor. Proc Natl Acad Sci U S A. 1993;90:6345–6349. doi: 10.1073/pnas.90.13.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pisegna JR, Wank SA. Cloning and characterization of the signal transduction of four splice variants of the human pituitary adenylate cyclase activating polypeptide receptor (hPACAP-R): evidence for dual coupling to adenylate cyclase and phospholipase C. J Biol Chem. 1996;271:17267–17274. doi: 10.1074/jbc.271.29.17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reglodi D, Kiss P, Lubics A, Tamas A. Review on the protective effects of PACAP in models of neurodegenerative diseases in vitro and in vivo. Curr Pharm Des. 2011;17:962–972. doi: 10.2174/138161211795589355. [DOI] [PubMed] [Google Scholar]
- 47.Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–467. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reubi JC, Laemmli UK, Waser B, Gebbers J-O, Robberecht P, Laissue JA. Vasoactive intestinal peptide/pituitary adenylate cyclase-activating peptide receptor subtypes in human tumors and their tissues of origin. Cancer Res. 2000;60:3105–3112. [PubMed] [Google Scholar]
- 49.Robberecht P, Gourlet P, DeNeef P, Woussen-Colle MC, Vandermeers-Piret MC, Vandermeers A, et al. Receptor occupation and adenylate cyclase activation in AR42J rat pancreatic acinar cell membranes by analogs of pituitary adenylate cyclase-activating peptides amino-terminally shortened or modified at position 1,2,3,20 or 21. Mol Pharmacol. 1992;42:347–355. [PubMed] [Google Scholar]
- 50.Rowley WH, Sato S, Huang SC, Collado-Escobar DM, Beaven MA, Wang LH, et al. Cholecystokinin-induced formation of inositol phosphates in pancreatic acini. Am J Physiol. 1990;259:G655–G665. doi: 10.1152/ajpgi.1990.259.4.G655. [DOI] [PubMed] [Google Scholar]
- 51.Spengler D, Waeber C, Pantalonl C, Holsboer F, Bockaert J, Seeburg PH, et al. Differential signal transduction by five splice variants of the PACAP receptor. Nature. 1993;365:170–175. doi: 10.1038/365170a0. [DOI] [PubMed] [Google Scholar]
- 52.Sun C, Song D, vis-Taber RA, Barrett LW, Scott VE, Richardson PL, et al. Solution structure and mutational analysis of pituitary adenylate cyclase-activating polypeptide binding to the extracellular domain of PAC1-RS. Proc Natl Acad Sci U S A. 2007;104:7875–7880. doi: 10.1073/pnas.0611397104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uchida D, Tatsuno I, Tanaka T, Hirai A, Saito Y, Moro O, et al. Maxadilan is a specific agonist and its deleted peptide (M65) is a specific antagonist for PACAP type 1 receptor. Ann N Y Acad Sci. 1998;865:253–258. doi: 10.1111/j.1749-6632.1998.tb11185.x. [DOI] [PubMed] [Google Scholar]
- 54.Vaudry D, Falluel-Morel A, Bourgault S, Basille M, Burel D, Wurtz O, et al. Pituitary adenylate cyclase-activating polypeptide and its receptors: 20 years after the discovery. Pharmacol Rev. 2009;61:283–357. doi: 10.1124/pr.109.001370. [DOI] [PubMed] [Google Scholar]
- 55.Vaudry D, Gonzalez BJ, Basille M, Yon L, Fournier A, Vaudry H. Pituitary adenylate cyclase-activating polypeptide and its receptors: from structure to functions. Pharmacol Rev. 2000;52:269–324. [PubMed] [Google Scholar]
- 56.Ward BP, Ottaway NL, Perez-Tilve D, Ma D, Gelfanov VM, Tschop MH, et al. Peptide lipidation stabilizes structure to enhance biological function. Mol Metab. 2013;2:468–479. doi: 10.1016/j.molmet.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waschek JA. VIP and PACAP: neuropeptide modulators of CNS inflammation, injury, and repair. Br J Pharmacol. 2013;169:512–523. doi: 10.1111/bph.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wray V, Kakoschke C, Nokihara K, Naruse S. Solution structure of pituitary adenylate cyclase activating polypeptide by nuclear magnetic resonance spectroscopy. Biochemistry (Mosc) 1993;32:5832–5841. doi: 10.1021/bi00073a016. [DOI] [PubMed] [Google Scholar]
- 59.Wray V, Kiyoshi N, Naruse S. Solution structure comparison of the VIP/PACAP family of peptides by NMR spectroscopy. Ann N Y Acad Sci. 1998;865:37–44. doi: 10.1111/j.1749-6632.1998.tb11160.x. [DOI] [PubMed] [Google Scholar]
- 60.Xia M, Sreedharan SP, Bolin DR, Gaufo GO, Goetzl EJ. Novel cyclic peptide agonist of high potency and selectivity for the type II vasoactive intestinal peptide receptor. J Pharmacol Exp Ther. 1997;281:229–633. [PubMed] [Google Scholar]
- 61.Yung LS, Dela Cruz F, Hamren S, Zhu J, Tsutsumi M, Bloom JW, et al. Generation of highly selective VPAC2 receptor agonists by high throughput mutagenesis of vasoactive intestinal peptide and pituitary adenylate cyclase-activating peptide. J Biol Chem. 2003;278:10273–10281. doi: 10.1074/jbc.M211945200. [DOI] [PubMed] [Google Scholar]
- 62.Zhou ZC, Gardner JD, Jensen RT. Interaction of peptides related to VIP and secretin with guinea pig pancreatic acini. Am J Physiol. 1989;256:G283–G290. doi: 10.1152/ajpgi.1989.256.2.G283. [DOI] [PubMed] [Google Scholar]