Abstract
Pyrethroids are the major class of insecticides used for mosquito control. Excessive and improper use of insecticides, however, has resulted in pyrethroid resistance, which has become a major obstacle for mosquito control. The development of pyrethroid resistance is a complex process involving many genes, and information on post-transcription regulation of pyrethroid resistance is lacking. In this study, we extracted RNA from mosquitoes in various life stages (fourth-instar larvae, pupae, male and female adult mosquitoes) from deltamethrin-sensitive (DS) and resistant (DR) strains. Using Illumina sequencing, we obtained 13760296 and 12355472 reads for DS-strains and DR-strains, respectively. We identified 100 conserved miRNAs and 42 novel miRNAs derived from 21 miRNA precursors in Culex pipiens. After normalization, we identified 28 differentially expressed miRNAs between the two strains. Additionally, we found that cpp-miR-71 was significant down regulated in female adults from the DR-strain. Based on microinjection and CDC Bottle Bioassay data, we found that cpp-miR-71 may play a contributing role in deltamethrin resistance. The present study provides the firstly large-scale characterization of miRNAs in Culex pipiens and provides evidence of post-transcription regulation. The differentially expressed miRNAs between the two strains are expected to contribute to the development of pyrethroid resistance.
Keywords: Culex pipiens, pyrethroid resistance, expression profile, differentially expressed microRNA, microinjection, American CDC Bottle Bioassay
1. Introduction
Mosquito-borne diseases, such as malaria, dengue fever, yellow fever, filariasis and encephalitis, dramatically jeopardize public health and impede economic development (Nkya et al., 2013). Between 2006 and 2009, there were 5794 cases of Japanese encephalitis reported in China and India each year, with the majority of cases occurring in children aged 0–14 years (Campbell et al., 2011).
Historically, chemical compounds have been the principal method of mass prevention for mosquito-borne diseases (Sun et al., 2013). Pyrethroid, fourth generation synthetic insecticide, modifies the function of voltage sensitive sodium channels (Soderlund and Knipple, 2003) which disrupts the nervous system (Chen et al., 2010). Deltamethrin, a representative pyrethroid insecticide, is commonly used for the indoor residual spraying and impregnation of bed nets due to its low toxicity (Xu et al., 2014). Excessive and improper application of pyrethroids, however, has caused resistance, and mosquito-borne diseases are now resurgent (Hart et al., 2014).
Insecticide resistance results from polygenic inheritance (David et al., 2005). Mutation of sodium channels, activation of detoxification enzymes and upregulation of other genes could contribute to the development of pyrethroid resistance (Hemingway et al., 2004; Hemingway and Ranson, 2000). Expression of these genes is regulated at both transcriptional and post-transcriptional levels. MicroRNAs (miRNAs) are known to be a key component in post-transcriptional gene expression regulation in many species.
MiRNAs are endogenous noncoding RNAs with lengths of 19–23 nucleotides. Upon Dcr-1 cleavage of pre-miRNA to form a mature miRNA/miRNA* duplex, one strand is loaded into RNA-induced silencing complex (Wostenberg et al., 2012) and the other strand is degraded (Leclercq et al., 2013). MiRNAs regulate gene expression at the post-transcription level via base pairing to target sites within messenger RNAs (mRNAs) (Lucas and Raikhel, 2013). In most cases, the miRNA “seed sequence” (nucleotides 2–8 at the 5′ end of miRNA) binds to the 3′untranslated regions (3′UTR) of mRNA resulting in regulation of mRNA translation or mRNA degradation (Bartel, 2004). Since miRNAs were first discovered in Caenorhabditis elegans, multiple miRNAs have been shown to play considerable roles in regulating cellar differentiation, proliferation, apoptosis and development in vertebrates, invertebrates, plants and viruses (Ameres and Zamore, 2013). In Anopheles gambiae, the expression profile of miRNAs was examined and four miRNAs were found to be affected by the presence of Plasmodium (Winter et al., 2007). In An. stephensi, 27 miRNAs have been identified and distinct patterns of miRNAs expression were revealed from embryo to adult stages (Gregory et al., 2011). Sixty-five miRNAs in the Aedes albopictus C7/10 cell line and 77 miRNAs in Cx. quinquefasciatus were also detected and miR-92 and miR-989 showed significant changes following WNV infection (Skalsky et al., 2010).
The role of miRNAs in insecticide resistance in mosquito has received little attention. Next-generation sequencing technology makes it possible to precisely identify non-conserved or weakly expressed miRNAs (Yamamoto et al., 2014). The identification and comparison of miRNA expression in deltamethrin-sensitive strains (DS) and deltamethrin–resistant strains (DR) of Cx. pipiens would contribute to a better understanding of the mechanism of pyrethroid resistance. In addition, we provide the first evidence that cpp-miR-71 participates in pyrethroid resistance in mosquitoes and confirme interactions between cpp-miR-71 and the 3′UTR of its target gene cytochrome P450 325BG3 (CYP325BG3).
2. Materials and Methods
2.1. Mosquito strains
A DS-strain of Cx. pipiens (LC50 was 0.03mg/l) which was never been exposed to any insecticides, was collected from Tang Kou, ShanDong province. Two DR-strains (DR1-strain, DR2-strain) were used in this study. DR1-strain was selected with deltamethrin for more than 10 generations and the resistance index reaches 28.3(LC50 was 0.85mg/l). DR2-strain was subjected to deltamethrin selection for more than 60 generations, the LC50 was up to 7mg/l. All the strains were reared in a humidified insectary at 28–30 °C on a 16 h light/8 h dark cycle.
2.2. Cx. pipiens sample preparation for illumina sequencing
Total RNA was extracted using RNAiso Plus reagent (TaKaRa, Dalian, China) from mixed life stages (including fourth-instar larvae, pupae, male and female adults) from DS and DR1-strains. Pupae were collected from varied ages. Male and female adults aged 3–5 days post emergence were collected. Approximately 30 μg of the total RNA was extracted and sent to the Beijing Genome Institute Inc. for sequencing and analysis. About 10 μg total RNA was isolated on a 15% denaturing polyacrylamide gel. Small RNAs ranging from 18 to 30 nt in length were excised and ligated sequentially from 5′ to 3′ ends. Then adding the adaptor primers to amplify RNAs and about 90 nt fragments were isolated from agarose gels. The extracted DNA was sequenced by Illumina Hi Seq™ 2000. Clean reads were processed after removing contaminated reads and adaptor sequences.
2.3. Small RNA sequence analysis
Small RNA clean reads were mapped to the Cx. quinquefasciatus genome with Short Oligonucleotide Alignment Programs (SOAP) (Li et al., 2008), which was closely related mosquito in the same subgenus with Cx. pipiens. Only perfect matches were accepted and retained for the next analysis. We aligned all small RNA reads to mature miRNAs and blasted miRNA hairpins in miRBase18.0 and filtered them using an E-value threshold of 0.01. We annotated the other non-coding RNAs to rRNA, scRNA, snoRNA, snRNA and tRNA from Genbank and Rfam (9.1). We also annotated small RNAs according to exon and intron of mRNAs from Vector Base. The remaining unannotated small RNAs were tested by MIREAP (https://sourceforge.net/projects/mireap/) for miRNA-coding potential. We took the predominant small RNA from “MIREAP” as the novel representative miRNA. When miRNA or miRNA* had at least 15 reads in the two samples, we classified it into the expressed miRNA.
2.4. Amplification of the miRNA precursors
We extracted genomic DNA from the female adults using the method as previously described (Collins et al., 1987). We designed primers for 36 conserved miRNAs and 12 novel miRNAs according to the sequences of the mature miRNA and miRNA* or the sequences in Cx. quinquefasciatus genome using Primer Premier 5.0 (Premier Biosoft International, PaloAlto, CA, USA). The sequences of the primers were in the Table S1. Fragments were amplified by PCR and the products were examined by 2.5% agarose gels.
2.5. Stem-loop quantitative RT-PCR assay
Stem-loop RT-PCR was used to verify the expression levels of miRNAs between the two strains (Tang et al., 2006). Briefly, total RNA from embryos, instar larvae, pupae, male and female adults was extracted by RNAiso Plus reagent, respectively. 2 μg total RNA was reverse-transcribed to cDNA using AMV transcriptase (TaKaRa, Dalian, China) and looped antisense primer. The mixture was incubated at 42°C for 60 minutes and 85 °C for 5 minutes. qRT-PCR was performed on the ABI Prism 7300 HT Sequence Detection system (Applied Biosystem, CA, USA) using FastStart® SYBR Green (Applied Biosystem, CA, USA). Reactions were incubated in a 96-well optical plate at 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1min. A melting curve program was run immediately after the PCR and the data was analyzed by 7300 System SDS Software v1.2.1 (Applied Biosystems). Table S2 was the primers for the stem-loop quantitative RT-PCR. The raw threshold cycle (Ct) values were normalized against U6 standard to obtain normalized Ct values, which were used to calculate relative expression levels in samples using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
2.6. Target prediction
It was important to predict miRNA targets in understanding miRNA function. The rules used for target prediction were based on those suggested by Allen et al. (Allen et al., 2005) and Schwab et al. (Schwab et al., 2005). Unfortunately, there was no 3′UTR database available for Cx. quinquefasciatus, making it difficult to predict targets for cpp-miRNAs using computational prediction tools. Cytochrome P450 monooxygenase (CYP450) genes have proven particularly important in resistance to insecticides (Schuler and Berenbaum, 2013). Based on transcripts of CYP450 from the Cx. quinquefasciatus, the 3′UTR sequence of CYP325BG3 contained a target site for cpp-miR-71. The partial 3′UTR sequence of the Cx. pipiens CYP325BG3 gene was obtained by PCR using RACE cDNA. The RACE cDNA was reversed using SMART RACE cDNA Amplification kit (Clontech, Tokyo, Japan). The PCR primer was: forward primer, 5′-ATCGGAATCCATTTTCGTTTT-3′, reverse primer, 5′-TTGTAGTCTCCATTTGCCACG-3′.
2.7. Vector construction and luciferase assay
The CYP325BG3 3′UTR sequence was amplified from cDNAs from female adults by PCR using the following primers: forward primer, 5′-CCAAGCTTGAAGAGCCTTAAGTCTAATTT-3′; reverse primer, 5′-CGAGCTCATCGTCTTATGAAAGACAATA-3′. For its mutagenisis, the sequence complementary to the binding site of cpp-miR-71 seed sequence in its 3′UTR (GTCTTTC) was replaced by CTGTATG. The wild type and muted 3′UTR of CYP325BG3 were both cloned into PMIR-Report miRNA reporter vector using the Hind III and Sac I sites. The constructs were validated by sequencing.
The sequence of cpp-miR-71 was: 5′-AGAAAGACATGGGTAGTGAGAT-3′. The miRNA mimic was designed and procured from GenePharma (GenePharma, Shanghai, China) at a concentration of 20 μM. HEK-293 cells were seeded into a 6-well plate for 24h. The cells were divided into four groups, UTR/miR-71mimic, UTR/NC mimic, MUT/miR-71 mimic and MUT/NC mimic. The plasmids were transfected into cells using FuGENE® HD Transfection Reagent (Promega, WI, USA). The solutions were mixed gently and incubated at room temperature for 15 min. The mixture was layered onto cells gently. 48h after transfection, cells were lysed and subjected to luciferase assay that was performed using the Dual Luciferase Reporter Assay System (Promega, WI, USA).
2.8. Microinjection of female adult mosquito
The cpp-miR-71 mimic was designed and procured from GenePharma (GenePharma, Shanghai, China) at a concentration of 20 μM. The details of the sequences were included in Table S3. Unfed female adults mosquitoes from the DR2-strain at one day post emergence were used for microinjection. The mosquitoes were anaesthetized in the −20 °C refrigerator for 6 to 8 minutes and injected in the thorax with 0.5 μl (500 nl) of cpp-miR-71 mimic at a final concentration of 20 pmol /μl on ice (Kumar and Puttaraju, 2012; Puthiyakunnon et al., 2013). After microinjection, mosquitoes were placed to small plastic tubes immediately and were allowed to recover with sugar water in a humidified insectary at 28–30 °C on a 16 h light/8 h dark cycle. The negative control (NC) mimic was injected the same volume into mosquitoes and the control group was injected 0.5 μl water pretreated with diethypyrocarbonate (DEPC-water) at the same time. Three days after recovery, the efficiency of the miRNA mimic microinjection was confirmed using stem-loop quantitative RT-PCR. The sequences of primer of cpp-miR-71 for stem-loop quantitative RT-PCR were in Table S2. The expression levels of target gene CYP325BG3 were confirmed by qRT-PCR. Its primers were: forward primer, 5′-ATTCTTGCTGAAGGAAGTGGC-3′; reverse primer, 5′-AATCGTGATCGCGTGGTGT-3′.
2.9. American CDC Bottle Bioassay
Unfed female adults of one day post emergence from the DR2-strain were used for microinjection. Then we microinjected miRNA mimic, control mimic and DEPC-water as above. Mosquitoes were put in the recovery tubes after microinjection. Three days later, we examined them using American CDC Bottle Bioassay, a tool for detecting the resistance to insecticides (Aizoun et al., 2013). According to the laboratory guideline of the CDC bottle bioassay, the recommended dosage of deltamethrin did not kill individual mosquito in our first bioassay, we decided to increase the dosage. The diagnostic dose of deltamethrin applied in this study was 4 mg per bottle (250 ml). For each bioassay, 15 to 20 mosquitoes per bottle were exposed to deltamethrin. The control bottle was coated with acetone only and tested with cpp-miR-71 mimic microinjected mosquitoes. The number of dead mosquitoes was recorded every 15 minutes, up to 2 hours. At last the mortality rate was calculated at the diagnostic time.
2.10. siRNA microinjection
Short interfering RNAs (siRNA) targeting CYP325BG3 were designed and procured from GenePharma (GenePharma, Shanghai, China) at a concentration of 20 μM (The sequences were in Table S4). 0.07μl of the siRNA and negative control (NC) RNA were microinjected into female adults from the DR2-strain at a final concentration of 5 μg/μl (The procedures of microinjection were described in section 2.8.). The control group was microinjected the same volume of DEPC-water at the same time. Three days after microinjection, the expression levels of target gene CYP325BG3 were confirmed by qRT-PCR. The relationship between deltamethrin resistance and CYP325BG3 was confirmed by American CDC Bottle Bioassay.
3. Results and Discussion
3.1. Small RNA expression profiling in Cx. pipiens
In order to examine the role of small RNAs in pyrethroid resistance in Cx. pipiens, we collected samples from fourth-instar larvae, pupae, male and female adults from DS and DR1 strains for sequencing. Small RNAs of less than 30 nt were isolated and sequenced. After filtering out sequences shorter than 18 nt and lower quality reads, 13760296 and 12355472 reads of the DS-strain and DR1-strain remained for further analysis. More than 50% of the reads were expected to be insect miRNA. The percentage of miRNA isolated from the DR1-strain was found to be 62.94% while in the DS-strain the percentage of miRNA was 55.35%. The length distribution of these clean reads was summarized to illustrate the compositions of small RNA samples (Fig. S1A, B in Supplementary data). miRNA is normally ~19–23 nt (Bartel, 2004), endogenous siRNA is ~21 nt (Czech et al., 2008), and piRNA is ~27–30 nt (Siomi et al., 2011). MiRNA and endogenous siRNA are similar lengths, however, miRNA has a tendency to begin with uracil (Fig. S1C, D in Supplementary data). We analyzed the first nucleotide bias of small RNAs. The results suggested that majority of the ~19–22 nt small RNAs were miRNA rather than endogenous siRNA. The proportion of small RNAs with a length of ~27–30 nt was much higher in the DS-strain than that in DR1-strain, which suggested that piRNA may also play a role in pyrethroid resistance. Further investigation of these small RNAs could be helpful in improving our understanding of mechanisms of insecticide resistance.
3.2. Conserved and novel miRNAs in Cx. pipiens
We aligned small RNA sequences to the miRNA of corresponding species in miRBase (http://www.mirbase.org/) and the characteristic hairpin structure of miRNA was predicted by MIREAP. In total, we detected 100 conserved miRNA genes. Among these, 71 genes were present as a single copy in the Cx. quinquefasciatus genome, while others had multiple copies distributed on different loci that produced identical mature miRNAs (Table 1). The Cx. pipiens miRNA and miRNA* strands mapped with 100% identity to the Cx. quinquefasciatus. However, the strand selection differed between the two closely-related mosquito species. In Cx. quinquefasciatus, cqu-miR-11, cqu-miR-308, and cqu-miR-998 had more 3p reads than 5p reads. In Cx. pipiens, cpp-miR-11 had the similar 5p and 3p reads, while cpp-miR-308, cpp-miR-998 had more 5p reads. One reason for this strand selection difference may be that the miRNAs came from different subgenus. Another potential reason was that samples used for sequencing came from different sources. The samples used for sequencing of Cx. quinquefasciatus were from female adults, while the samples from Cx. pipiens were obtained from mixed growth stages (fourth-instars larvae, pupae, male and female adults).
Table 1.
Information on the Cx. pipiens miRNA examined in this study
| miRBase Name | Sequence | Location in genome |
|---|---|---|
| Previously known miRNAs that are also found in Culex quinquefasciatus | ||
| cpp-bantam | UGAGGUAGUUGGUUGUAUAGU | supercont3.65: 199694-199763:− |
| cpp-let-7 | UGAGGUAGUUGGUUGUAUAGU | supercont3.4:280600:280681:+ |
| cpp-miR-1-3p | UGGAAUGUAAAGAAGUAUGGAG | supercont3.78:246202:246281:+ |
| cpp-miR-2-3p-1 | UAUCACAGCCAGCUUUGAAGAGC | supercont3.366:116579:116662: − |
| cpp-miR-2-3p-2 | UAUCACAGCCAGCUUUGAAGAGC | supercont3.366:116853:116935: − |
| cpp-miR-2-3p-3 | UAUCACAGCCAGCUUUGAAGAGC | supercont3.366:117351:117432: − |
| cpp-miR-7 | UGGAAGACUAGUGAUUUUGUUGU | supercont3.1:3357343:3357422: − |
| cpp-miR-8 | UAAUACUGUCAGGUAAAGAUGUC | supercont3.40:815854:815935: − |
| cpp-miR-9 | UCUUUGGUUAUCUAGCUGUA | supercont3.1009:114890:114972: − |
| cpp-miR-10-3p | CAAAUUCGGUUCUAGAGAGGUUU | supercont3.12:95990:96072: − |
| cpp-miR-11-3p | CAUCACAGUCUGAGUUCUUGCU | supercont3.153:639659:639743: − |
| cpp-miR-12-3p | UGAGUAUUACAUCAGGUACUGGU | supercont3.153: 639666-639741: − |
| cpp-miR-13 | UAUCACAGCCAUUUUGACGAGUU | supercont3.366:116983:117072: − |
| cpp-miR-14-3p | UCAGUCUUUUUCUCUCUCCUAU | supercont3.676:52241:52327: − |
| cpp-miR-31 | UGGCAAGAUGUUGGCAUAGCUGA | supercont3.559:256532:256609: − |
| cpp-miR-33 | GUGCAUUGUAGUUGCAUUGCA | supercont3.1258: 69328-69403: − |
| cpp-miR-71 | AGAAAGACAUGGGUAGUGAGAU | supercont3.366:117493:117583: − |
| cpp-miR-79-3p | UAAAGCUAGAUUACCAAAGCAU | supercont3.83:80579:80662:+ |
| cpp-miR-87 | GUGAGCAAAUUUUCAGGUGUGU | supercont3.431:379743:379819:+ |
| cpp-miR-92a | UAUUGCACUUGUCCCGGCCUAU | supercont3.722:174902:174982: − |
| cpp-miR-92b | AAUUGCACUUGUCCCGGCCUGC | supercont3.722:164904:164984: − |
| cpp-miR-100 | AACCCGUAGAUCCGAACUUGUG | supercont3.4:271404:271484:+ |
| cpp-miR-124 | UAAGGCACGCGGUGAAUGCCAA | supercont3.8:2074719:2074803:+ |
| cpp-miR-125 | UCCCUGAGACCCUAACUUGUGA | supercont3.4:280965:281043:+ |
| cpp-miR-133-3p | UUGGUCCCCUUCAACCAGCUGU | supercont3.1189: 55672-55785:+ |
| cpp-miR-137-3p | UAUUGCUUGAGAAUACACGUAG | supercont3.1714:27556:27636: − |
| cpp-miR-184-3p | UGGACGGAGAACUGAUAAGGGC | supercont3.567:240302:240382: − |
| cpp-miR-190-1 | AGAUAUGUUUGAUAUUCUUGGUUG | supercont3.181:347902:347986: − |
| cpp-miR-190-2 | AGAUAUGUUUGAUAUUCUUGGUUG | supercont3.1806:19610:19690: − |
| cpp-miR-210-3p | CUUGUGCGUGUGACAACGGCUAU | supercont3.549:157647:157724: − |
| cpp-miR-252-1 | CUAAGUACUAGUGCCGCAGGAG | supercont3.1787:6793:6867: − |
| cpp-miR-252-2 | CUAAGUACUAGUGCCGCAGGAG | supercont3.174:422878:422969: − |
| cpp-miR-263 | AATGGCACTGGAAGAATTCACGG | supercont3.219:351799:351880: − |
| cpp-miR-275 | TCAGGTACCTGAAGTAGCGCG | supercont3.291:329765:329845:+ |
| cpp-miR-276-3p-1 | UAGGAACUUCAUACCGUGCUCU | supercont3.136:340857:340942:+ |
| cpp-miR-276-3p-2 | UAGGAACUUCAUACCGUGCUCU | supercont3.136:541142:541223:+ |
| cpp-miR-276-3p-3 | UAGGAACUUCAUACCGUGCUCU | supercont3.2457:876:961:+ |
| cpp-miR-277 | UAAAUGCACUAUCUGGUACGACA | supercont3.36:1153726:1153817:+ |
| cpp-miR-278-3p | UCGGUGGGACUUUCGUCCGUUU | supercont3.16:1026152:1026243:+ |
| cpp-miR-279-3p | UGACUAGAUCCACACUCAUUAA | supercont3.19:1441114:1441197: − |
| cpp-miR-281-1 | AAGAGAGCUAUCCGUCGACAGU | supercont3.1661:13247:13325:+ |
| cpp-miR-281-2 | AAGAGAGCUAUCCGUCGACAGU | supercont3.640:99734:99812:+ |
| cpp-miR-283 | CAAUAUCAGCUGGUAAUUCUGGG | supercont3.57: 559440-559549 :+ |
| cpp-miR-285-3p | UAGCACCAUUCGAAAUCAGUAC | supercont3.98:262280:262362: − |
| cpp-miR-305 | ATTGTACTTCATCAGGTGCTC | supercont3.291:339122:339204:+ |
| cpp-miR-306 | UCAGGUACUGAGUGACUCUCAG | supercont3.83: 80431-80502:+ |
| cpp-miR-308 | CGCGGTATATTCTTGTGGCTTG | supercont3.98:764123:764204:+ |
| cpp-miR-309-3p | UCACUGGGCAUAGUUUGUCGC | supercont3.145: 66026-66116: − |
| cpp-miR-315 | UUUUGAUUGUUGCUCAGAAAGC | supercont3.438:61916:61999:+ |
| cpp-miR-316 | UGUCUUUUUCCGCUUACUGCCG | supercont3.496:152458:152539: − |
| cpp-miR-317-3p-1 | UGAACACAGCUGGUGGUAUCU | supercont3.36:1133161:1133241:+ |
| cpp-miR-317-3p-2 | UGAACACAGCUGGUGGUAUCU | supercont3.36:1134827:1134907:+ |
| cpp-miR-375-1 | UUUGUUCGUUUGGCUCGAGUUA | supercont3.4:25124:25212: − |
| cpp-miR-375-2 | UUUGUUCGUUUGGCUCGAGUUA | supercont3.455:42526:42614:+ |
| cpp-miR-932-3p | UGCAAGCAAUGUGGAAGUGA | supercont3.261:301401:301481: − |
| cpp-miR-957-3p | UGAAACCGUCCAAAACUGAGGC | supercont3.787:29537:29622:+ |
| cpp-miR-965-3p | UAAGCGUAUAGCUUUUCCCAUU | supercont3.48:484122:484208:+ |
| cpp-miR-970-3p | UCAUAAGACACACGCGGCUAU | supercont3.495: 35917-36003:+ |
| cpp-miR-980-3p | TAGCTGCCTTGTGAAGGGCTTA | supercont3.263:352870:352954:+ |
| cpp-miR-981 | UUCGUUGUCGACGAAACCUGCA | supercont3.431: 151310-151408:+ |
| cpp-miR-988-3p | CCCUUGUUGCAAACCUCACGC | supercont3.791:14282:14361:+ |
| cpp-miR-989-3p | UGUGAUGUGACGUAGUGGUAC | supercont3.315:321306:321394:+ |
| cpp-miR-993 | UACCCUGUAGUUCCGGGCUUUU | supercont3.12:55477:55565:+ |
| cpp-miR-996 | UGACUAGAUUACAUGCUCGU | supercont3.19:1437000:1437085: − |
| cpp-miR-998 | ACTGAATTCTCGTGGGTCTGCA | supercont3.153:639515:639597: − |
| cpp-miR-999-3p | UGUUAACUGUAAGACUGUGUCU | supercont3.14:96865:96948:+ |
| cpp-miR-1000 | AUAUUGUCCUGUCACAGCAGU | supercont3.153: 102798-102891: − |
| cpp-miR-1174 | CUGGGUAUUUUAGAUCAUCGGC | supercont3.86:865891:865964:+ |
| cpp-miR-1175 | AAGUGGAGUAGUGGUCUCAUCG | supercont3.86:866106:866187:+ |
| cpp-miR-1889 | UAAUCUCAAAUUGUAACAGUGG | supercont3.57: 562539-562677:+ |
| cpp-miR-1890 | UGAAAUCUUUGAUUAGGUCU | supercont3.64:982778:982862: − |
| cpp-miR-1891 | UGAGGAGUUAAUUUGCGUGUUU | supercont3.829: 180333-180418: − |
| cpp-miR-2941-3p-1 | UAGUACGGCUAGAACUCCACGG | supercont3.5:753632:753715: − |
| cpp-miR-2941-3p-2 | UAGUACGGCUAGAACUCCACGG | supercont3.5:753786:753871: − |
| cpp-miR-2951-1 | GAAGAGCTCAGCACGCAGGGGTG | supercont3.1464: 4408-4477:+ |
| cpp-miR-2951-2 | GAAGAGCTCAGCACGCAGGGGTG | supercont3.1464: 13130-13199:+ |
| cpp-miR-2951-3 | GAAGAGCTCAGCACGCAGGGGTG | supercont3.1464: 21931-22000 :+ |
| cpp-miR-2951-4 | GAAGAGCTCAGCACGCAGGGGTG | supercont3.1464: 30698-30767:+ |
| cpp-miR-2951-5 | GAAGAGCTCAGCACGCAGGGGTG | supercont3.1464: 39499-39568:+ |
| cpp-miR-2951-6 | GAAGAGCTCAGCACGCAGGGGTG | supercont3.1464: 48275-48344:+ |
| cpp-miR-2951-7 | GAAGAGCTCAGCACGCAGGGGTG | supercont3.1464: 56986-57055:+ |
| cpp-miR-2951-8 | GAAGAGCTCAGCACGCAGGGGTG | supercont3.679: 54985-55054:+ |
| cpp-miR-2952-3p | UAGUACGGCCAUGACUGAGGGC | supercont3.5: 753920-753983: − |
| cpp-miR-iab-4 | ACGUAUACUGAAUGUAUCCUGA | supercont3.12:681153:681228:+ |
| Previously known miRNAs that are also found in non-Culex quinquefasciatus species | ||
| cpp-miR-2779-1 | AUAUCCGGCUCGAAGGACCA | supercont3.1527:60074:60167:+ |
| cpp-miR-2779-2 | AUAUCCGGCUCGAAGGACCA | supercont3.2199:21971:22064: − |
| cpp-miR-2779-3 | AUAUCCGGCUCGAAGGACCA | supercont3.806:109239:109311: − |
| cpp-miR-2796-3p | GUAGGCCGGCGGAAACUACUUGC | supercont3.1234:55494:55572: − |
| cpp-miR-2840 | UAGGAACUGGAAGAAGAGGAGG | supercont3.830:78060:78155: − |
| cpp-miR-2942 | UAUUCGAGACUUCACGAGUUAAU | supercont3.277:323072:323166:+ |
| cpp-miR-2944b | GAAGGAACUCCCGGUGUGAUAUA | supercont3.145:66345:66420: − |
| cpp-miR-2945 | UGACUAGAGGCAGACUCGUUUA | supercont3.4:184409:184491:+ |
| cpp-miR-2981 | CCGGGCCGGGCGGGCGGG | supercont3.772:7842:7925: − |
| cpp-miR-3781 | UAAGUGAUUGAUCGAUCGUGGAU | supercont3.1005:38090:38187: − |
| cpp-miR-4682 | UCUGAGUUCCUGGAGCCUGGUCU | supercont3.1:1608156:1608242:+ |
| cpp-miR-4448 | GGCUCCUUGGUCUAGGGGUA | supercont3.461:292111:292200: − |
| cpp-miR-307 | UCACAACCUCCUUGAGUGAGCGA | supercont3.16:157813:157891: − |
| cpp-miR-493-3p | UGAAGGUCCUACUGUGUGCCAGG | supercont3.147:509902:509990:+ |
| cpp-miR-929-3p | CUCCCUAACGGAGUCAGAUUG | supercont3.60:858168:858245: − |
| cpp-miR-1290 | UGGAUUUUUGGAUCAGAGA | supercont3.383:279453:279536:+ |
| Novel miRNAs found in this study | ||
| cpp-novel-miR1 | AGAGCTAATTGGAGACTTCTTG | supercont3.64:982778:982859: − |
| cpp-novel-miR2-1 | GTGTCCTGTCACGGTCGCCA | supercont3.360:171826:171903: − |
| cpp-novel-miR2-2 | GTGTCCTGTCACGGTCGCCA | supercont3.88:405605:405682: − |
| cpp-novel-miR3-3p | GTTTGAACTTGATCCGCGGCTGA | supercont3.112:3529:3623: − |
| cpp-novel-miR4 | GTGCTTTTCGTTGGAACTTG | supercont3.153:639486:639575: − |
| cpp-novel-miR5 | AATTAGAAATCACACAAACGTT | supercont3.316:156915:156993:+ |
| cpp-novel-miR6-3p | TAGGGAAACAGATTGGCCAATG | supercont3.1403:25947:26028:+ |
| cpp-novel-miR7-3p-1 | ACATCGCGTGTCGTTGGCAT | supercont3.1002:30216:30294: − |
| cpp-novel-miR7-3p-2 | ACATCGCGTGTCGTTGGCAT | supercont3.1002:37099:37177: − |
| cpp-novel-miR7-3p-3 | ACATCGCGTGTCGTTGGCAT | supercont3.1002:45236:45314: − |
| cpp-novel-miR7-3p-4 | ACATCGCGTGTCGTTGGCAT | supercont3.1002:54195:54273: − |
| cpp-novel-miR8-3p | ACACGTCCATTAACTCTGGTAC | supercont3.829:180337:180416: − |
| cpp-novel-miR9-1 | AATCGGAATTCTAAAACGGAA | supercont3.156:169435:169530: − |
| cpp-novel-miR9-2 | AATCGGAATTCTAAAACGGAA | supercont3.213:211573:211659: − |
| cpp-novel-miR9-3 | AATCGGAATTCTAAAACGGAA | supercont3.664:19971:20057:+ |
| cpp-novel-miR9-4 | AATCGGAATTCTAAAACGGAA | supercont3.924:122727:122815:+ |
| cpp-novel-miR10 | TGATCTTGTATTTTGATGCTCC | supercont3.329:155673:155749: − |
| cpp-novel-miR11-3p | CAGTGCATGGCCAACACGGTTT | supercont3.1328:35831:35912:+ |
| cpp-novel-miR12 | GGTGTTCACTGCCGGCCTGTATG | supercont3.8:2074722:2074801:+ |
| cpp-novel-miR13 | GTTAGTTTTGGGCGGGTTTTAGT | supercont3.787:29538:29623:+ |
| cpp-novel-miR14-3p | GGCGCGAGCGTGTGTTATTC | supercont3.196:659156:659228:+ |
| cpp-novel-miR15 | ATTTGTGGTATATGTCGGACGAG | supercont3.829:126138:126232:+ |
| cpp-novel-miR16-3p | AAGGAGTGGAACTTGGTCGCGGA | supercont3.112:109706:109789:+ |
| cpp-novel-miR17-3p | CGGGATTCCAACTGATATCCAC | supercont3.57:559449:559540:+ |
| cpp-novel-miR18-3p-1 | TTGCAGTGGATGGTCGTTTGACG | supercont3.1369:75814:75893: − |
| cpp-novel-miR18-3p-2 | TTGCAGTGGATGGTCGTTTGACG | supercont3.91:950407:950490: − |
| cpp-novel-miR19-3p-1 | CAGGAGTTGATTTGGAGGACACCA | supercont3.1422:16129:16229:+ |
| cpp-novel-miR19-3p-2 | CAGGAGTTGATTTGGAGGACACCA | supercont3.830:159415:159515: − |
| cpp-novel-miR20-1 | TGATTGTTTACACTCGATCGTTGG | supercont3.533:194362:194441:+ |
| cpp-novel-miR20-2 | TGATTGTTTACACTCGATCGTTGG | supercont3.816:142480:142580:+ |
| cpp-novel-miR20-3 | TGATTGTTTACACTCGATCGTTGG | supercont3.843:44076:44155:+ |
| cpp-novel-miR21-1 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.1761:8353:8427: − |
| cpp-novel-miR21-2 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.1761:10406:10480: − |
| cpp-novel-miR21-3 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.207:246904:246978: − |
| cpp-novel-miR21-4 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.207:248944:249018: − |
| cpp-novel-miR21-5 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.2777:15561:15635: − |
| cpp-novel-miR21-6 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.46:879159:879234:+ |
| cpp-novel-miR21-7 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.65:252545:252619:+ |
| cpp-novel-miR21-8 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.65:254717:254791:+ |
| cpp-novel-miR21-9 | TTGTCAGTGACGGGTAGTTAGGTT | upercont3.69:203697:203771: − |
| cpp-novel-miR21-10 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.69:205631:205705: − |
| cpp-novel-miR21-11 | TTGTCAGTGACGGGTAGTTAGGTT | supercont3.78:292181:292255: − |
In order to identify novel miRNAs, we used MIREAP software to identify potential stem-loop structures in sequences flanking the remaining unannotated reads. The criteria to predict the authenticity of miRNAs was set forth by Berezikov et al. (Berezikov et al., 2011). This method yielded 42 potential miRNAs derived from 21 miRNA precursors (Table 1). We found the levels of novel miRNAs were far lower than the known conserved miRNAs. This phenomenon was also found in Honey bees and Ae. albopictus (Guo et al., 2013; Kim et al., 2013). The low expression levels suggested that the novel miRNAs were expressed in specific developmental stages and organs.
The precursors of conserved and novel miRNAs were all predicted from the Cx. quinquefasciatus genome. We confirmed these precursors using PCR. We designed 48 pairs of primers for the precursors, 36 pairs for conserved miRNAs and 12 for novel miRNAs. As expected, we amplified products shorter than 100 bp from the Cx. pipiens genome (Fig. S2, in Supplementary data). The results showed that the precursors of miRNAs existed in the Cx. pipiens genome.
3.3. Differentially expressed miRNAs and determination of candidate miRNA
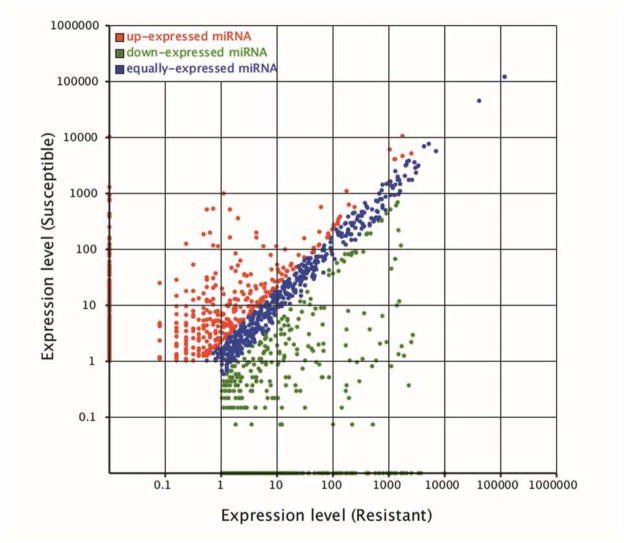
In total, 13760296 and 12355472 miRNAs reads were obtained in the DS-strain and DR1-strain. We first normalized the expression of miRNAs in two samples and calculated the fold-change and P-value based on the normalized expression. Next, we generated the log2 ratio figure and scatter plot (Fig. 1). The expression levels of 26 conserved miRNAs and 2 novel miRNAs differed between the two strains. Nineteen miRNAs showed higher expression levels in the DS-strain and nine miRNAs showed higher expression level in the DR1-strain (Table 2).
Figure 1. Scatter plot of differentially expressed miRNA in the DS-strain and DR1-strain.
Each plot represents a miRNA. Red points indicate a ratio >2. Blue points indicate a 1/2<ratio ≤2. Green points indicate a ratio<1/2. Ratio: standard expression level in DR1-strain/ standard expression level in DS-strain.
Table 2.
Differential expression of miRNAs in Culex pipiens fold-chang
| Susceptible-expressed | Resistant-expressed | Susceptible-std | Resistant-std | (log2 Susceptible/Resistant) | p-value | |
|---|---|---|---|---|---|---|
| cpp-bantam | 3330 | 3181 | 242.0006 | 257.4568 | -0.08931975 | 0.012522392 |
| cpp-let-7 | 23296 | 19387 | 1692.9868 | 1569.1023 | 0.10963131 | 5.10E-15 |
| cpp-miR-1-3p | 1670635 | 1467552 | 121409.8156 | 118777.4939 | 0.03162357 | 1.16E-83 |
| cpp-miR-2-3p-1 | 4293 | 1532 | 311.9846 | 123.9936 | 1.33120916 | 2.27E-93 |
| cpp-miR-7 | 856 | 614 | 62.208 | 49.6946 | 0.32401103 | 2.00E-05 |
| cpp-miR-8 | 26618 | 17744 | 1934.4061 | 1436.1248 | 0.42970957 | 8.75E-211 |
| cpp-miR-9 | 5397 | 3660 | 392.2154 | 296.225 | 0.40495278 | 8.29E-40 |
| cpp-miR-10-3p | 3020 | 2353 | 219.472 | 190.4419 | 0.20468597 | 2.33E-07 |
| cpp-miR-11-3p | 625 | 424 | 45.4205 | 34.3168 | 0.40442856 | 7.33E-06 |
| cpp-miR-12-3p | 506 | 422 | 36.7725 | 34.1549 | 0.10653469 | 0.263488572 |
| cpp-miR-13 | 3510 | 1297 | 255.0817 | 104.9737 | 1.28093148 | 2.71E-183 |
| cpp-miR-14-3p | 6029 | 2816 | 438.1446 | 227.9152 | 0.94290993 | 5.04E-192 |
| cpp-miR-31 | 10745 | 9409 | 780.8698 | 761.5249 | 0.03619081 | 0.075629556 |
| cpp-miR-33 | 889 | 464 | 64.6062 | 37.5542 | 0.78269836 | 3.46E-22 |
| cpp-miR-71 | 7535 | 5245 | 547.59 | 424.5083 | 0.36730334 | 4.74E-46 |
| cpp-miR-79-3p | 671 | 383 | 48.7635 | 30.9984 | 0.65360793 | 6.62E-13 |
| cpp-miR-87 | 677 | 308 | 49.1995 | 24.9282 | 0.98086494 | 1.49E-24 |
| cpp-miR-92a | 2238 | 991 | 162.6419 | 80.2074 | 1.01989172 | 2.79E-82 |
| cpp-miR-92b | 339 | 213 | 24.6361 | 17.2393 | 0.51507269 | 3.77E-05 |
| cpp-miR-100 | 4049 | 2899 | 294.2524 | 234.6329 | 0.32664886 | 8.50E-21 |
| cpp-miR-124 | 5419 | 4123 | 393.8142 | 333.6983 | 0.2389708 | 9.01E-16 |
| cpp-miR-125 | 935 | 439 | 67.9491 | 35.5308 | 0.93538427 | 6.81E-31 |
| cpp-miR-133-3p | 5212 | 1670 | 378.7709 | 135.1628 | 1.48662735 | 0 |
| cpp-miR-137-3p | 119 | 105 | 8.6481 | 8.4983 | 0.02520893 | 0.900201288 |
| cpp-miR-184-3p | 93422 | 53214 | 6789.2435 | 4306.9176 | 0.65659511 | 0 |
| cpp-miR-190-1 | 1319 | 808 | 95.8555 | 65.3961 | 0.55165661 | 4.61E-18 |
| cpp-miR-210-3p | 4563 | 9404 | 331.6062 | 761.1203 | -1.19865352 | 0 |
| cpp-miR-252-1 | 2677 | 1521 | 194.5452 | 123.1034 | 0.66023478 | 1.33E-47 |
| cpp-miR-263 | 55760 | 16302 | 4052.2384 | 1319.4154 | 1.6188202 | 0 |
| cpp-miR-275 | 52109 | 28270 | 3786.9098 | 2288.055 | 0.72689933 | 0 |
| cpp-miR-276-3p-1 | 6372 | 4239 | 463.0714 | 343.0869 | 0.43266062 | 1.53E-52 |
| cpp-miR-277 | 2614 | 1779 | 189.9668 | 143.9848 | 0.39983079 | 1.08E-19 |
| cpp-miR-278-3p | 52 | 21 | 3.779 | 1.6997 | 1.15272439 | 0.001378883 |
| cpp-miR-279-3p | 2658 | 4041 | 215.1274 | 293.671 | 0.44900952 | 3.08E-36 |
| cpp-miR-281-1 | 340 | 181 | 24.7088 | 0.75418337 | 6.84E-09 | |
| cpp-miR-283 | 1795 | 1929 | 130.4478 | 156.1252 | -0.25923081 | 4.22E-08 |
| cpp-miR-285-3p | 1667 | 717 | 121.1456 | 58.031 | 1.06184631 | 7.84E-66 |
| cpp-miR-305 | 6083 | 4461 | 442.069 | 361.0546 | 0.29205455 | 6.13E-25 |
| cpp-miR-306 | 31595 | 38138 | 2296.0989 | 3086.7295 | -0.42689428 | 0 |
| cpp-miR-308 | 151 | 155 | 10.9736 | 12.545 | -0.19307558 | 0.241100194 |
| cpp-miR-309-3p | 25 | 32023 | 0.01 | 2591.8071 | -17.98360325 | 0 |
| cpp-miR-315 | 225 | 187 | 16.3514 | 15.135 | 0.11152549 | 0.436931367 |
| cpp-miR-316 | 660 | 285 | 47.9641 | 23.0667 | 1.05614336 | 7.34E-27 |
| cpp-miR-317-3p-1 | 33482 | 16531 | 2433.2325 | 1337.9497 | 0.8628503 | 0 |
| cpp-miR-375-1 | 235 | 172 | 17.0781 | 13.921 | 0.29488463 | 0.041400308 |
| cpp-miR-932-3p | 759 | 507 | 55.1587 | 41.0345 | 0.42675108 | 2.05E-07 |
| cpp-miR-957-3p | 549 | 360 | 39.8974 | 29.1369 | 0.45344734 | 3.00E-06 |
| cpp-miR-965-3p | 11 | 6299 | 0.7994 | 509.8146 | -9.3168394 | 0 |
| cpp-miR-970-3p | 4604 | 3366 | 334.5858 | 272.4299 | 0.29649117 | 8.93E-20 |
| cpp-miR-980-3p | 113 | 45 | 8.212 | 3.5612 | 1.17296309 | 1.51E-06 |
| cpp-miR-981 | 749 | 226 | 54.432 | 18.2915 | 1.57328166 | 1.48E-54 |
| cpp-miR-988-3p | 57 | 18 | 4.1424 | 1.4568 | 1.50766404 | 3.81E-05 |
| cpp-miR-989-3p | 2507 | 4928 | 182.1908 | 411.8013 | -1.13040196 | 5.93E-238 |
| cpp-miR-993 | 1321 | 882 | 96.0008 | 71.3854 | 0.42741739 | 6.64E-12 |
| cpp-miR-996 | 11712 | 10931 | 851.1445 | 884.7092 | -0.05579924 | 0.003638387 |
| cpp-miR-998 | 593 | 324 | 43.095 | 26.2232 | 0.71667675 | 2.38E-13 |
| cpp-miR-999-3p | 1430 | 1137 | 103.9222 | 92.024 | 0.17542181 | 0.002182576 |
| cpp-miR-1000 | 228 | 92 | 16.5694 | 7.4461 | 1.15396446 | 1.36E-11 |
| cpp-miR-1174 | 5216 | 2385 | 379.0616 | 193.0319 | 0.97359303 | 2.16E-175 |
| cpp-miR-1175 | 13407 | 8399 | 974.325 | 679.7798 | 0.51933559 | 7.76E-151 |
| cpp-miR-1889 | 21 | 19 | 1.5261 | 1.5378 | -0.01101838 | 0.972158615 |
| cpp-miR-1890 | 234 | 144 | 17.0054 | 11.6548 | 0.54506869 | 0.000316285 |
| cpp-miR-1891 | 2235 | 1136 | 162.4238 | 91.9431 | 0.82094983 | 1.15E-57 |
| cpp-miR-2941-3p-1 | 1199 | 2750 | 87.1348 | 222.5734 | -1.35296026 | 6.70E-177 |
| cpp-miR-2951-1 | 43267 | 24845 | 3144.3364 | 2010.8499 | 0.64495018 | 0 |
| cpp-miR-2952-3p | 68 | 134 | 4.9418 | 10.8454 | -1.13397473 | 5.48E-08 |
| cpp-miR-iab-4 | 596 | 630 | 43.313 | 50.9896 | -0.23540292 | 0.004279915 |
| cpp-miR-2779-1 | 3907 | 4295 | 283.9328 | 347.6193 | -0.29195866 | 5.17E-20 |
| cpp-miR-2796-3p | 19851 | 19927 | 1442.6289 | 1612.8077 | -0.1608742 | 1.03E-28 |
| cpp-miR-2840 | 2 | 398 | 0.1453 | 32.2124 | -7.79243765 | 9.08E-126 |
| cpp-miR-2942 | 251 | 104 | 18.2409 | 8.4173 | 1.11574747 | 4.92E-12 |
| cpp-miR-2944b | 20 | 12 | 1.4535 | 0.9712 | 0.58169074 | 0.277420799 |
| cpp-miR-2945 | 38776 | 37021 | 2817.9626 | 2996.3242 | -0.08854127 | 3.08E-17 |
| cpp-miR-2981 | 81697 | 13083 | 5937.1543 | 1058.8831 | 2.48722828 | 0 |
| cpp-miR-3781 | 1262 | 563 | 91.7131 | 45.5669 | 1.00914159 | 2.16E-46 |
| cpp-miR-4682 | 2 | 73 | 0.01 | 5.9083 | -9.2065993 | 1.77E-24 |
| cpp-miR-4448 | 143222 | 21921 | 10408.3517 | 1774.1937 | 2.55250619 | 0 |
| cpp-miR-307 | 56 | 21 | 4.0697 | 1.6997 | 1.25964232 | 0.00036701 |
| cpp-miR-493-3p | 122 | 872 | 8.8661 | 70.576 | -2.99280613 | 1.06E-158 |
| cpp-miR-929-3p | 32 | 27 | 2.3255 | 2.1853 | 0.08970959 | 0.82070986 |
| cpp-miR-1290 | 4988 | 7443 | 362.4922 | 602.4052 | -0.73278427 | 1.80E-173 |
| cpp-novel-miR1 | 1071 | 759 | 77.8326 | 61.4303 | 0.34142412 | 5.37E-07 |
| cpp-novel-miR2-1 | 326 | 178 | 23.6914 | 14.4066 | 0.71763356 | 5.52E-08 |
| cpp-novel-miR3-3p | 115 | 66 | 8.3574 | 5.3418 | 0.64572823 | 0.003377852 |
| cpp-novel-miR4 | 80 | 65 | 5.8138 | 5.2608 | 0.14419924 | 0.554453216 |
| cpp-novel-miR5 | 76 | 54 | 5.5231 | 4.3705 | 0.33767991 | 0.190166848 |
| cpp-novel-miR6-3p | 78 | 38 | 5.6685 | 3.0756 | 0.88209913 | 0.001598861 |
| cpp-novel-miR7-3p-1 | 416 | 40 | 30.2319 | 3.2374 | 3.22316412 | 1.07E-71 |
| cpp-novel-miR8-3p | 61 | 46 | 4.433 | 3.723 | 0.25181775 | 0.376102001 |
| cpp-novel-miR9-1 | 192 | 144 | 13.9532 | 11.6548 | 0.25967178 | 0.102701359 |
| cpp-novel-miR10 | 43 | 32 | 3.1249 | 2.5899 | 0.27091363 | 0.427849104 |
| cpp-novel-miR11-3p | 62 | 11 | 4.5057 | 0.8903 | 2.3393878 | 8.04E-09 |
| cpp-novel-miR12 | 36 | 33 | 2.6162 | 2.6709 | -0.02985313 | 0.925384881 |
| cpp-novel-miR13 | 42 | 21 | 3.0523 | 1.6997 | 0.84461664 | 0.026542101 |
| cpp-novel-miR14-3p | 30 | 22 | 2.1802 | 1.7806 | 0.29209703 | 0.479901119 |
| cpp-novel-miR15 | 26 | 18 | 1.8895 | 1.4568 | 0.37520169 | 0.405654141 |
| cpp-novel-miR16-3p | 21 | 16 | 1.5261 | 1.295 | 0.2368974 | 0.633309487 |
| cpp-novel-miR17-3p | 20 | 15 | 1.4535 | 1.214 | 0.25976265 | 0.611351264 |
| cpp-novel-miR18-3p-1 | 34 | 28 | 2.4709 | 2.2662 | 0.12476143 | 0.743660138 |
| cpp-novel-miR19-3p-1 | 22 | 28 | 1.5988 | 2.2662 | -0.50328571 | 0.220271427 |
| cpp-novel-miR20-1 | 33 | 33 | 2.3982 | 2.6709 | -0.15537398 | 0.658290585 |
| cpp-novel-miR21-1 | 99 | 110 | 7.1946 | 8.9029 | -0.30736087 | 0.123569093 |
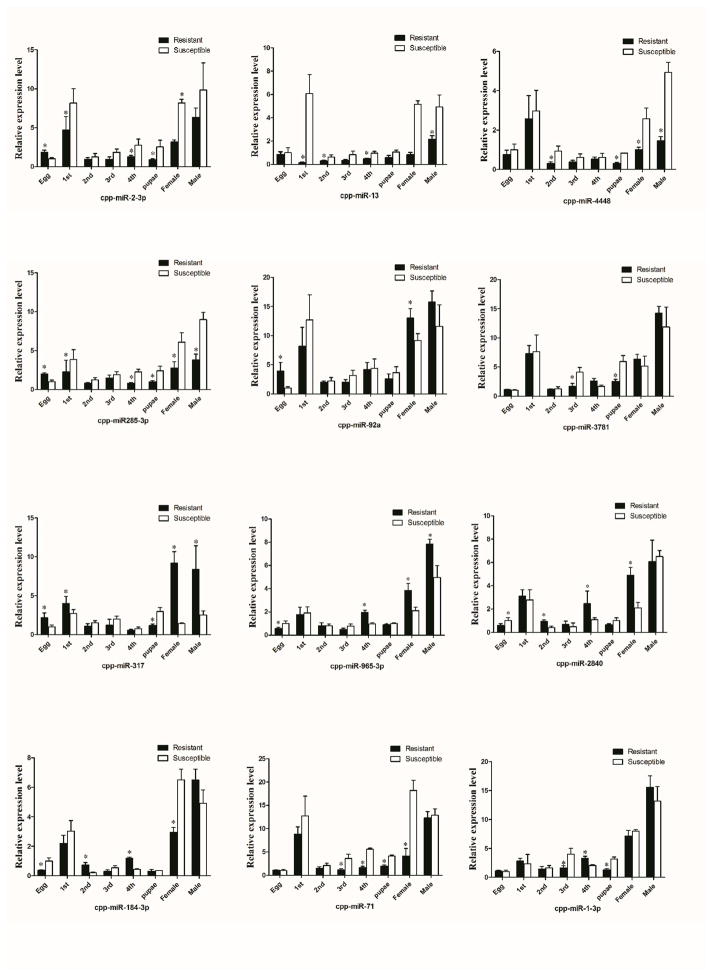
Stem-loop RT-PCR was used to verify the expression levels of candidate differentially expressed miRNAs (Fig. 2). The results suggested that cpp-miR-2-3p, cpp-miR-13, cpp-miR-285-3p, cpp-miR-4448 and cpp-miR-71 were up-regulated in the DS-strain at most stages. Cpp-miR-965-3p, cpp-miR-317 and cpp-miR-2840 were up-regulated in the DR1-strain at most stages. Cpp-miR-92a, cpp-miR-3781, cpp-miR-184-3p and cpp-miR-1-3p had different expression levels at different life stages.
Figure 2. The expression levels of miRNAs in the DS-strain and DR1-strain measured by stem-loop RT-PCR.
Expression levels of cpp-miR-2-3p, cpp-miR-13, cpp-miR-285-3p, cpp-miR-71 and cpp-miR-4448 were up-regulated in the DS-strain, while cpp-miR-965-3p, cpp-miR-317 and cpp-miR-2840 were up-regulated in the DR1-strain at most stages. The results were presented as mean± standard error (SE) of three independent experiments (*p<0.05).
The differentially expressed miRNAs in the two strains provide a basis for investigation of pyrethroid resistance. MiR-1-3p was specifically expressed in musle-rich tissues and organs (Yan et al., 2012). It had been implicated in the determination of differentiated stages of muscle cells and in myogenesis (Chen et al., 2006).
MiR-184-3p was the most highly expressed miRNA in Cx. quinquefasciatus described to date, and had been identified in over 39 organisms, although it has no defined role in insects (Skalsky et al., 2010). MiR-92 was significantly up-regulated miRNA in WNV-infected Cx. quinquefasciatus and may play a role in mediating flavivirus infection in mosquito host (Skalsky et al., 2010). Cpp-miR-71 was highly conserved in the miRNA encoding region (He et al., 2008). In Drosophila, the miR-13 and miR-2 family shared target genes, targeting pro-apoptotic genes and conserving target properties in distant species (Stark et al., 2003). Cpp-miR-71 which clustered with cpp-miR-13 and cpp-miR-2-3p (de Souza Gomes et al., 2013), had higher expression levels in most stages in DS-strains compared with DR1-strain. In Caenorhabdities elegans, miR-71 was required for life span extension (Boulias and Horvitz, 2012). Taken together, cpp-miR-71 may be involved in the control of apoptosis resulting in insecticides resistance and warrants further further investigation.
3.4. Target prediction
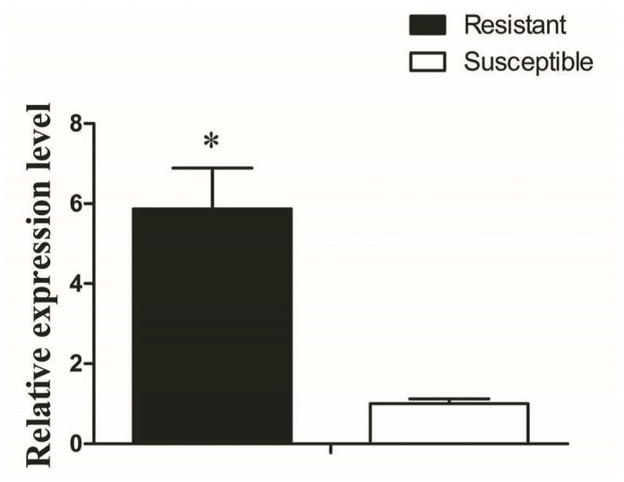
MiRNA works through imperfect base-paring to target seed region sties in the 3′UTR of mRNAs resulting in translational inhibition or mRNA degradation. Using the CYP450 sequence from Cx. quinquefasciatus, we predicted the target gene of cpp-miR-71 using miRNA seed regions binding to the 3′UTR of mRNAs. We found the 3′UTR sequence of CYP325BG3 contained a target site for cpp-miR-71(Fig. 3A). We confirmed the expression level of CYP325BG3 using qRT-PCR and found that CYP325BG3 was up-regulated in DR2-strain (Fig. 4).
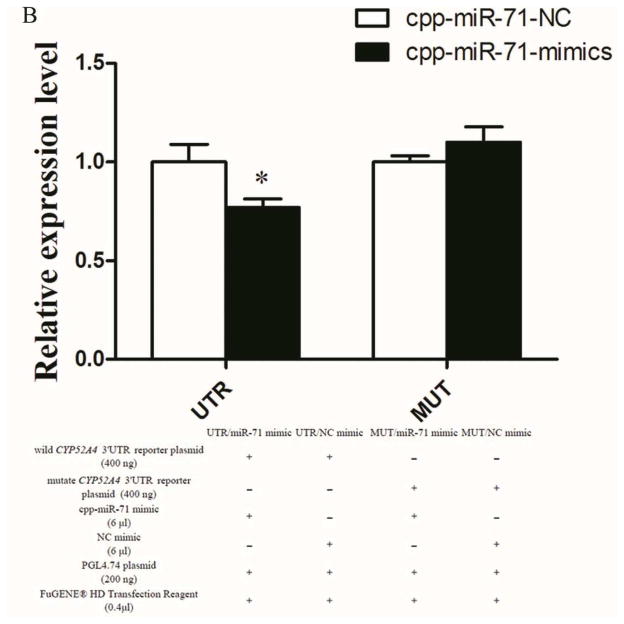
Figure 3. Interaction between cpp-miR-71 and CYP325BG3 using a dual fluorescent reporter system.
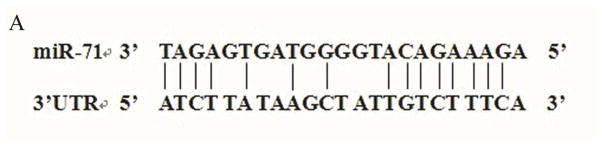
(A) Base pairing between the seed sequence and the 3′UTR of CYP325BG3. (B) Luciferase activity decreased 23% compared to the control after overexpression of cpp-miR-71.The results were presented as mean± standard error (SE) of three independent experiments (*p<0.05).
Figure 4. Expression level of CYP325BG3 in the DR2-strain.
The results were representative of three independent experiments (*p<0.05).
To verify that cpp-miR-71 could regulate CYP325BG3 by directly binding to the 3′UTR of the gene, we used a dual fluorescent reporter system. The miRNA mimic worked as endogenously expressed miRNA. The PGL4.74 plasmid served as an internal control for transfection efficiency using FuGENE® HD Transfection Reagent (Mottahedin et al., 2013). CYP325BG3 targeting by cpp-miR-71 resulted in CYP325BG3 degradation. As predicted, the cpp-miR-71 mimic decreased luciferase activity by approximately 23% in the CYP325BG3 UTR PMIR-reporter vector, compared to the negative mimic control (Fig. 3B). This result indicated that cpp-miR-71 can directly regulate CYP325BG3 in vitro. To confirm whether cpp-miR-71 specifically inhibited CYP325BG3 by binding to the seed sequence, the mutated reporter of the cpp-miR-71 binding site was constructed. Overexpression of cpp-miR-71 did not affect the CYP325BG3 MUT PMIR-report vector activity (Fig. 3B). These results confirmed that CYP325BG3 was a direct target of cpp-miR-71 with the specific binding site at the seed sequence.
3.5. Effects of cpp-miR-71 in female mosquito
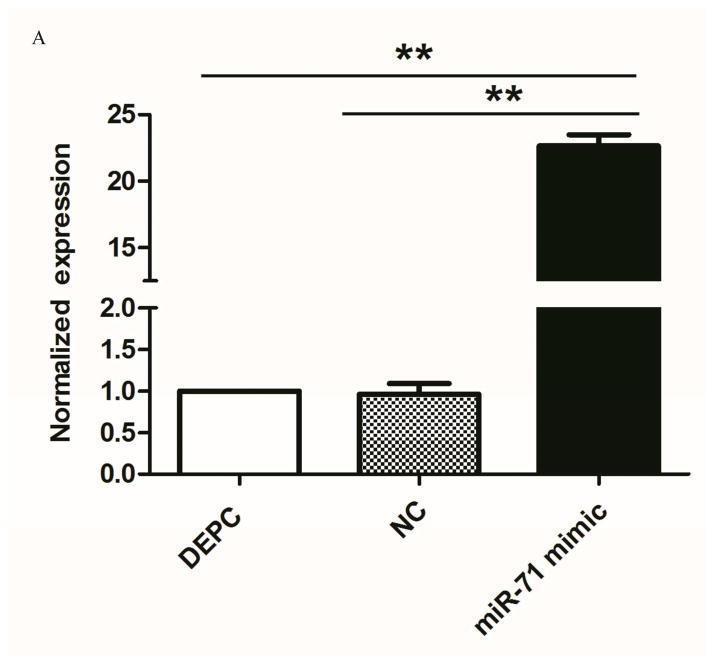
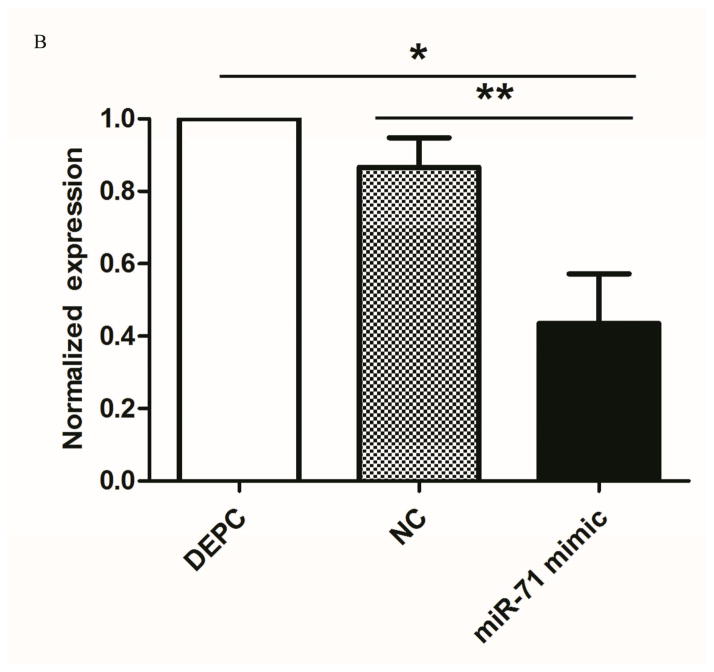
To determine whether the cpp-miR-71 mimic could knockdown the target gene in vivo, the cpp-miR-71 mimic was injected into female adults from the DR2-strain. Compared to the control group, mosquitoes injected with the cpp-miR-71 mimic had much higher expression levels of miR-71 (Fig. 5A) and lower levels of CYP325BG3 (Fig. 5B). These results indicated that overexpression of cpp-miR-71 would cause the down-regulation (almost 57%) of CYP325BG3 in female mosquitoes compared with control group.
Figure 5. Microinjection of the cpp-miR-71 mimic in female adult mosquitoes.
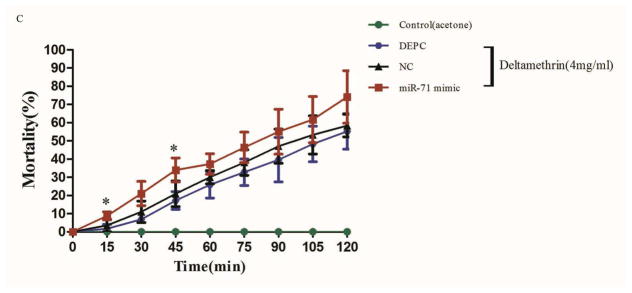
(A) The expression of cpp-miR-71 as analysed by stem-loop quantitative RT-PCR after microinjection the negative control mimic and cpp-miR-71 mimic and DEPC. In the cpp-miR-71 mimic microinjection group, the expression level was 22.63 fold higher than with the DEPC microinjection. (B) qRT-PCR analysis of CYP325BG3 after microinjection. Compared with DEPC-water group, the target gene CYP325BG3 was knocked down almost 57% with the cpp-miR-71 microinjection. (C) Mortality of microinjected mosquitoes observed after a two hour exposure to CDC bottles treated with deltamethrin (4 mg/ml). The results were presented as mean± standard error (SE) of three independent experiments (*p<0.05, **p<0.01). There was no statistically significant difference between the DEPC and negative control (NC) mimic microinjection group.
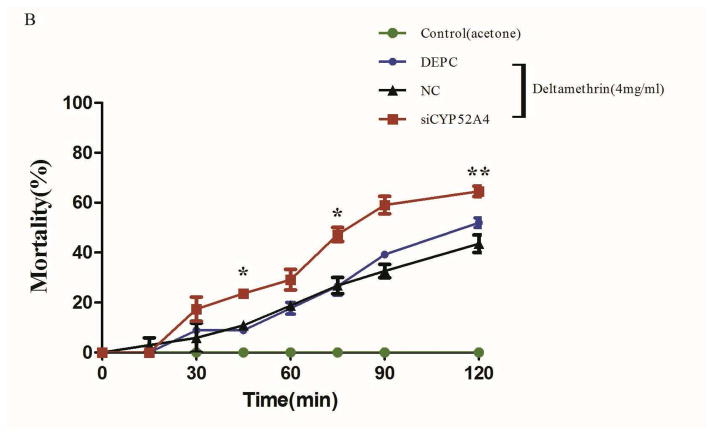
The American CDC Bottle Bioassay was applied after cpp-miR-71 injection to illustrate its contribution to pyrethroid resistance (Fig. 5C). The results showed that cpp-miR-71 injected group had a much higher mortality rate than other groups at each diagnostic time point. At 45 minutes, the mortality rate of the cpp-miR-71 injected group was 33.97%, while the NC group was 20.92% and the DEPC group was 17.24%. There was a statistically significant difference between the cpp-miR-71 injected group and the non cpp-miR-71 injected groups. At 120 minutes, the mortality of the cpp-miR-71 injected group was up to 74.13%. The control group injected with cpp-miR-71 exposed to acetone was all alive after two hours. The results showed that overexpression of cpp-miR-71 in female adults from the DR2-strain decreased resistance to deltamethrin.
Cpp-miR-71 was identical to cel-miR-71, it has been reported that cel-miR-71 may function in pathways that impacted the life span of C.elegans (de Lencastre et al., 2010). Cel-miR-71 increased during the aging process and promoted longevity. Moreover, research showed that cel-miR-71 interacted with the DNA damage response pathway. In our study, cpp-miR-71 was up-regulated in the DS-strain and down-regulated in the DR-strain. The CDC Bottle Bioassay showed that cpp-miR-71 played a negative role in promoting pyrethroid resistance. However, the mechanism of cpp-miR-71 in deltamethrin resistance needs to be further investigation.
3.6. Silencing of CYP325BG3 in vivo
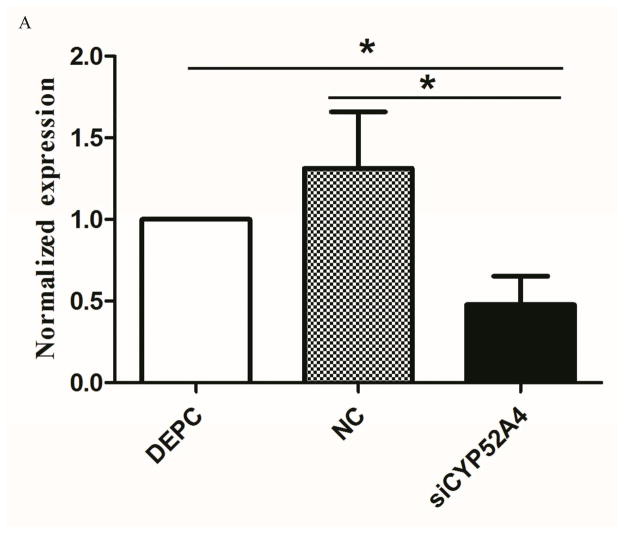
RNA interference (RNAi) is a genetic tool that allows researchers to rapidly modulate genes of interest (Song et al., 2010). We tested the function of CYP325BG3 in vivo by injecting specific siRNA into female adults. qRT-PCR was used to quantify the expression level of CYP325BG3 relative to the control group (Fig. 6A). The group injected with siRNA showed a reduction in CYP325BG3 expression of 57% compared to the control group. Based on the American CDC Bottle Assay, the group injected with siCYP325BG3 had a higher mortality rate than the other two groups (Fig. 6B). The control group was injected with siCYP325BG3 exposed to acetone. The results showed that a lack of CYP325BG3 made female adults more sensitized to deltamethrin.
Figure 6. Silencing of CYP325BG3 in female mosquitoes.
(A) qRT-PCR analysis of the knockdown efficiency of CYP325BG3 in female mosquitoes. Compared with the DEPC-water group, the target gene CYP325BG3 was knocked down almost 60% in the treated group. (B) Mortality of microinjected mosquitoes observed following a two hour exposure to CDC bottles treated with deltamethrin (4 mg/ml). The results are presented as mean± standard error (SE) of three independent experiments (*p<0.05, **p<0.01). There was no statistically significant difference between the DEPC and NC mimic microinjection groups.
CYP325BG3 belongs to the CYP4 clade (Nelson, 2011). P450 enzymes from each clades have been shown to be associated with resistance to insecticides (Feyereisen, 2006). Knockdown of CYP325BG3 made female adults more sensitive to deltamethrin. The results demonstrated that CYP325BG3 contributed to deltamethrin resistance in mosquitoes.
4. Conclusion
In summary, we identified 100 distinct conserved miRNAs and 42 novel miRNAs from mixed growth stages (fourth-instars larvae, pupae, male and female adults) of Cx. pipiens. Our study provides the first large-scale miRNAs profiling of Cx. pipiens and identifies the precursors of these miRNAs. We also compared miRNA expression differences in DS and DR-strains and validated the differences in expression by qRT-PCR. These differentially expressed miRNAs gives us clues to further study the mechanisms of pyrethroid resistance. Cpp-miR-71 is down-regulated at most life stages in DR-strains, especially in female adult mosquitoes. MiRNAs regulate target genes by binding to their 3′UTR and we find that cpp-miR-71 could directly regulate CYP325BG3. This is the first study to show the important role cpp-miR-71 plays in pyrethroid resistance. In addition, our study is the first to provide evidence that the target gene CYP325BG3 participates in pyrethroid resistance. The signaling pathway involving cpp-miR-71 and CYP325BG3, however, requires further study.
Supplementary Material
Analysis of small RNAs. The length distribution in the DS-strain (A) and DR-strain (B). Analysis of the first nucleotide in small RNA sequences from the DS-strain (C) and DR-strain (D).
PCR analysis of the miRNAs precursors in Culex pipiens. Agarose gel electrophoresis of 48 miRNAs precursors. M: Marker 2000. 1-48: PCR product of the 48 miRNAs precursors. N: PCR control, control reaction system: forward primer, reverse primer and enzyme.
Supplementary Table 1 Primers for 36 conserved miRNAs and 12 candidate miRNAs
Supplementary Table 2 Primers for the Stem-loop quantitative RT-PCR
Supplementary Table 3 The sequence of miRNA mimic and negative control of cpp-miR-71
Supplementary Table 4 The sequence of siCYP325BG3 and negative control of siCYP325BG3
Highlights.
We identified 100 conserved miRNAs and 42 novel miRNAs in Culex pipiens
Expression of 28 miRNAs differed in deltamethrin-sensitive and -resistant strains
Cpp-miR-71 mediates pyrethroid resistance via cytochrome P450 325BG3 regulation
Overexpression of cpp-miR-71 in female mosquitoes reduced resistance to deltamethrin
Acknowledgments
This work was supported by the National Institutes of Health of US (NIH) [Grant No. 2R01AI075746], the National Natural Science Foundation of China [Grant Nos. 81171900 and 81101279], the National S & T Major Program [Grant Nos.2012ZX10004-219 and 2012ZX10004-220], the Specialized Research Fund for the Doctoral Program of Higher Education of China [Grant No.20113234120007], and the Natural Science Foundation of Jiangsu Province [Grant No.81101279].
Abbreviations
- DS-strain
deltamethrin-sensitive strain
- DR-strain
deltamethrin–resistant strain
- Cpp-
Culex pipiens-
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Guideline for Evaluating Insecticide Resistance in Vectors Using the CDC Bottle Bioassay.
- Aizoun N, Aikpon R, Padonou GG, Oussou O, Oke-Agbo F, Gnanguenon V, Osse R, Akogbeto M. Mixed-function oxidases and esterases associated with permethrin, deltamethrin and bendiocarb resistance in Anopheles gambiae s.l. in the south--north transect Benin, West Africa. Parasites & vectors. 2013;6:223. doi: 10.1186/1756-3305-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nature reviews Molecular cell biology. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Robine N, Samsonova A, Westholm JO, Naqvi A, Hung JH, Okamura K, Dai Q, Bortolamiol-Becet D, Martin R, Zhao Y, Zamore PD, Hannon GJ, Marra MA, Weng Z, Perrimon N, Lai EC. Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome research. 2011;21:203–215. doi: 10.1101/gr.116657.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulias K, Horvitz HR. The C. elegans MicroRNA mir-71 Acts in Neurons to Promote Germline-Mediated Longevity through Regulation of DAF-16/FOXO. Cell Metab. 2012;15:439–450. doi: 10.1016/j.cmet.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM, Marfin AA, Solomon T, Tsai TF, Tsu VD, Ginsburg AS. Estimated global incidence of Japanese encephalitis: a systematic review. Bull World Health Organ. 2011;89:766–774. 774A–774E. doi: 10.2471/BLT.10.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhong D, Zhang D, Shi L, Zhou G, Gong M, Zhou H, Sun Y, Ma L, He J, Hong S, Zhou D, Xiong C, Chen C, Zou P, Zhu C, Yan G. Molecular ecology of pyrethroid knockdown resistance in Culex pipiens pallens mosquitoes. PLoS One. 2010;5:e11681. doi: 10.1371/journal.pone.0011681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am J Trop Med Hyg. 1987;37:37–41. doi: 10.4269/ajtmh.1987.37.37. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David JP, Strode C, Vontas J, Nikou D, Vaughan A, Pignatelli PM, Louis C, Hemingway J, Ranson H. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4080–4084. doi: 10.1073/pnas.0409348102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A, Pincus Z, Zhou K, Kato M, Lee SS, Slack FJ. MicroRNAs Both Promote and Antagonize Longevity in C-elegans. Current Biology. 2010;20:2159–2168. doi: 10.1016/j.cub.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Gomes M, Donoghue MT, Muniyappa M, Pereira RV, Guerra-Sa R, Spillane C. Computational identification and evolutionary relationships of the microRNA gene cluster miR-71/2 in protostomes. Journal of molecular evolution. 2013;76:353–358. doi: 10.1007/s00239-013-9563-2. [DOI] [PubMed] [Google Scholar]
- Feyereisen R. Evolution of insect P450. Biochem Soc T. 2006;34:1252–1255. doi: 10.1042/BST0341252. [DOI] [PubMed] [Google Scholar]
- Fonseca-Gonzalez I, Quinones ML, Lenhart A, Brogdon WG. Insecticide resistance status of Aedes aegypti (L.) from Colombia. Pest Manag Sci. 2011;67:430–437. doi: 10.1002/ps.2081. [DOI] [PubMed] [Google Scholar]
- Gregory R, Darby AC, Irving H, Coulibaly MB, Hughes M, Koekemoer LL, Coetzee M, Ranson H, Hemingway J, Hall N, Wondji CS. A de novo expression profiling of Anopheles funestus, malaria vector in Africa, using 454 pyrosequencing. PLoS One. 2011;6:e17418. doi: 10.1371/journal.pone.0017418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Su S, Skogerboe G, Dai S, Li W, Li Z, Liu F, Ni R, Guo Y, Chen S, Zhang S, Chen R. Recipe for a Busy Bee: MicroRNAs in Honey Bee Caste Determination. PLoS One. 2013;8:e81661. doi: 10.1371/journal.pone.0081661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart J, Tillman G, Kraut MA, Chiang HS, Strain JF, Li YF, Agrawal AG, Jester P, Gnann JW, Whitley RJ Stud NCA Team WNVP. West Nile virus neuroinvasive disease: neurological manifestations and prospective longitudinal outcomes. BMC Infect Dis. 2014;14 doi: 10.1186/1471-2334-14-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He PA, Nie Z, Chen J, Lv Z, Sheng Q, Zhou S, Gao X, Kong L, Wu X, Jin Y, Zhang Y. Identification and characteristics of microRNAs from Bombyx mori. BMC Genomics. 2008;9:248. doi: 10.1186/1471-2164-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemingway J, Hawkes NJ, McCarroll L, Ranson H. The molecular basis of insecticide resistance in mosquitoes. Insect biochemistry and molecular biology. 2004;34:653–665. doi: 10.1016/j.ibmb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Hemingway J, Ranson H. Insecticide resistance in insect vectors of human disease. Annu Rev Entomol. 2000;45:371–391. doi: 10.1146/annurev.ento.45.1.371. [DOI] [PubMed] [Google Scholar]
- Kim S, Nguyen TD, Lee J, Hong MK, Pham TV, Ahn YJ, Lee BM, Han YS, Kim DE, Kim JG, Kang LW. Homologous expression and T3SS-dependent secretion of TAP-tagged Xo2276 in Xanthomonas oryzae pv. oryzae induced by rice leaf extract and its direct in vitro recognition of putative target DNA sequence. J Microbiol Biotechnol. 2013;23:22–28. doi: 10.4014/jmb.1207.07039. [DOI] [PubMed] [Google Scholar]
- Kumar SS, Puttaraju HP. Improvised microinjection technique for mosquito vectors. Indian J Med Res. 2012;136:971–978. [PMC free article] [PubMed] [Google Scholar]
- Leclercq M, Diallo AB, Blanchette M. Computational prediction of the localization of microRNAs within their pre-miRNA. Nucleic Acids Res. 2013;41:7200–7211. doi: 10.1093/nar/gkt466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lucas K, Raikhel AS. Insect microRNAs: biogenesis, expression profiling and biological functions. Insect biochemistry and molecular biology. 2013;43:24–38. doi: 10.1016/j.ibmb.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottahedin A, Paidikondala M, Cholleti H, Baule C. NF-kappaB activation by equine arteritis virus is MyD88 dependent and promotes viral replication. Arch Virol. 2013;158:701–705. doi: 10.1007/s00705-012-1515-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR. Progress in tracing the evolutionary paths of cytochrome P450. Biochimica et biophysica acta. 2011;1814:14–18. doi: 10.1016/j.bbapap.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Nkya TE, Akhouayri I, Kisinza W, David JP. Impact of environment on mosquito response to pyrethroid insecticides: facts, evidences and prospects. Insect biochemistry and molecular biology. 2013;43:407–416. doi: 10.1016/j.ibmb.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Puthiyakunnon S, Yao Y, Li Y, Gu J, Peng H, Chen X. Functional characterization of three MicroRNAs of the Asian tiger mosquito, Aedes albopictus. Parasit Vectors. 2013;6:230. doi: 10.1186/1756-3305-6-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA, Berenbaum MR. Structure and function of cytochrome P450S in insect adaptation to natural and synthetic toxins: insights gained from molecular modeling. J Chem Ecol. 2013;39:1232–1245. doi: 10.1007/s10886-013-0335-7. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- Skalsky RL, Vanlandingham DL, Scholle F, Higgs S, Cullen BR. Identification of microRNAs expressed in two mosquito vectors, Aedes albopictus and Culex quinquefasciatus. BMC Genomics. 2010;11:119. doi: 10.1186/1471-2164-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund DM, Knipple DC. The molecular biology of knockdown resistance to pyrethroid insecticides. Insect biochemistry and molecular biology. 2003;33:563–577. doi: 10.1016/s0965-1748(03)00023-7. [DOI] [PubMed] [Google Scholar]
- Song C, Gallup JM, Day TA, Bartholomay LC, Kimber MJ. Development of an in vivo RNAi protocol to investigate gene function in the filarial nematode, Brugia malayi. PLoS pathogens. 2010;6:e1001239. doi: 10.1371/journal.ppat.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS biology. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Ye Y, Sun H, Yu J, Zhang L, Sun Y, Zhang D, Ma L, Shen B, Zhu C. Identification of proteasome subunit beta type 6 (PSMB6) associated with deltamethrin resistance in mosquitoes by proteomic and bioassay analyses. PLoS One. 2013;8:e65859. doi: 10.1371/journal.pone.0065859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Hajkova P, Barton SC, Lao K, Surani MA. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 2006;34:e9. doi: 10.1093/nar/gnj009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter F, Edaye S, Huttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic acids research. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wostenberg C, Lary JW, Sahu D, Acevedo R, Quarles KA, Cole JL, Showalter SA. The role of human Dicer-dsRBD in processing small regulatory RNAs. PLoS One. 2012;7:e51829. doi: 10.1371/journal.pone.0051829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Zhong D, Tang L, Chang X, Fu F, Yan G, Zheng B. Anopheles sinensis mosquito insecticide resistance: comparison of three mosquito sample collection and preparation methods and mosquito age in resistance measurements. Parasites & vectors. 2014;7:54. doi: 10.1186/1756-3305-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Watanabe Y, Maehata T, Morita R, Yoshida Y, Oikawa R, Ishigooka S, Ozawa S, Matsuo Y, Hosoya K, Yamashita M, Taniguchi H, Nosho K, Suzuki H, Yasuda H, Shinomura Y, Itoh F. An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa. World journal of gastroenterology : WJG. 2014;20:3927–3937. doi: 10.3748/wjg.v20.i14.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan XC, Ding L, Li YC, Zhang XF, Liang Y, Sun XW, Teng CB. Identification and Profiling of MicroRNAs from Skeletal Muscle of the Common Carp. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of small RNAs. The length distribution in the DS-strain (A) and DR-strain (B). Analysis of the first nucleotide in small RNA sequences from the DS-strain (C) and DR-strain (D).
PCR analysis of the miRNAs precursors in Culex pipiens. Agarose gel electrophoresis of 48 miRNAs precursors. M: Marker 2000. 1-48: PCR product of the 48 miRNAs precursors. N: PCR control, control reaction system: forward primer, reverse primer and enzyme.
Supplementary Table 1 Primers for 36 conserved miRNAs and 12 candidate miRNAs
Supplementary Table 2 Primers for the Stem-loop quantitative RT-PCR
Supplementary Table 3 The sequence of miRNA mimic and negative control of cpp-miR-71
Supplementary Table 4 The sequence of siCYP325BG3 and negative control of siCYP325BG3