Abstract
Objective
Runx1, the hematopoietic lineage determining transcription factor, is present in perichondrium and chondrocytes. Here we addressed Runx1 functions, by examining expression in cartilage during mouse and human osteoarthritis (OA) progression and in response to mechanical loading.
Methods
Spared and diseased compartments in knees of OA patients and in mice with surgical destabilization of the medial meniscus were examined for changes in expression of Runx1 mRNA (Q-PCR) and protein (immunoblot, immunohistochemistry). Runx1 levels were quantified in response to static mechanical compression of bovine articular cartilage. Runx1 function was assessed by cell proliferation (Ki67, PCNA) and cell type phenotypic markers.
Results
Runx1 is enriched in superficial zone (SZ) chondrocytes of normal bovine, mouse, and human tissues. Increasing loading conditions in bovine cartilage revealed a positive correlation with a significant elevation of Runx1. Runx1 becomes highly expressed at the periphery of mouse OA lesions and in human OA chondrocyte ‘clones’ where Runx1 co-localizes with Vcam1, the mesenchymal stem cell (MSC) marker and lubricin (Prg4), a cartilage chondroprotective protein. These OA induced cells represent a proliferative cell population, Runx1 depletion in MPCs decreases cell growth, supporting Runx1 contribution to cell expansion.
Conclusion
The highest Runx1 levels in SZC of normal cartilage suggest a function that supports the unique phenotype of articular chondrocytes, reflected by upregulation under conditions of compression. We propose Runx1 co-expression with Vcam1 and lubricin in murine cell clusters and human ‘clones’ of OA cartilage, participate in a cooperative mechanism for a compensatory anabolic function.
Introduction
The Runx family of transcription factors have been characterized as master regulatory factors for the differentiation of specific cell phenotypes [1]. Both null mutations in mice and human mutations have established Runx1 is essential for hematopoiesis [2]. Runx2 is essential for vascularization and ossification of the hyperytrophic zones and bony elements [3, 4]. Runx3 is necessary for nerve and gut development [1], but is also expressed in prehypertrophic chondrocytes and its loss of functions causes delayed chondrocyte maturation [5]. Remarkably, Runx2 target genes include matrix metalloproteinases (MMPs), growth factors (VEGF) and extracellular matrix proteins which are important for endochondral development and are deregulated in chondro-osseous diseases [1, 3]. However, the specific functions of Runx1 during chondrogenesis in supporting post-natal cartilage homeostasis and in contributing to disease states are less well understood.
Many studies show Runx1, 2 and 3 cooperate for development of the skeleton and in degeneration of the intervertebral disc (IVD) [6–9]. For chondrogenesis to proceed from mesenchyme Runx2 must be downregulated in mesenchymal progenitors with a concomitant upregulation of Sox9[10, 11], while Runx1 expression is retained in mature chondrocytes [12]. Runx2 is highly expressed in hypertrophic chondrocytes and with Runx3 drives hypertrophic cartilage ossification [9, 13]. Runx2 appears to keep Runx1 and Runx3 repressed during development of the intervertebral disc [7]. These observations suggest both overlapping and non-redundant functions of Runx2 and Runx3 and that Runx1 has independent functions in different cartilage tissues [14].
Runx1 expression is robust in mesenchymal condensations, resting and proliferating zone chondrocytes and is the only Runx factor that is expressed in permanent cartilage structures including the xyphoid process, articular and hyoid cartilages [12]. Mice deficient for Runx1 in non-hematopoietic lineages develop normal skeletons but their sterna fail to mineralize, delayed endochondral development of sternal vertebrae and non-fusion of the supraoccipital bone [15]. In mesenchymal specific Runx1 knockout mice, mesenchymal cells condense normally but have delayed commitment to the chondrocyte lineage [6]. These findings suggest that Runx1 is involved in, but not required for commitment to the chondrocyte lineage.
In the present study, we examined a role for Runx1 supporting cartilage homeostasis, by determining Runx1 expression in chondrocyte populations, under varying loading conditions, during late stages of human knee OA and throughout induced stages of experimental osteoarthritis (OA). It has been reported that Runx2 and Runx2 target genes that degrade cartilage matrix (e.g., MMPs) are upregulated in OA tissue [16]. Consistent with these findings, haploinsufficiency of Runx2 leads to a reduced severity of OA in mice challenged with which induces OA in mice [17]. Our key findings support the hypothesis that the highest Runx1 level in the SZ contributes to the stability of the phenotype of these cells and that elevated Runx1 expression in chondrocytes at the periphery of OA lesions may be an adaptive response of articular cartilage to OA induced cartilage damage.
Materials and Methods
Samples from Human Osteoarthritic Patients
Knee joints were obtained from osteoarthritic patients with varus malalignments undergoing total knee replacements, in accordance with Institutional policy on discarded samples. The tibial plateau and medial and lateral condyles of the femur were removed and immediately frozen. Full thickness articular cartilage samples were removed from the tibial plateau and the medial and lateral femoral condyles and each sample divided in half for both RNA and protein analysis (Table 1).
Table 1.
Summery of Runx1 Experimental and Patient Data from Human OA Knees
| Pt # | Age | Gender | Malalignment | Lateral Femur | Medial Femur | Tibia | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Anterior Femur | Posterior Femur | Combined LF | Anterior Femur | Posterior Femur | Combined MF | Lateral Tibia | Medial Tibia | ||||
|
|
|
|
|||||||||
| K1 | 52 | F | Varus | 0.17 (0.1) | 1.37 (0.31) | NC | NC | ||||
| K2 | 81 | F | Varus | 0.92 (0.25) | 0.11 (0.06) | 0.14 (0.05) | 0.50 (0.11) | 0.24 (0.04) | 0.22 (0.08) | 0.52 (0.05) | 0.74 (0.28) |
| K3 | 52 | F | Varus | 0.05 (0.01) | 1.18 (0.2) | 1.71 (0.42) | 1.35 (0.48) | 1.28 (0.11) | 1.60 (0.65) | 0.55 (0.09) | 0.95 (0.09) |
| K4 | 66 | F | Varus | 0.68 (0.09) | 1.24 (0.44) | NC | NC | ||||
| K5 | 64 | F | Varus | 0.01 (0.08) | 0.55 (0.25) | 0.25 (0.08) | 1.18 (0.15) | 1.02 (0.13) | 1.01 (0.51) | 0.44 (0.08) | 0.63 (0.06) |
NC, Not collected. Combined samples of anterior with posterior tissue not separated
Densitometry measurements of Runx1 protein levels by immunoblot are expressed as mean (+/− SEM)
Bovine Studies
Full-thickness articular cartilage was isolated from three 10-day old bovine knee joints. Cartilage was processed into 3 x 1mm discs for compression (n=12 discs/group). 12 cartilage discs were cultured in either one well of a 6-well plate (free swelling control) or under conditions of 25% or 50% compression for 3 days (2 media changes) in Dulbecco’s Modified Eagle Media (DMEM) supplemented with insulin-transferrin-sodium selenite (ITS) (Sigma). For compression experiments, discs were cultured in a previously described compression apparatus [18] that holds 12 discs in individual compartments under conditions of constant compression. Repetitive experiments were performed with 36 discs from 3 calves. Statistics were performed in 24 samples comparing 0%, 25%, and 50% compression groups.
Mouse Osteoarthritis Model
129S6/SvEv mice were obtained from Taconic Farms (Germantown, NY). Using a UMass approved animal IACUC protocol, osteoarthritis was surgically induced in the right leg of 10-week old male 129 S6/SvEv mice by destabilization of the medial meniscus (DMM) as previously described in detail [19]. In sham-operated animals, the skin and joint capsule were surgically opened but the medial meniscotibial ligament was left intact. Tissue from n=5 mice per group were harvested at each time point from all DMM animals and one mouse was harvested at each time point for Sham-operated animals.
Immunohistochemistry and Immunofluorescense
Samples from human and mouse were fixed in freshly prepared 4% paraformaldehyde in cacodylic buffer for 24 hours, dehydrated through a series of graded ethanols to citrusol (Fisher Scientific, then paraffin embedded for 6μm sectioning. Safranin O staining was performed as previously described [20]. Runx1 and PCNA by immunohistochemistry (IHC) was performed on rehydrated, paraffin sections. Hydrogen peroxide (3% in methanol) was used to block endogenous peroxidase. Antigen retrieval was performed using Dako (Carpinteria, CA) Antigen Retrieval reagent in a 2100 Retriever apparatus, according to the manufacturer’s specifications (Pick Cell Laboratories). Protein blocking was carried out in 5% BSA in PBS for 1 hour, sections were then incubated in primary antibody overnight, followed by three washes in buffer (0.1% BSA, 0.2% Gelatin, 0.05% Saponin in PBS), then incubating in secondary antibody for 2 hours, and washed three times in wash buffer. Staining was visualized using the DAB detection system (Dako). Negative control sections consisted of overnight incubation using nonimmune serum from the same species as the secondary antibody.
Immunofluorescense (IF) was used to detect Runx1, Vcam1, and Ki-67. Paraffin sections were rehydrated through a series of graded alcohols to phosphate buffered saline. Sections were visualized on a Zeiss Axioplan Fluorescence microscope.
Immunoblot Analysis
Cartilage samples from human and bovine knee joints to be used for immunoblotting were flash-frozen in liquid nitrogen, pulverized with a Bessman tissue pulverizer (Fischer Scientific), chilled in liquid Nitrogen, and put into Lysis Buffer optimized for cartilage protein extraction as previously described [18]. Proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed for the presence of Runx1 and Gapdh. We analyzed 58 lateral and 52 human cartilage samples from n=5 patients which were compiled for statistical analysis. For bovine experiments, 12 cartilage discs were harvested from the same compression chamber and pooled together for protein isolation. Densitometry analysis (HP Scanjet 7400c using transilluminator XPA attachment) of immunoblots were performed for the Runx1 and corresponding GAPDH for normalization of each individual sample.
Antibodies
For human, mouse and bovine samples for either IF or IHC the following specific antibodies were used for IF: AML1/Runx1 (cat#4334, Cell Signaling Technologies), 1 (Cat# 1504, Santa Cruz), Ki-67 (Cat# ab8191, Abcam) and PCNA (Cat# ab29, Abcam). Secondary antibodies used for IHC included Goat anti-Mouse HRP conjugate (Cat# SC-2005, Santa Cruz), and Goat anti-Rabbit HRP conjugate (Cat# SC-2004, Santa Cruz). Secondary antibodies used for immunofluorescense included Alexa Fluor 488 Donkey anti-Rabbit (Cat# A21206, Invitrogen), Alexa Fluor 594 Donkey anti-Goat (Cat# A11058, Invitrogen), and Alexa Fluor 594 Donkey anti-Mouse (Cat# A21203, Invitrogen). For immunoblots, we used GAPDH (Cat# SC-31915 Santa Cruz) and the same AML1 antibody as stated above which has been validated [21, 22].
RT-PCR
Bovine cartilage samples (n=12 plugs/sample) to be used for RNA isolation were flash-frozen in liquid nitrogen, and pulverized as above. Each sample was homogenized in 1mL of Trizol using a Polytron PT1200 homogenizer for 20 seconds on level 1. RNA isolation was carried out by phenol/chloroform extraction followed by ethanol precipitation. DNA contamination was removed using the DNase-free RNA kit (Zymo) according to the manufacturer’s specifications. Nucleic acid quantification was carried out using a Nano-drop spectrophotometer, and RNA purity was assessed using the 260/280 ratio. One μg of RNA was reverse transcribed into cDNA with the First Strand cDNA synthesis kit (Invitrogen) using random hexamers according to the manufacturer’s protocol. RT-PCR was performed on a 7500 fast RT-PCR machine (Applied Biosystems). Samples were run in duplicate using 20ng of cDNA, 5μM forward and reverse primer, and SybrGreen Master Mix (BioRad) in a 20 μL reaction. Values were normalized to B-actin. Primers used were Runx1 Forward: 5′ AACCCTCAGCCTCAGAGTCA 3′; Runx1 Reverse: 5′ GCGATGGATCCCAGGTACT 3′; B-actin Forward: 5′CTGCGGCATTCACGAAACTA 3′; B-actin Reverse: 5′ ACCGTGTTGGCGTAGAGGTC 3′
Runx1 depletion in mouse embryo fibroblasts
Excision of Runx1 in mouse embryo fibroblasts from Runx1f1/f1 mice was accomplished by transfection with adenovirus-Cre and replated under chondrogenic conditions for 24 hours [25]. The mRNA levels of both Runx1 and Histone H4, a marker of cell proliferation were normalized to Gapdh. Expression of these markers relative to the control (empty adenovirus vector) was then compared.
Results
Runx1 is upregulated in human osteoarthritis
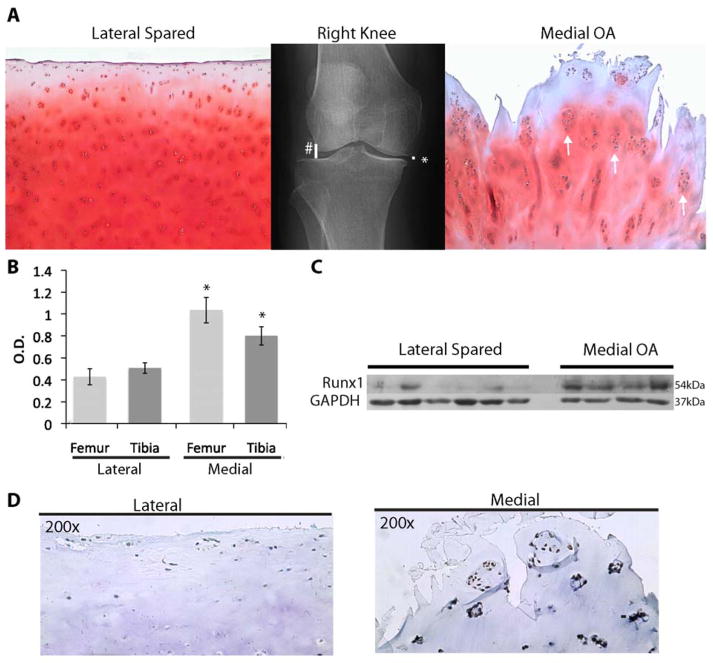
In patients with varus knee the knee than on the lateral side (spared) as can be seen in Figure 1A by the loss of Safranin O staining, severe fibrillation, and chondrocyte cloning in the medial compartment of this malaligned knee. Also observed are the typical clustered chondrocytes, or “clones”, in the affected OA area (arrows in Figure 1A, right panel). Since Runx1 expression predominates in articular cartilage, we hypothesized that its expression would be modified in human OA tissue. Immunoblot analysis of cartilage from both compartments using a Runx1 specific antibody revealed an upregulation of Runx1 protein levels from n=5 OA patients (Figures 1. B & C). Densitometry analysis of immunoblots of cartilage samples (58 lateral; 52 medial) shows that the medial tibial and femoral compartments are increased 2-fold and 2.5-fold, when compared to the lateral tibial and femoral compartments, respectively (p<0.05, Table 1 and Figure 1B). A representative immunoblot from a single patient examines multiple samples (each lane) from the two compartments for Runx1 protein (Figure 1C). The findings reveal barely detectable levels on the lateral side, but robust expression in the medial OA tissue samples.
Figure 1.
Runx1 is upregulated in Human Osteoarthritis. (A) X-ray from a patient with a varus malalignment of the right knee showing compression in the medial compartment (*) of the knee and a normal joint space in the lateral compartment (#) (Patient K1 in Table 1). Safranin O staining shows loss of extracellular matrix on the medial side of the joint versus normal extracellular matrix on the lateral side. White arrows indicate “OA clones”. (B) Graph shows mean +/− SEM quantification of immunoblots stained for Runx1 using 110 cartilage samples from both tibial and femur locations from n=5 patients (see Table 1 and Methods). Data from the medial and lateral compartments were compared using Tukey’s test, *=p<0.05. (C) A representative immunoblot from a human OA knee (Table 1, patient K2) using a Runx1-specific antibody shows overall increased Runx1 in n=4 medial cartilage samples compared to n=6 lateral cartilage samples. (D) Immunohistochemical staining of tissue samples from the same patient as shown in Figure 1C, using a Runx1 specific antibody on the lateral (spared) and medial (OA) compartments (20x).
The cellular localization of Runx1-positive cells in articular cartilage was identified by IHC analysis of OA knees. Runx1 protein is detected in normal chondrocytes of the spared tissue and in cells located mainly in the SZ (Figure 1D, lateral compartment). However, in this spared compartment, a few clusters of Runx1-positive cells chondrocytes are found, indicating evidence of some disease on the spared side of an OA joint. These clusters, characteristic of fibrillated OA cartilage, are enriched in the medial diseased compartment of the joint and show the presence of Runx1 (Figure 1D, medial compartment). Taken together, these findings suggest that Runx1 is upregulated in chondrocyte clones in response to damaged cartilage in human osteoarthritis.
Runx1 is upregulated in response to mechanical compression
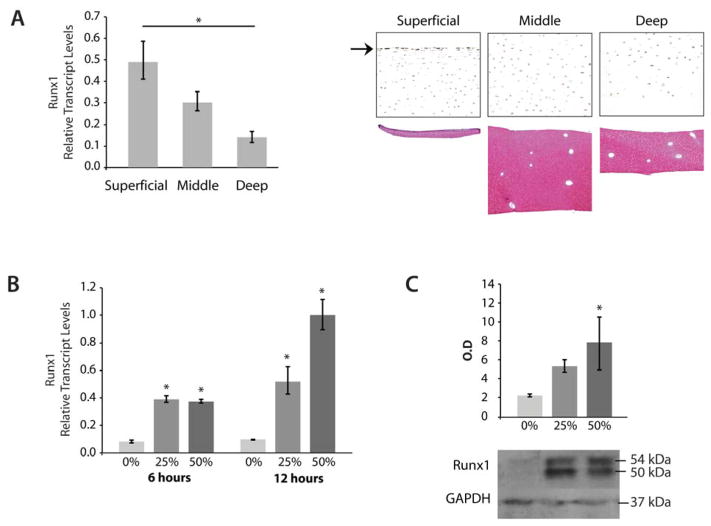
To address a functional role for Runx1 in articular cartilage, we examined its physiological responsiveness to load distribution on articular surfaces. First, Runx1 expression was determined in chondrocytes that define the superficial, middle, and deep zones of bovine articular cartilage (Figure 2A), with thickness of each zone shown in lower panel). QPCR analysis of the dissected zones shows that levels of Runx1 are highest in the SZ and diminish through the two deeper zones (Figure 2A, upper panel). Runx1 mRNA expression is significantly different between the superficial and deep zones (p<0.05). Immunohistochemical analysis also shows that Runx1 protein throughout bovine articular cartilage has the most intense staining in surface cells of the SZ (Figure 2A, middle panel, arrow) and is uniformly and robustly expressed in middle zone chondrocytes (Figure 2A, middle panel,), but in fewer chondrocytes in the deep zone.
Figure 2.
Runx1 is Upregulated in Bovine Cartilage in Response to Mechanical Compression. (A) Left Panel: qPCR analysis of Runx1 mRNA in the superficial, middle and deep zones of normal bovine articular cartilage were normalized to beta-actin levels and then compared using Tukey’s test, *=p<0.05. Right Panel: Immunohistochemistry showing the zonal distribution of Runx1 in bovine articular cartilage (10x). The arrow indicates Runx1 in the upper SZ. Lower Right Panel: Safranin O staining showing isolation of cartilage zones. (B) qPCR of Runx1 mRNA in cartilage samples from n=3 bovine knees undergoing 0%, 25%, or 50% compression for 6 or 16 hours (n=12 pooled samples per group; duplicate assay of samples). Data from 25% or 50% compression was normalized to beta-actin levels and then compared to 0% compression using Tukey’s test, *=p< 0.05. (C) Upper: Quantification of immunoblot analysis shows a significant increase of Runx1 protein levels in response to 25% or 50% compression when compared to uncompressed samples (Tukey’s test, *=p<0.05) from n=3 bovine knees. Lower: Representative immunoblots of Runx1 protein and Gapdh (loading control) in compressed cartilage samples. The Runx1 immunoblot revealed a clear Runx1 doublet band, both of which are included in the densitometry measurements.
Since OA is a disease that results from chronic overloading of articular cartilage, it is of interest to determine if Runx1 responds directly to mechanical compression. Bovine cartilage plugs (see Methods) that underwent compression loading began to show loss of extracellular matrix as evidenced by loss of Safranin-O staining, indicating that compression of these plugs mimics the disease process of OA (data not shown). Runx1 mRNA levels are upregulated in response to graded degrees of tissue strain after as little as 6 hours of mechanical compression (Figure 2B). Runx1 levels continued to increase through 12 hours of compression with significant differences when compared to 0% loading and with a positive correlation to load. Runx1 protein is also upregulated in cartilage plugs after undergoing three days of either 25% or 50% strain when normalized to GAPDH protein levels and then compared to the plugs that remained uncompressed during culture (Figure 2C, upper panel, n=3, p <0.05). This elevated Runx1 expression that occurs in normal bovine cartilage in response to compression, suggests that the increased Runx1 observed in human OA samples is a consequence of compression of the diseased joints.
Runx1 is expressed in a subpopulation of chondrocytes in a mouse model of OA
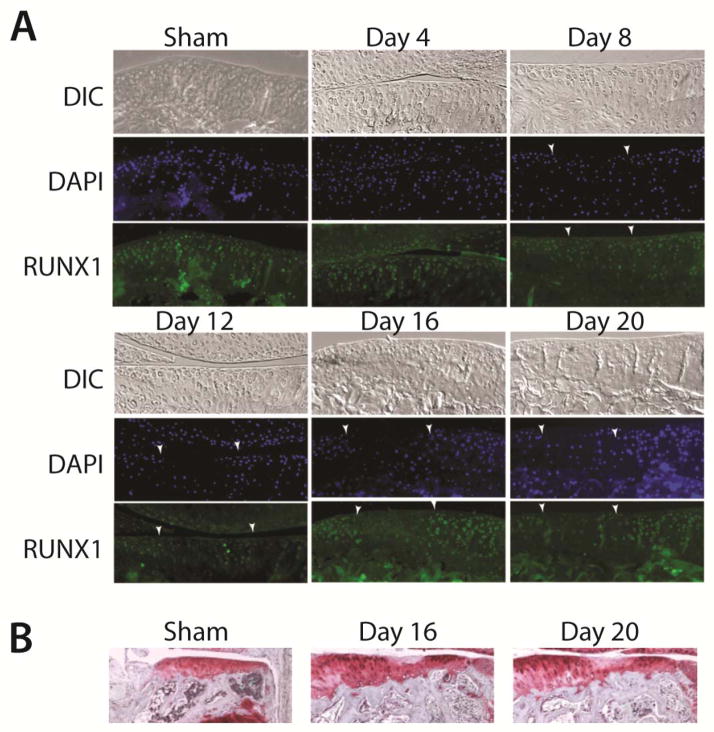
To better characterize the changes in distribution and cellular activity of Runx1 during progression of OA, we generated OA in the DMM mouse model [23]. This surgery creates a focal osteoarthritic lesion on the medial side of the joint, while sparing the lateral side of the joint, allowing for examination of Runx1 during the progressive degenerative stages of articular cartilage over a 3 week. In sham-operated animals, no arthritic lesions were observed and Runx1 was detected in the population of chondrocytes that are present near the joint surface (Figure 3A, sham & day 4). We also observe Runx1-positive cells in subchondral bone marrow spaces indicating hematopoietic lineage cells that express Runx1 (Figure 3A,B, sham). By day 8, live cells (DAPI-positive) are observed; however, Runx1 immunostaining began to decrease between the white arrowheads (Figure 3A). In this region, degenerating cartilage is indicated by weaker DAPI staining on day 8. On day 12, the loss of DAPI and Runx1 within the OA lesion boundaries was more pronounced. This is in striking contrast to robust expression on day 16. On day 16, a clear increase in Runx1 expression is observed in chondrocytes clustered at the periphery of the OA lesions, in contrast to the absence of Runx1 expression in the developing OA lesion between the arrows. This pattern remained throughout the time course to day 21. Thus in the mouse, Runx1 is expressed in SZ chondrocytes of normal articular cartilage and is depleted within the progressing OA lesion by loss of cells (confirmed by absence of DAPI staining). Significantly, Runx1 induced expression occurs in chondrocyte clusters appearing at lesion boundaries. This expression pattern of Runx1 present in normal tissue of the SZ and in lesion boundary cells in response to damaged cartilage surfaces, suggests a linkage of Runx1 to an important boundary function.
Figure 3. Runx1 expression during progression of OA lesions in the DMM Mouse Model of OA.
(A) Top row of each set: DIC (differential interference contrast); middle row: DAP1 nuclei staining; lower panel: immunofluorescence shows robust Runx1-positive cells (middle: DAPI; lower: Runx1), Runx1 in SZ (shown day 4), and a decline during progression to a fibrillated surface (day8, 12, 16, 20) indicated by arrows with strong staining at the boundary of OA and normal tissue on day 20 (40x). Note that the areas appear aggregated of positive cells below the articular cartilage represents Runx1-positive marrow cells in subchondral bone trabecular spaces. Nuclei are stained using DAPI (blue). (B) Safranin O – hematoxylin at the indicated days to show area in subchondral bone coinciding with Runx1 expressing cells in the bone marrow compartment.
Runx1 functions with markers of mesenchymal progenitor cells in normal and osteoarthritic cartilage
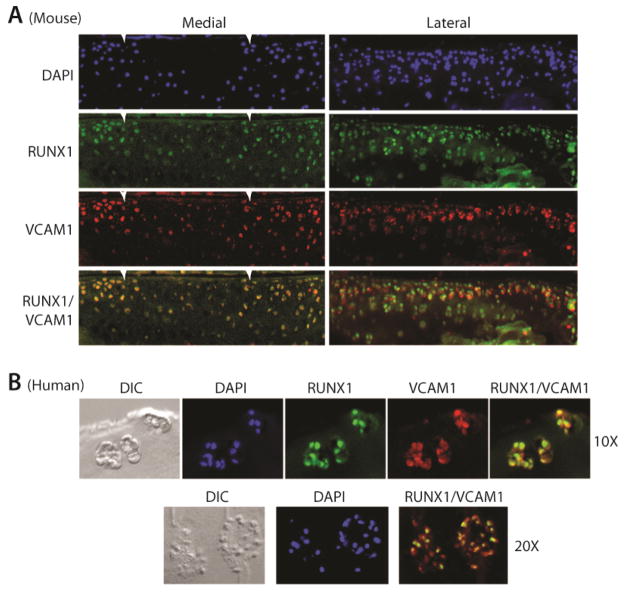
It is recognized that aggregations of chondrocytes in human ‘OA clones’ in damaged cartilage express MSC markers [8, 12]. We thus tested a functional role for Runx1 in association with these MSC-expressing chondroprogenitor cells. Analysis of normal and osteoarthritic mouse cartilage revealed that Runx1 and Vcam1 are found in the same population of chondrocytes at the surface of normal articular cartilage (Figure 4A, lateral compartment), as well as in the characteristic clustered cells at the periphery of osteoarthritic lesions (Figure 4A, medial). We also observed in human OA clones of damaged cartilage that 70% (over a range of 63–85%) of the chondrocytes which are Vcam1-positive, also co-express Runx1 (Figure 4B lower panel quantifying n=6 clusters in n=2 OA patients.
Figure 4.
Runx1 and cam1 Co-localize in Normal and Osteoarthritic Cartilage. (A) Immunofluorescense for Runx1 (green), Vcam1 (red), and DAPI (blue) in articular cartilage on the lateral (spared) and medial (OA) side of the knee joint after induction of osteoarthritis in a mouse (40x). White arrows indicate lesion boundaries. (B) Immunofluorescense for Runx1 (green), Vcam1 (red), and DAPI (blue) on sections from the medial (OA) condyle of a human osteoarthritic knee joint (10x, upper panel). (C) Higher magnification of merged images of Runx1- and Vcam1-positive cells in the OA clones which reveals that both proteins co-localize in the majority of clone cells.
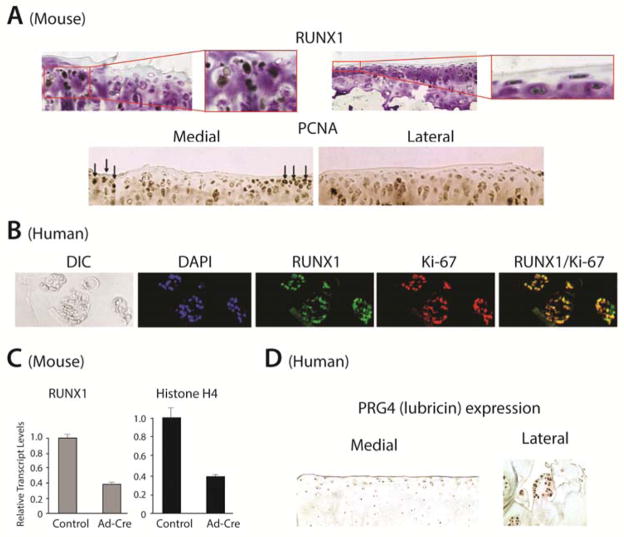
The possible role of Runx1 in expanding the population of MSC clustered cells by proliferation, in an attempt to re-populate damaged osteoarthritic cartilage, was addressed. We find chondrocyte clusters at the periphery of OA lesions are PCNA positive (Figure 5A, lower panel) in the mouse model of osteoarthritis. In human sections both Runx1 and PCNA overlapped in expression (data not shown). Ki-67 is an additional proliferation marker for human specimens (Figure 5B) and demonstrates co-expression with Runx1 in nearly all cells in the human OA clones. For further validation Runx1 is functionally related to proliferation, we conditionally deleted Runx1 in mouse embryo fibroblasts from Runx1fl/fl mouse [24]. This population of cells, derived from MSCs, form chondroid nodules [25]. Proliferation was monitored following Adenovirus-Cre excision of Runx1 in vitro which resulted in a 61% reduction in expression and a similar reduction in Histone H4 expression, a marker of cell proliferation, compared to virus-treated control cells (Figure 5 C). Taken together, the multiple proliferation markers expressed in the same population of cells that are Runx1 positive in human and mouse OA lesions and the contribution of Runx1 to the proliferative capacity of MEFs suggest Runx1 supports cell proliferation. Our findings are consistent with delayed cartilage callus formation during fracture healing in mice with conditional deletion of Runx1 MSCs [26].
Figure 5.
Putative mechanisms for the function of chondrocyte clones in OA. Runx1 and proliferation markers are expressed in the same population of cells in OA lesions. (A) Immunohistochemistry for Runx1 (upper) and PCNA (lower) in articular cartilage of a mouse 14days following OA-inducing surgery show expression in the same cells (10x left; 20x right inset; for Runx1). PCNA is shown at 10x (positive nuclei are denoted by black arrows). (B) Human chondrocyte clones: immunofluorescence for Runx1 (green), Ki-67 (red) and DAPI (blue) and Runx1/Ki-67 co-localization (yellow) in human OA cartilage from the medial OA knee compartment (40x). (C) Evidence that Runx1 is associated with proliferation. Excision of Runx1 from Runx1f1/f1 mouse embryo fibroblasts showed decreased mRNA levels of Runx1 and Histone H4, a marker of cell proliferation relative to the control (empty adenovirus vector) both were normalized to Gapdh. (D) Human OA clones express lubricin in the medial compartment (40x); lateral compartment shows lubricin enriched mainly in the SZ (20x). Note that these lubricin-positive clones were not restricted to the surface of the articular cartilage and the pattern of lubricin expression within each clone is peripherally organized at the outer edges of each clone.
Lubricin, a chondroprotective protein, is expressed in human OA clones
To date, it remains unclear if chondrocyte clones are a consequence of the tissue degeneration or a compensatory response to the damage [27, 28]. We tested the hypothesis that the clonal aggregates in OA cartilage are being formed to repopulate the damaged surface. To support this hypothesis we examined expression of a key articular surface component Lubricin (Prg4), recognized as a chondroprotective protein critical for SZ integrity [23, 27]. Examination of human OA knee cartilage revealed that chondrocytes in the SZ of normal articular cartilage have enriched lubricin expression (lateral tibial condyle; spared compartment) (Figure 5D). The chondrocyte clones present in the OA-damaged cartilage regions (medial tibial condyle; non-spared) robustly express lubricin. It is notable that within each clone, these lubricin-positive cells exhibit a pattern of peripherally-organized expression. This observation suggests the presence of lubricin in the clones together with its known function contributes an anabolic function to the clone.
Discussion
Here we demonstrate a novel biological association of the transcription factor Runx1 to the integrity of articular cartilage. The enrichment of Runx1 expression in the SZ cells that is further increased in response to mechanical loading, suggests a function that supports the phenotype of these unique cells. Our findings further demonstrate Runx1 and lubricin are both robustly expressed in SZ chondrocytes and in human OA clones that form in response to damaged cartilage and the margins of OA lesions in the DMM mouse model. These findings provide insight into the functional activity of OA cell clusters. The expression of Runx1 and Vcam1 in proliferating OA ‘clone’ cells, together reflect both a mesenchymal and chondroprogenitor cell phenotype with regenerative potential. Consistent with known properties of Runx1 in other tissues, we propose that the robust expression of Runx1 with PGR4 and other components of OA ‘clonal’ cells appears to be an attempt to repopulate the surface with viable cells.
The normal articular cartilage surface offers a high degree of protection from mechanical insults. Enrichment of Runx1 in normal superficial articular cartilage is analogous to the ubiquitous high expression of Runx1 in protective epithelium of many tissues and in periosteum and perichondrium linings of the skeleton [12]. The SZ represents only a 1–2 cell layer that is closest to the articular cartilage surface and differs from the middle and deep zones in morphology, organization of collagen fibrils and gene expression [29]. When cultured, SZ cells have higher rates of proliferation than cells of the deep and middle zones [30]. Lubricin is expressed exclusively by SZ chondrocytes [31]. In addition, SZ cells have been shown to express markers of MSCs such as Notch-1, Endoglin (CD105), ALCAM (CD166), and Vcam1 (CD106) [29, 32], Stro-1 [29, 33]. Thus, Runx1 may function as a chondroprotective factor supporting the SZ cell phenotypic properties. A compelling question is whether Runx1 directly regulates genes in the clones that produce proteins which can reconstitute the articular surface. We explored the possibility that Runx1 may regulate lubricin and used TRANSFAC analysis which revealed a Runx consensus recognition motif within the lubricin promoter at chr1 : 186265215–186265220, 185 bp upstream of the TSS in the lubricin human gene promoter and 9 putative binding sites in the ACAN gene [34, 35]. Runx factors have recently been identified in binding to the ACAN promoter particularly Runx3 in hypertrophic chondrocytes [36]. Thus high Runx1 levels in OA chondrocyte clones OA, Runx1 is likely regulating aggrecan and lubricin as well as other Runx target genes.
The exact function of the human OA ‘clones’ is unknown. One study links the clones to migration of [28] and other studies to cells that secrete a specialized pericellular matrix [37]. Human OA clones are well documented to be comprised of cells with characteristic features of MSCs and chondroprogenitors [28, 37]. A key finding here is the evidence linking the function of Runx1 in OA clones to the proliferative capacity of these cells, and to the expression of lubricin, a chondroprotective molecule [23, 27]. Several studies have reported properties of cells in clones following flow cytometry separation using various MSC markers [28]. Our studies have identified the organization of cells expressing lubricin, Runx1 and Vcam1 in OA ‘clones’, further implicating these cells in an adaptive function in OA cartilage. Although others have recognized that formation of the OA clones may provide a compensatory mechanism for degenerating articular cartilage surface [38], ultimately end-stage OA occurs. Nonetheless, our data support a growing concept that human OA clones are involved in a reparative process rather than a degenerative process [28, 37]. Recent studies have established Runx1 activity in promoting stem cell proliferation of the hair follicle [39]. A similar function may occur in the SZ chondrocytes of normal cartilage and the clones in OA, to increase MSC/chondroprogenitor cells. The OA ‘clones’ expressing Runx1 representing proliferating progenitor cells and lubricin, together may function to limit the OA lesion or re-establish a subpopulation of cells that express markers of SZ chondrocytes [28].
The mouse DMM model was used to follow the contribution of Runx1 expression from the onset to final stages of OA. Runx1 expression is not observed in the SZ at the site of the initial lesion formation. Although, clones as observed in the very late stages of human OA are absent in this mouse model, there is an induction of Runx1-positive cells in small clusters of cells at lesion peripheries. Although the spatial localization of the clones differ between the mouse and the human OA tissue, the enrichment of Runx1 in clonal aggregates of both models, is supported by high expression of Runx1 in human OA cartilage (Figure 1), suggesting that the functional activity is similar in both models and is related to the presence of Runx1 in the clones. The mouse cell aggregates have all of the properties of the human OA clonal cells in that they exhibit MSC markers and Runx1 and Vcam1 are co-expressed in the same cells and the cells that are PCNA-positive and express lubricin. Taken together our novel findings support a growing concept [28, 37] that OA clones or mouse OA clusters are involved in a reparative rather than a degenerative process.
Insight into a role for Runx1 in the SZ zone was revealed by the striking increase in Runx1 expression in response to physiologic loading of bovine articular cartilage. We show that Runx1 expression positively correlates with increasing loading conditions. In human OA cartilage, it has been shown that varying degrees of chronic varus malalignment result in significant overloading differences in medial and lateral stress distribution [40]. Because we observed an increase in Runx1 protein levels on the medial side of a varus joint suggests Runx1 function increases under compressive loading. It is known that under loading conditions, cell proliferation is increased [41]. This finding is consistent with Runx1 being present in proliferating cells and activating genes that support cell differentiation and phenotype stability [42]. Many genes become differentially expressed in response to articular cartilage compression [43, 44]. Under loading conditions, the pro-inflammatory cytokines and matrix metalloproteinases (MMPs) are upregulated [45]. MMPs are direct transcriptional targets of Runx proteins [16, 46]. This raises the possibility that compression-stimulated upregulation of Runx1 under load could contribute either to cartilage turnover or potentially promote expansion of a progenitor cell population in an attempt to repopulate damaged cartilage. In support of this concept, Sox9 and Runx1 have been reported to be expressed in the periosteal callus during fracture repair [47]. Also, the presence of Runx1 in SZ cells is consistent with its expression in other mesenchymal proliferative tissues, including suture lines of the calvaria, perichondrium and periosteum, all sources of the chondroid and osteo-lineage cells [8, 12, 48].
Of further biological significance, our studies provide a mechanistic basis for Runx1 as a potential therapeutic approach for OA. Two recent papers have identified Runx1 in association with cartilage repair. The first study implicated increased Runx1 activity in response to the chondroprotective agent, kartogenin [49]. Furthermore, this study identified in human MSCs an upregulation of lubricin by Runx1 activation in response to Kartogenin [49]. This small molecule resulted in amelioration of OA in surgically induced OA in mice. The second report identified Runx1 as a molecular target in response to TD-198946, a novel OA disease modifying drug [50]. Neither study intended to discern the functional role of Runx1 in OA, a priori. Rather Runx1 appeared in their profiling studies as a candidate. Because Runx1 is capable of responding to mechanical load and cartilage damage, like lubricin, we propose a potential function of Runx1 in SZ cells with other factors in maintaining the surface integrity of the tissue by either stimulating proliferation or promoting expression of surface proteins.
Acknowledgments
Supported by:
NIH Grants: R01 AR039588 (GSS, JBL); (GSS, JBL); R37 DE012528 (JBL); P01 AR048818 (GSS, JBL) Institutional Support, Department of Orthopedics and the Musculoskeletal Center of Excellence (PJF).
We thank Steven Baker for help with statistics, Judy Rask and Marta DeSourdis for editorial assistance, and Sylvie Puig and Charlene Baron for assistance with Figure preparation. A component of this research was performed by Kirti Basil in fulfillment of thesis requirements in the Undergraduate Research Program at The College of the Holy Cross, Worcester, MA USA.
References
- 1.Chuang LS, Ito K, Ito Y. RUNX family: Regulation and diversification of roles through interacting proteins. Int J Cancer. 2013;132:1260–71. doi: 10.1002/ijc.27964. [DOI] [PubMed] [Google Scholar]
- 2.Swiers G, de Bruijn M, Speck NA. Hematopoietic stem cell emergence in the conceptus and the role of Runx1. Int J Dev Biol. 2010;54:1151–63. doi: 10.1387/ijdb.103106gs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Komori T. Regulation of skeletal development by the Runx family of transcription factors. J Cell Biochem. 2005;95:445–53. doi: 10.1002/jcb.20420. [DOI] [PubMed] [Google Scholar]
- 4.Takarada T, Hinoi E, Nakazato R, Ochi H, Xu C, Tsuchikane A, Takeda S, Karsenty G, Abe T, Kiyonari H, et al. An analysis of skeletal development in osteoblast-specific and chondrocyte-specific runt-related transcription factor-2 (Runx2) knockout mice. J Bone Miner Res. 2013;28:2064–69. doi: 10.1002/jbmr.1945. [DOI] [PubMed] [Google Scholar]
- 5.Soung do Y, Dong Y, Wang Y, Zuscik MJ, Schwarz EM, O’Keefe RJ, Drissi H. Runx3/AML2/Cbfa3 regulates early and late chondrocyte differentiation. J Bone Miner Res. 2007;22:1260–70. doi: 10.1359/jbmr.070502. [DOI] [PubMed] [Google Scholar]
- 6.Kimura A, Inose H, Yano F, Fujita K, Ikeda T, Sato S, Iwasaki M, Jinno T, Ae K, Fukumoto S, et al. Runx1 and Runx2 cooperate during sternal morphogenesis. Development. 2010;137:1159–67. doi: 10.1242/dev.045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato S, Kimura A, Ozdemir J, Asou Y, Miyazaki M, Jinno T, Ae K, Liu X, Osaki M, Takeuchi Y, et al. The distinct role of the Runx proteins in chondrocyte differentiation and intervertebral disc degeneration: findings in murine models and in human disease. Arthritis Rheum. 2008;58:2764–75. doi: 10.1002/art.23805. [DOI] [PubMed] [Google Scholar]
- 8.Smith N, Dong Y, Lian JB, Pratap J, Kingsley PD, van Wijnen AJ, Stein JL, Schwarz EM, O’Keefe RJ, Stein GS, et al. Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J Cell Physiol. 2005;203:133–43. doi: 10.1002/jcp.20210. [DOI] [PubMed] [Google Scholar]
- 9.Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, et al. Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 2004;18:952–63. doi: 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dy P, Wang W, Bhattaram P, Wang Q, Wang L, Ballock RT, Lefebvre V. Sox9 directs hypertrophic maturation and blocks osteoblast differentiation of growth plate chondrocytes. Dev Cell. 2012;22:597–609. doi: 10.1016/j.devcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lengner CJ, Hassan MQ, Serra RW, Lepper C, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Nkx3. 2-mediated repression of Runx2 promotes chondrogenic differentiation. J Biol Chem. 2005;280:15872–79. doi: 10.1074/jbc.M411144200. [DOI] [PubMed] [Google Scholar]
- 12.Lian JB, Balint E, Javed A, Drissi H, Vitti R, Quinlan EJ, Zhang L, Van Wijnen AJ, Stein JL, Speck N, et al. Runx1/AML1 hematopoietic transcription factor contributes to skeletal development in vivo. J Cell Physiol. 2003;196:301–11. doi: 10.1002/jcp.10316. [DOI] [PubMed] [Google Scholar]
- 13.Stricker S, Fundele R, Vortkamp A, Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- 14.Levanon D, Brenner O, Negreanu V, Bettoun D, Woolf E, Eilam R, Lotem J, Gat U, Otto F, Speck N, et al. Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1 (Aml1) indicates non-redundant functions during mouse embryogenesis. Mech Dev. 2001;109:413–17. doi: 10.1016/s0925-4773(01)00537-8. [DOI] [PubMed] [Google Scholar]
- 15.Liakhovitskaia A, Lana-Elola E, Stamateris E, Rice DP, van ‘t Hof RJ, Medvinsky A. The essential requirement for Runx1 in the development of the sternum. Dev Biol. 2010;340:539–46. doi: 10.1016/j.ydbio.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Selvamurugan N, Jefcoat SC, Kwok S, Kowalewski R, Tamasi JA, Partridge NC. Overexpression of Runx2 directed by the matrix metalloproteinase-13 promoter containing the AP-1 and Runx/RD/Cbfa sites alters bone remodeling in vivo. J Cell Biochem. 2006;99:545–57. doi: 10.1002/jcb.20878. [DOI] [PubMed] [Google Scholar]
- 17.Kamekura S, Kawasaki Y, Hoshi K, Shimoaka T, Chikuda H, Maruyama Z, Komori T, Sato S, Takeda S, Karsenty G, et al. Contribution of runt-related transcription factor 2 to the pathogenesis of osteoarthritis in mice after induction of knee joint instability. Arthritis Rheum. 2006;54:2462–70. doi: 10.1002/art.22041. [DOI] [PubMed] [Google Scholar]
- 18.Fanning PJ, Emkey G, Smith RJ, Grodzinsky AJ, Szasz N, Trippel SB. Mechanical regulation of mitogen-activated protein kinase signaling in articular cartilage. J Biol Chem. 2003;278:50940–48. doi: 10.1074/jbc.M305107200. [DOI] [PubMed] [Google Scholar]
- 19.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage. 2007;15:1061–69. doi: 10.1016/j.joca.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz N, Laverty S, Kraus VB, Aigner T. Basic methods in histopathology of joint tissues. Osteoarthritis Cartilage. 2010;18 (Suppl 3):S113–116. doi: 10.1016/j.joca.2010.05.026. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson AC, Ballabio E, Geng H, North P, Tapia M, Kerry J, Biswas D, Roeder RG, Allis CD, Melnick A, et al. RUNX1 is a key target in t(4;11) leukemias that contributes to gene activation through an AF4-MLL complex interaction. Cell Rep. 2013;3:116–27. doi: 10.1016/j.celrep.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markova EN, Kantidze OL, Razin SV. Transcriptional regulation and spatial organisation of the human AML1/RUNX1 gene. J Cell Biochem. 2011;112:1997–2005. doi: 10.1002/jcb.23117. [DOI] [PubMed] [Google Scholar]
- 23.Jay GD, Fleming BC, Watkins BA, McHugh KA, Anderson SC, Zhang LX, Teeple E, Waller KA, Elsaid KA. Prevention of cartilage degeneration and restoration of chondroprotection by lubricin tribosupplementation in the rat following anterior cruciate ligament transection. Arthritis Rheum. 2010;62:2382–91. doi: 10.1002/art.27550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lengner CJ, Lepper C, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Primary mouse embryonic fibroblasts: a model of mesenchymal cartilage formation. J Cell Physiol. 2004;200:327–33. doi: 10.1002/jcp.20118. [DOI] [PubMed] [Google Scholar]
- 26.Soung do Y, Talebian L, Matheny CJ, Guzzo R, Speck ME, Lieberman JR, Speck NA, Drissi H. Runx1 dose-dependently regulates endochondral ossification during skeletal development and fracture healing. J Bone Miner Res. 2012;27:1585–97. doi: 10.1002/jbmr.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flannery CR, Zollner R, Corcoran C, Jones AR, Root A, Rivera-Bermudez MA, Blanchet T, Gleghorn JP, Bonassar LJ, Bendele AM, et al. Prevention of cartilage degeneration in a rat model of osteoarthritis by intraarticular treatment with recombinant lubricin. Arthritis Rheum. 2009;60:840–7. doi: 10.1002/art.24304. [DOI] [PubMed] [Google Scholar]
- 28.Koelling S, Kruegel J, Irmer M, Path JR, Sadowski B, Miro X, Miosge N. Migratory chondrogenic progenitor cells from repair tissue during the later stages of human osteoarthritis. Cell Stem Cell. 2009;4:324–35. doi: 10.1016/j.stem.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 29.Dowthwaite GP, Bishop JC, Redman SN, Khan IM, Rooney P, Evans DJ, Haughton L, Bayram Z, Boyer S, Thomson B, et al. The surface of articular cartilage contains a progenitor cell population. J Cell Sci. 2004;117:889–97. doi: 10.1242/jcs.00912. [DOI] [PubMed] [Google Scholar]
- 30.Hidaka C, Cheng C, Alexandre D, Bhargava M, Torzilli PA. Maturational differences in superficial and deep zone articular chondrocytes. Cell Tissue Res. 2006;323:127–35. doi: 10.1007/s00441-005-0050-y. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher BL, Block JA, Schmid TM, Aydelotte MB, Kuettner KE. A novel proteoglycan synthesized and secreted by chondrocytes of the superficial zone of articular cartilage. Arch Biochem Biophys. 1994;311:144–52. doi: 10.1006/abbi.1994.1219. [DOI] [PubMed] [Google Scholar]
- 32.Pretzel D, Linss S, Rochler S, Endres M, Kaps C, Alsalameh S, Kinne RW. Relative percentage and zonal distribution of mesenchymal progenitor cells in human osteoarthritic and normal cartilage. Arthritis Res Ther. 2011;13:R64. doi: 10.1186/ar3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grogan SP, Miyaki S, Asahara H, D’Lima DD, Lotz MK. Mesenchymal progenitor cell markers in human articular cartilage: normal distribution and changes in osteoarthritis. Arthritis Res Ther. 2009;11:R85. doi: 10.1186/ar2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitfield TA. Putative Runx binding sites in human PRG4 gene. 2012 http://zlabumassmededu/~whitfiet/FIMO/PRG4/fimohtml.
- 35.Whitfield TA. Putative Runx binding sites in human ACAN gene. 2012 http://zlabumassmededu/~whitfiet/FIMO/ACAN/finohtml.
- 36.Wigner NA, Soung do Y, Einhorn TA, Drissi H, Gerstenfeld LC. Functional role of Runx3 in the regulation of aggrecan expression during cartilage development. J Cell Physiol. 2013;228:2232–42. doi: 10.1002/jcp.24396. [DOI] [PubMed] [Google Scholar]
- 37.Lotz MK, Otsuki S, Grogan SP, Sah R, Terkeltaub R, D’Lima D. Cartilage cell clusters. Arthritis Rheum. 2010;62:2206–18. doi: 10.1002/art.27528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hiraoka K, Grogan S, Olee T, Lotz M. Mesenchymal progenitor cells in adult human articular cartilage. Biorheology. 2006;43:447–54. [PubMed] [Google Scholar]
- 39.Scheitz CJ, Tumbar T. New insights into the role of Runx1 in epithelial stem cell biology and pathology. J Cell Biochem. 2013;114:985–93. doi: 10.1002/jcb.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner FW, Ayers DC, Maletsky LP, Rullkoetter PJ. The effect of valgus/varus malalignment on load distribution in total knee replacements. J Biomech. 2005;38:349–55. doi: 10.1016/j.jbiomech.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 41.Ryan JA, Eisner EA, DuRaine G, You Z, Reddi AH. Mechanical compression of articular cartilage induces chondrocyte proliferation and inhibits proteoglycan synthesis by activation of the ERK pathway: implications for tissue engineering and regenerative medicine. J Tissue Eng Regen Med. 2009;3:107–16. doi: 10.1002/term.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaidi SK, Young DW, Montecino M, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Bookmarking the genome: maintenance of epigenetic information. J Biol Chem. 2011;286:18355–61. doi: 10.1074/jbc.R110.197061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitzgerald JB, Jin M, Chai DH, Siparsky P, Fanning P, Grodzinsky AJ. Shear- and compression-induced chondrocyte transcription requires MAPK activation in cartilage explants. J Biol Chem. 2008;283:6735–43. doi: 10.1074/jbc.M708670200. [DOI] [PubMed] [Google Scholar]
- 44.Knobloch TJ, Madhavan S, Nam J, Agarwal S, Jr, Agarwal S. Regulation of chondrocytic gene expression by biomechanical signals. Crit Rev Eukaryot Gene Expr. 2008;18:139–50. doi: 10.1615/critreveukargeneexpr.v18.i2.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li NG, Shi ZH, Tang YP, Wang ZJ, Song SL, Qian LH, Qian DW, Duan JA. New hope for the treatment of osteoarthritis through selective inhibition of MMP-13. Curr Med Chem. 2011;18:977–1001. doi: 10.2174/092986711794940905. [DOI] [PubMed] [Google Scholar]
- 46.Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–91. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shintaku Y, Murakami T, Yanagita T, Kawanabe N, Fukunaga T, Matsuzaki K, Uematsu S, Yoshida Y, Kamioka H, Takano-Yamamoto T, et al. Sox9 expression during fracture repair. Cells Tissues Organs. 2011;194:38–48. doi: 10.1159/000322557. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Belflower RM, Dong YF, Schwarz EM, O’Keefe RJ, Drissi H. Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J Bone Miner Res. 2005;20:1624–36. doi: 10.1359/JBMR.050516. [DOI] [PubMed] [Google Scholar]
- 49.Johnson K, Zhu S, Tremblay MS, Payette JN, Wang J, Bouchez LC, Meeusen S, Althage A, Cho CY, Wu X, et al. A stem cell-based approach to cartilage repair. Science. 2012;336:717–21. doi: 10.1126/science.1215157. [DOI] [PubMed] [Google Scholar]
- 50.Yano F, Hojo H, Ohba S, Fukai A, Hosaka Y, Ikeda T, Saito T, Hirata M, Chikuda H, Takato T, et al. A novel disease-modifying osteoarthritis drug candidate targeting Runx1. Ann Rheum Dis. 2013;72:748–53. doi: 10.1136/annrheumdis-2012-201745. [DOI] [PubMed] [Google Scholar]