Highlights
-
•
Adaptive cellular responses to 1- or 2-day fasting differ among tissues.
-
•
Ubiquitin–proteasome and autophagy–lysosome pathways are activated in thymus/lung/heart/muscle.
-
•
The amino acid response is activated mainly in thymus.
-
•
An Nrf2-mediated antioxidant system is activated in thymus, heart, and kidney.
-
•
Expression of amino acid transporter genes is activated in a tissue-specific manner.
Abbreviations: AL, ad libitum-fed; AST, aspartate aminotransferase; BUN, blood urea nitrogen; CPK, creatine phosphokinase; CT, computed tomography; ER, endoplasmic reticulum; LDH, lactate dehydrogenase; qPCR, quantitative PCR; TG, triglyceride
Keywords: Amino acid response, Amino acid transport, Autophagy, Nrf2, Proteasome, Ubiquitin
Abstract
Dietary or caloric restriction confers various clinical benefits. Short-term fasting of mice is a common experimental procedure that may involve systemic metabolic remodeling, which may significantly affect experimental outputs. This study evaluated adaptive cellular responses after 1- or 2-day fasting in 13 mouse tissues by quantitative PCR using 15 marker primer sets for the activation of ubiquitin–proteasome (Atrogin-1 and MuRF1), autophagy–lysosome (LC3b, p62 and Lamp2), amino acid response (Asns, Trib3, Herpud1, xCT, and Chop), Nrf2-mediated antioxidant (HO-1 and Gsta1), and amino acid transport (Slc38a2, Slc7a5, and Slc7a1) systems. Differential activation profiles obtained in seven highly (thymus, liver, spleen, and small intestine) or mildly (stomach, kidney, and colon) atrophied tissues as well as in six non-atrophied tissues (brain, eye, lung, heart, skeletal muscle, and testis) suggested tissue-specific active metabolic remodeling.
1. Introduction
Fasting has been practiced for millennia to prevent obesity, hypertension, asthma, rheumatoid arthritis, seizures, and seizure-associated brain damage [1,2]. Well-controlled investigations of clinical studies and experimental animals indicate that intermittent fasting or caloric restriction imparts various clinical benefits to protect against cardiovascular disease, diabetes, cancers, and neurodegeneration [1]. In addition, short-term dietary restriction or single (preoperative) fasting is known to confer various clinical benefits in humans and rodents [3–5]. Importantly, fasting of mice is a standard protocol performed in association with many different types of experiments in order to reduce feeding-associated variability in investigative parameters (e.g. blood glucose and insulin levels) or to facilitate surgical procedures [6], but fasting may also substantially modify the outcome of experimental studies. However, the effects of fasting not directly related to investigated parameters or tissues of interest are often ignored. The aim of this study was to clarify tissue-specific adaptive responses against short-term (1- or 2-day) fasting using quantitative PCR (qPCR) in mice. Here we comparatively analyzed the expression of 15 genes involved in various cellular responses/functions in 13 different tissues.
2. Materials and methods
2.1. Mice
C57BL/6J male mice were purchased from Japan SLC (Shizuoka, Japan). Eight to nine-week-old mice were group-housed (four mice per cage) in an air-conditioned room kept on a 12-h dark/light (8 pm–8 pm) cycle, and allowed free access to water and the CE-2 standard dry rodent diet (CLEA Japan, Tokyo, Japan). In fasting experiments, mice were deprived of the diet for 1 or 2 day(s) between 2 pm and 2 pm. After anesthetization of mice by isoflurane inhalation, blood samples were collected from the heart, and tissues were quickly dissected out for weight measurement or total RNA preparation. All animal procedures conformed to the Guide for the Care and Use of Laboratory Animals, Eighth Edition published by the US National Research Council, and were approved by the Animal Care Committee of Keio University (No. 09187-(4)).
2.2. qPCR
Total RNA was isolated from tissues using TRI Reagent (Molecular Research Center, Cincinnati, OH, USA) and further purified with a PureLink RNA mini kit (Ambion, Carlsbad, CA, USA). RNA (1 μg) was used to produce first-strand cDNA with a ReverTra Ace qPCR RT kit (Toyobo, Tokyo, Japan). A total of 5 ng cDNA from each sample was amplified via real-time PCR using a SYBR Green Realtime PCR Master Mix (Toyobo), 16 primer sets (Table 1), and a CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) [7]. Each mRNA level was quantified using the comparative CT method with housekeeping gene hypoxanthine guanine phosphoribosyl transferase (Hprt) levels used for normalization, and the relative expression in ad libitum-fed (AL) mice was set at 1.
Table 1.
Primer sets for quantitative PCR.
| Gene | Primer sequence | Size (bp) |
|---|---|---|
| Ubiquitin–proteasome system | ||
| Atrogin-1 (MAFbx) | 5′-AGGAGCGCCATGGATACTGT-3′ (Forward) | 161 |
| 5′-GAAGTTCTTTTGGGCGATGC-3′ (Reverse) | ||
| MuRF1 (Trim63) | 5′-ACCTGCTGGTGGAAAACATC-3′ (Forward) | 96 |
| 5′-CTTCGTGTTCCTTGCACATC-3′ (Reverse) | ||
| Autophagy–lysosome system | ||
| LC3b (Map1lc3b) | 5′-CGATACAAGGGGGAGAAGCA-3′ (Forward) | 185 |
| 5′-ACTTCGGAGATGGGAGTGGA-3′ (Reverse) | ||
| p62 (Sqstm1) | 5′-TGCTCTTCGGAAGTCAGCAA-3′ (Forward) | 122 |
| 5-CCCGACTCCATCTGTTCCTC-3′ (Reverse) | ||
| Lamp2 | 5′-TAGGAGCCGTTCAGTCCAAT-3′ (Forward) | 158 |
| 5′-GTGTGTCGCCTTGTCAGGTA-3′ (Reverse) | ||
| Amino acid response | ||
| Asns | 5′-CTGTACGGATGAACCATTGC-3′ (Forward) | 210 |
| 5′-GCCTCCTTGAGTTGCTTCA-3′ (Reverse) | ||
| Trib3 | 5′-TCTTCCGGCAGATGGCTAGT-3′ (Forward) | 123 |
| 5′-GGTTCTCCAGCACCAGCTTC-3′ (Reverse) | ||
| Herpud1 | 5′-ATGGTGGTCCTCGAGATGCT-3′ (Forward) | 139 |
| 5′-ATAAACGAGGGGCTGGTGTG-3′ (Reverse) | ||
| xCT (Slc7a11) | 5′-GTCATCGGATCAGGCATCTT-3′ (Forward) | 124 |
| 5′-CATAGGACAGGGCTCCAAAA-3′ (Reverse) | ||
| Chop (Ddit3) | 5′-CATCCCCAGGAAACGAAGAG-3′ (Forward) | 163 |
| 5′-TCTTCCTCCTGGGCCATAGA-3′ (Reverse) | ||
| Nrf2-mediated antioxidant system | ||
| HO-1 | 5′-TGCTCGAATGAACACTCTGG-3′ (Forward) | 206 |
| 5′-TCTCTGCAGGGGCAGTATCT-3′ (Reverse) | ||
| Gsta1 | 5′-CCCCTTTCCCTCTGCTGAAG-3′ (Forward) | 148 |
| 5′-TTCACTGAATCTTGAAAGCC-3′ (Reverse) | ||
| Amino acid transport | ||
| Slc38a2 (Sat2) | 5′-TCACCGTGACCATCTTGTCC-3′ (Forward) | 166 |
| 5′-TGCACGGATCTCATTGGTTC-3′ (Reverse) | ||
| Slc7a5 (Lat1) | 5′-GCTTGTCTTGCTTCGGCTCT-3′ (Forward) | 113 |
| 5′-CTGTGGGTGGATCATGGAGA-3′ (Reverse) | ||
| Slc7a1 (Cat1) | 5′-CTGGAGTGCGACTTTTGACG-3′ (Forward) | 179 |
| 5′-TGTTGACCATGGCTGACTCC-3′ (Reverse) | ||
| Housekeeping gene (as a control) | ||
| Hprt | 5′-GACTGATTATGGACAGGACTG-3′ (Forward) | 211 |
| 5′-GACTGATCATTACAGTAGCTC-3′ (Reverse) | ||
2.3. Measurement of serum amino acids and biochemical parameters
Blood samples were allowed to clot at room temperature for 10 min. The clot was removed by centrifugation (2300g for 5 min and then 9300g for 2 min at 4 °C), and the resulting supernatant (serum) was isolated and stored at −80 °C until analysis. Free amino acids (except tryptophan) in the serum were measured using a high-performance liquid chromatography (HPLC) system as described previously; the amino acids (and homophenylalanine as an internal control) were derivatized using 4-fluoro-7-nitrobenzofurazan (NBD-F; Dojindo, Kumamoto, Japan) in accordance with the manufacturer’s instructions, and samples were injected onto a reversed-phase C18 column (CAPCELL PAK C18 MGII S5; Shiseido, Tokyo, Japan) for the separation of fluorescent-derivatized amino acids by acetonitrile gradient elution [7]. All gradient steps were controlled by a LaChrom Elite chromatography workstation (Hitachi, Tokyo, Japan), and peaks in fluorescence (Ex. 480 nm; Em. 530 nm) were measured using an L-2400 fluorescence detector (Hitachi). Tryptophan was independently quantified by its own fluorescence (Ex. 295 nm; Em. 340 nm) without NBD-F derivatization. Total cysteine/homocysteine/glutathione, which gives rise to the thiol cysteine/homocysteine/glutathione after reductive cleavage of disulfide bonds [8], were measured by the following procedure. The serum samples or standard reagents (20 μL) were mixed with 10 μL of phosphate-buffered saline (pH 7.4), 10 μL of 80 μM (2-mercaptopropionyl)glycine (an internal control), and 4 μL of 0.349 M (100 g/L) Tris(2-carboxyethyl)phosphine hydrochloride, and then incubated at room temperature for 30 min. Then the mixtures were added to 36 μL of 0.612 M (100 g/L) trichloroacetic acid (TCA)/1 mM EDTA (pH 8.0) solution and centrifuged at 13,000g at 20 °C for 10 min. The 50 μL supernatant was mixed with 185 μL of a mixture of 50 μg 4-fluoro-7-sulfobenzofurazan, ammonium salt (SBD-F, Dojindo) in 175 μL of 125 mM borate–HCl (pH 9.5)/4 mM EDTA (pH 8.0) and 10 μL of 1.55 M NaOH, and incubated at 60 °C for 1 h. The samples (10 μL) were injected onto the same column using the same HPLC system for the separation of fluorescent-derivatized thiol-containing amino acids. The gradient solutions consisting of A: 0.1 M sodium acetate (pH 4.5) and B: 50% (v/v) methanol, and the gradient program was initialized at 6% B for 8 min followed by a linear increase of B to 50% over 7 min, and held for 5 min followed by an immediate return to 6% B for 15 min. The peaks in fluorescence (Ex. 385 nm; Em. 515 nm) were measured. All amino acids were identified based on their retention times, and their concentrations were calculated relative to the calibrated amino acid standard solutions (Wako, Osaka, Japan). Serum biochemical parameters were measured using an AutoWako Total Ketone Bodies clinical assay kit (Wako) for ketone bodies and a Dri-Chem 7000i biochemistry analyzer (Fuji film, Tokyo, Japan) for all others.
2.4. X-ray computed tomography (CT) analysis
A third generation CT scanner, LaTheta LCT-200 (Hitachi-Aloka, Tokyo, Japan), was used for the non-invasive quantification of fat and muscle depots [7]. Prior to CT scanning, mice were anesthetized with an intraperitoneal injection of pentobarbital and remained anesthetized during the whole CT scan. The tube voltage was set at 50 kV and the current was constant at 0.5 mA. Mice were placed on their backs, face up, head front, and fore/hind limbs extended with tape in a 48-mm wide specimen holder. They were then scanned at a resolution of 96 μm. An overview scan of the whole mouse was created to allow the selection of regions of interest for the subsequent scans. Fat and muscle contents in the lower half of the body (the whole part under the diaphragm) were calculated using LaTheta software.
2.5. Western blotting
Each tissue aliquot (∼100 mg) was homogenized in Lysis buffer (50 mM Tris–HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% [w/v] Triton X-100, 0.1% [w/v] phenylmethylsulfonyl fluoride, and Complete EDTA-free Protease Inhibitor Cocktail [Roche Diagnostics, Tokyo, Japan]) using a MicroSmash-100R (MS-100R) homogenizing system (Tomy, Tokyo, Japan) and Zirconia beads (4100 rpm, 30 s × 3, 4 °C). Homogenates were centrifuged at 4 °C for 10 min at 10,000g, and then the supernatants were centrifuged at 4 °C for 10 min at 10,000g. The sample supernatants were mixed with equal volumes of 2× SDS/2-mercaptoethanol-containing sample buffer. After heating (100 °C, 3 min), samples (10 μg/lane) were resolved on 15% (for LC3b (microtubule-associated protein 1 light chain 3 beta [Map1lc3b]) and 10% (for p62 (Sequestosome 1 [Sqstm1]) and glyceraldehyde-3-phosphate dehydrogenase (Gapdh)) pre-cast PAGE gels using Tris–glycine–SDS electrophoresis buffer (25 mM Tris/192 mM glycine/0.1% SDS). Proteins were transferred to Immobilon PVDF membrane (Millipore) and subjected to western blotting. LC3b, p62, and Gapdh were detected using anti-LC3B rabbit polyclonal (1:2000; Sigma–Aldrich L7543), anti-p62/SQSTM1 rabbit polyclonal (1:2000; PM045 from MBL, Nagoya, Japan), and anti-GAPDH (14C10) rabbit monoclonal (1:2000; #2118 from Cell Signaling Technology) antibodies. Anti-rabbit IgG, horseradish peroxidase-linked donkey whole antibody (1:10,000; GE Healthcare NA934) was used as a secondary antibody. The ECL Prime western blotting detection reagent and an ImageQuant LAS 4000 image analyzer (GE Healthcare) were used for chemiluminescence detection.
2.6. Statistics
Data are expressed as mean ± S.D. (n: sample numbers). Statistical analysis was performed using two-tailed Mann–Whitney U-test with Prism ver. 5.0c software (GraphPad, La Jolla, CA, USA); P < 0.05 denoted a significant difference.
3. Results
3.1. Systemic influence of 1- or 2-day fasting in mice
Mice have a higher metabolic rate than humans; thus, 1- or 2-day fasting could have greater systemic effects. Indeed, body weight decreased by 16.7% at day 1 and 23.4% at day 2 (Table 2). Significant decreases in wet tissue weights were observed in thymus, spleen, pancreas, liver, and small intestine (31.6–54.7% decrease; designated as “highly” atrophied tissues) followed by kidney, stomach and colon (18.0–18.6% decrease; designated as “mildly” atrophied tissues) whereas brain, eye, lung, heart, skeletal (rectus femoris) muscle, and testis were not significantly affected (Table 2). Among these eight atrophied tissues, weight reduction was not apparent at day 1 only in colon (Table 2). X-ray CT analysis revealed a rapid loss of whole body fat but not muscle mass (Fig. 1). Serum biochemistry revealed that the blood glucose level had significantly decreased after 1-day fasting (from 351.5 to 116.2 mg/dL) but the minimal glucose level was thereafter maintained by day 2 (100.9 mg/dL; Table 3). Serum levels of triglyceride (TG) and albumin were maintained, and the levels of aspartate aminotransferase (AST), lactate dehydrogenase (LDH), creatine phosphokinase (CPK), and blood urea nitrogen (BUN) were elevated only slightly, indicating that the 2-day fasting elicited physiological (rather than pathophysiological) responses in mice (Table 3). Ketone bodies (acetoacetate and β-hydroxybutyric acid) were indeed highly accumulated in fasted mice (Table 3). Serum levels of Gly, Ala, Met, Pro, Phe, Trp, Ser, Thr, Asn/Asp, Tyr, Lys, Arg, Glu, (total) glutathione, ornithine, and citrulline were all decreased upon fasting whereas those of Val, Leu, Ile, Gln, and His were maintained and those of (total) Cys, (total) homocysteine, and taurine were rather elevated (Table 4).
Table 2.
Changes in wet tissue weights by 1- or 2-day of fasting.
| Weight at day 0 (mg tissue) | % decrease at day 1 | % decrease at day 2 | |
|---|---|---|---|
| Body weight (n = 13 each) | 23.9 ± 0.8 (g) | –16.7 ± 4.6∗∗∗ | –23.4 ± 2.9∗∗∗,### |
| Tissues | |||
| Brain (n = 13 each) | 427.9 ± 28.8 | –2.8 ± 5.3 | –2.7 ± 9.4 |
| Eye (n = 13 each) | 43.4 ± 4.5 | –6.3 ± 12.0 | (+)1.2 ± 7.6 |
| Thymus (n = 13 each) | 50.2 ± 12.2 | –33.5 ± 17.2∗∗ | –54.7 ± 11.7∗∗∗,## |
| Lung (n = 13 each) | 146.9 ± 32.4 | –1.6 ± 16.9 | –8.8 ± 19.9 |
| Heart (n = 13 each) | 120.0 ± 21.1 | –6.3 ± 15.4 | –8.2 ± 18.1 |
| Liver (n = 9 each) | 1197.7 ± 92.7 | –26.9 ± 6.4∗∗∗ | –32.2 ± 4.0∗∗∗ |
| Stomach (n = 13 each) | 174.0 ± 33.8 | –13.6 ± 16.8 | –18.0 ± 17.1∗ |
| Kidney (n = 13 each) | 322.3 ± 40.4 | –13.2 ± 13.5∗ | –18.6 ± 7.0∗∗∗ |
| Spleen (n = 13 each) | 74.4 ± 6.2 | –26.0 ± 10.6∗∗∗ | –41.2 ± 13.2∗∗∗,## |
| Pancreas (n = 6 each) | 122.2 ± 16.2 | –25.6 ± 8.8∗∗ | –37.4 ± 7.4∗∗,# |
| Small intestine (n = 6 each) | 780.6 ± 52.9 | –23.2 ± 6.0∗∗ | –31.6 ± 3.2∗∗,# |
| Colon (n = 6 each) | 119.4 ± 20.8 | (+)7.2 ± 5.6 | –18.0 ± 11.9∗,## |
| Skeletal muscle (n = 6 each) | 274.7 ± 26.8 | –5.6 ± 3.8 | –4.6 ± 12.0 |
| Testis (n = 13 each) | 208.6 ± 45.2 | –7.5 ± 18.6 | –9.2 ± 15.1 |
C57BL/6J male mice were fed ad libitum, or fasted for 1 or 2 day(s) between 2 pm and 2 pm with free access to water. Tissues were isolated after anesthetization and weighed. Blood in the heart and the contents of the stomach, small intestine, and colon, were washed out with ice-cold phosphate-buffered saline before weighing. Isolated rectus femoris muscles were weighed as skeletal muscle. Data are means ± S.D. (n: sample numbers); significant changes (decreases in weights) in ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus day 0; #p < 0.05 and ##p < 0.01 versus day 1 (Mann–Whitney U-test).
Fig. 1.
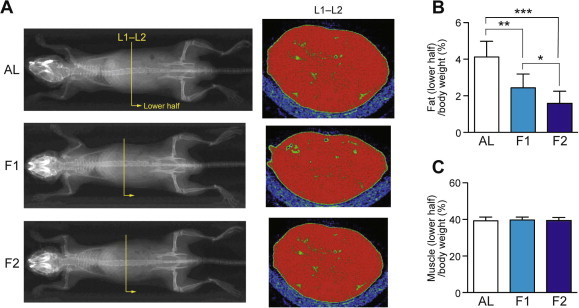
Fasting induced a rapid loss of fat but not muscle, as revealed by X-ray CT analysis. (A) Representative scanned whole body images of ad libitum-fed (AL), 1-day fasted, and 2-day fasted mice are shown. Representative lumber vertebra 1–2 (L1–L2) slice images are also shown, in which green and red regions represent fat and muscle (non-fat) depots, respectively. (B and C) Fat (B) and muscle (C) contents in the lower half of the body were estimated. Data are shown as the mean ± S.D. (n = 8), and significant changes were found in the fat tissues but not muscles; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (Mann–Whitney U-test).
Table 3.
Serum biochemical parameters after 1- or 2-day fasting in mice.
| Ad libitum-fed | 1-day fasting | 2-day fasting | |
|---|---|---|---|
| Glucose (mg/dL) | 351.5 ± 103.5 (15) | 116.2 ± 17.1 (10)∗∗∗ | 100.9 ± 29.5 (9)∗∗∗ |
| TG (mg/dL) | 127.9 ± 33.0 (13) | 166.6 ± 58.8 (13) | 119.0 ± 45.8 (13)# |
| Albumin (g/dL) | 2.66 ± 0.26 (15) | 2.63 ± 0.27 (12) | 2.81 ± 0.34 (9) |
| AST (IU/L) | 60.1 ± 31.5 (15) | 84.2 ± 33.5 (11)∗ | 118.2 ± 24.5 (10)∗∗∗,# |
| LDH (IU/L) | 182.4 ± 71.8 (15) | 207.6 ± 60.6 (11) | 283.3 ± 93.9 (9)∗∗ |
| CPK (IU/L) | 202.2 ± 148.4 (15) | 303.0 ± 186.6 (11) | 313.2 ± 120.4 (9)∗ |
| BUN (mg/dL) | 28.2 ± 8.5 (15) | 32.6 ± 5.7 (13)∗ | 38.3 ± 4.4 (10)∗∗,# |
| Ketone bodies (μM) | 91.6 ± 46.6 (8) | 1135 ± 259 (9)∗∗∗ | 4272 ± 608 (8)∗∗∗,### |
C57BL/6J male mice were fed ad libitum (day 0), or fasted for 1 or 2 day(s) between 2 pm and 2 pm with free access to water, and blood samples were collected after anesthetization. Serum biochemical parameters were measured using a clinical assay kit (for ketone bodies) or a biochemistry analyzer (for all others). Data are means ± S.D. (n: sample numbers); significant changes in ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus ad libitum; #p < 0.05 versus day 1 by Mann–Whitney U-test. TG, triglyceride; AST, aspartate aminotransferase; LDH, lactate dehydrogenase; CPK, creatine phosphokinase; BUN, blood urea nitrogen.
Table 4.
Serum amino acid concentrations after 1- or 2-day fasting in mice.
| Amino acid (μM) | Ad libitum-fed | 1-day fasting | 2-day fasting |
|---|---|---|---|
| Gly | 315.0 ± 44.3 | 210.6 ± 59.2∗ | 192.2 ± 21.7∗∗∗ |
| Ala | 397.1 ± 78.9 | 271.0 ± 79.0∗ | 227.8 ± 23.2∗∗∗ |
| Val | 261.6 ± 52.2 | 276.9 ± 53.2 | 223.7 ± 63.3 |
| Leu | 198.7 ± 50.0 | 230.0 ± 48.3 | 183.3 ± 57.4 |
| Ile | 115.4 ± 25.8 | 132.2 ± 24.4 | 104.9 ± 22.6 |
| Met | 71.2 ± 10.6 | 62.0 ± 9.4 | 53.2 ± 9.2∗∗ |
| Pro | 129.6 ± 37.5 | 85.5 ± 20.0∗ | 74.4 ± 12.3∗∗ |
| Phe | 94.6 ± 15.7 | 104.5 ± 14.5 | 84.9 ± 5.2## |
| Trp | 82.0 ± 18.0 | 56.9 ± 7.9∗ | 50.0 ± 5.5∗ |
| Ser | 134.1 ± 27.4 | 109.9 ± 30.6 | 83.6 ± 13.2∗∗,# |
| Thr | 159.7 ± 26.8 | 132.3 ± 16.9 | 126.5 ± 32.1∗ |
| Asn/Asp | 9.13 ± 3.11 | 5.83 ± 2.30 | 4.50 ± 1.54∗∗ |
| Gln | 1,695 ± 296 | 1,737 ± 401 | 1,569 ± 311 |
| Tyr | 134.8 ± 35.7 | 105.9 ± 40.8 | 82.2 ± 6.2∗∗ |
| (Total) Cys | 301.6 ± 28.9 | 342.0 ± 39.8 | 366.9 ± 54.8∗ |
| Lys | 382.0 ± 58.1 | 280.1 ± 68.3∗ | 229.0 ± 23.0∗∗∗ |
| Arg | 138.9 ± 20.0 | 103.2 ± 15.6∗∗ | 75.6 ± 7.2∗∗∗,## |
| His | 64.3 ± 13.7 | 76.2 ± 18.0 | 54.6 ± 8.8 |
| Glu | 17.5 ± 8.1 | 15.5 ± 6.3 | 9.36 ± 3.91∗,# |
| (Total) Homocysteine | 8.90 ± 0.76 | 8.76 ± 1.51 | 12.12 ± 1.98∗∗∗,## |
| (Total) Glutathione | 190.6 ± 25.1 | 73.7 ± 11.9∗∗∗ | 56.8 ± 5.9∗∗∗,# |
| Taurine | 392.2 ± 97.4 | 588.8 ± 153.3∗ | 530.5 ± 94.2∗ |
| Ornithine | 149.1 ± 52.0 | 84.1 ± 23.0∗∗ | 59.7 ± 12.7∗∗∗,# |
| Citrulline | 67.4 ± 12.4 | 51.2 ± 7.7∗ | 43.5 ± 4.7∗∗∗ |
C57BL/6J male mice were fed ad libitum (day 0), or fasted for 1 or 2 day(s) between 2 pm and 2 pm with free access to water, and blood samples were collected after anesthetization. Serum amino acid levels were measured using a HPLC system. The (total) levels of cysteine, homocysteine, and glutathione were measured after reductive cleavage of its disulfide bonds. Data are means ± S.D. (n = 7); significant changes in ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus ad libitum; #p < 0.05 and ##p < 0.01 versus day 1 by Mann–Whitney U-test.
3.2. Ubiquitin–proteasome system
Total RNA was prepared from 13 mouse tissues (rectus femoris muscle for skeletal muscle) and used for cDNA preparation/qPCR analysis; the preparation of high-quality RNA from pancreas was unsuccessful because of its high ribonuclease activity. The levels of mRNA expression were normalized by those of housekeeping Hprt because its expression was stable during fasting in all tested tissues. As the ubiquitin–proteasome system marker, we examined the expression of Atrogin-1 (also known as MAFbx [Muscle Atrophy F-box]) and MuRF1 (Muscle RING Finger 1; also known as Trim63 [Muscle Atrophy F-box]), the two E3 ubiquitin ligases involved in skeletal muscle atrophy [9,10]. The expression of Atrogin-1 was significantly upregulated upon fasting in skeletal muscle >> small intestine, thymus > heart, colon, lung > spleen, kidney > stomach (Fig. 2A). The expression of MuRF1 was upregulated in skeletal muscle, thymus >> lung, heart, and stomach although it was neither significantly upregulated nor expressed in other tissues (Fig. 2B). It is notable that the expression of both genes was more apparent at day 1 in skeletal muscle, lung, and stomach, but at day 2 in thymus (Fig. 2A and B).
Fig. 2.
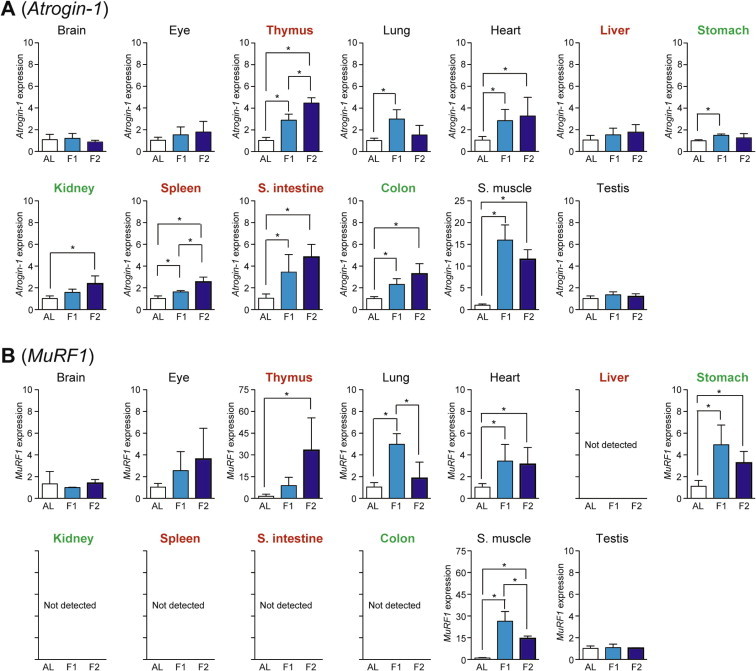
Fasting-induced tissue-specific expression changes in genes related to the ubiquitin–proteasome system. The expression of E3 ubiquitin ligases, Atrogin-1 (A) and MuRF1 (B), in various tissues was analyzed by qPCR and normalized to Hprt expression. The expression ratios in ad libitum-fed (AL) mice were set at 1. Data are shown as the mean ± S.D. (n = 4), and the differences were significant at ∗P < 0.05 (Mann–Whitney U-test). F1, 1-day fasting; F2, 2-day fasting; S. intestine, small intestine; S. muscle, skeletal (rectus femoris) muscle. Highly atrophied (>31% weight decrease), moderately atrophied (18–19% weight decrease), and non-atrophied tissues (no significant change) are labeled in red, green, and black, respectively.
3.3. Autophagy–lysosome system
As a measure of autophagy–lysosome system activity, we examined the expression of LC3b, p62, and Lamp2 (Lysosomal-associated membrane protein 2). LC3b is an essential component for the elongation of autophagosomal isolation membrane and p62 is a LC3-/ubiquitin-binding protein for selective autophagy of cytosolic ubiquitinated/aggregated proteins; therefore, both are involved in autophagosome-mediated macroautophagy [11,12]. The expression of LC3b was upregulated upon fasting in heart > skeletal muscle > thymus > lung, small intestine, testis, colon, and liver (Fig. 3A), and that of p62 was upregulated in heart > skeletal muscle > thymus > colon, lung, kidney, spleen, and small intestine (Fig.3B). Western blotting analyses of various tissue extracts revealed that the expression of LC3-II (membrane-bound lipidated form) was upregulated in heart, skeletal muscle, thymus, small intestine, and colon, and that p62 expression was upregulated in heart, skeletal muscle, thymus, and small intestine; these results were generally consistent with the qPCR results (Fig. 4). Lamp2 acts as a receptor for substrate proteins of chaperone-mediated autophagy in that vesicular traffic (autophagosome or invaginated lysosomal membrane for microautophagy) is not involved [13]. The expression of Lamp2 was increased only in thymus (Fig. 3C).
Fig. 3.
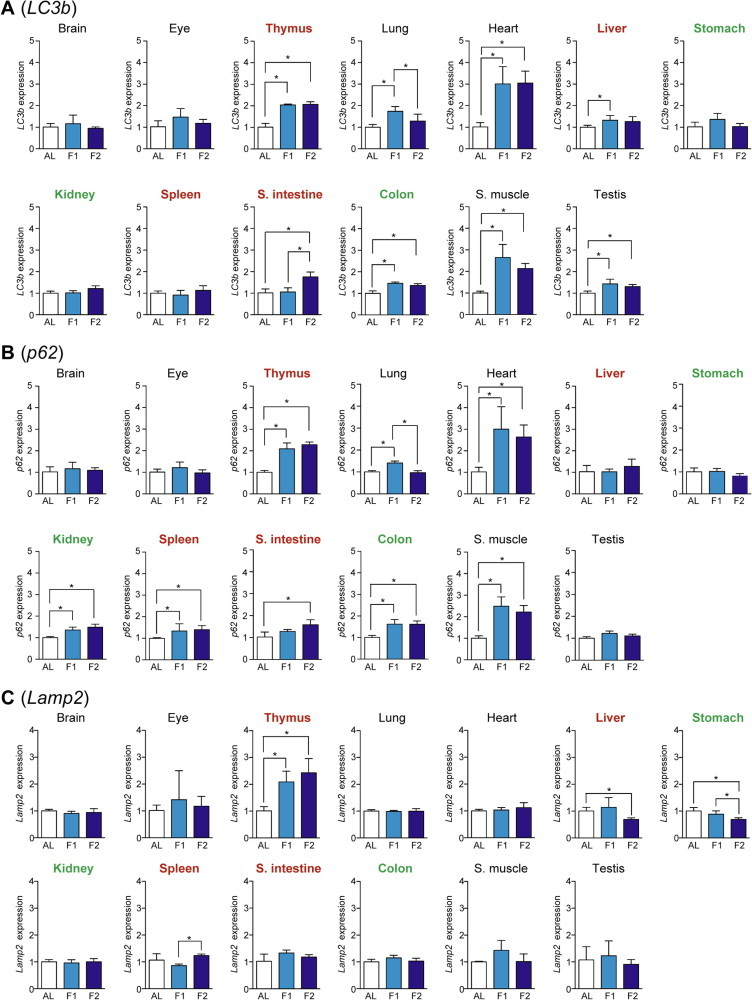
Fasting-induced tissue-specific expression changes in genes related to the autophagy–lysosome system. The expression of LC3b (A), p62 (B), and Lamp2 (C) in various tissues was analyzed by qPCR and normalized to Hprt expression. The expression ratios in ad libitum-fed (AL) mice were set at 1. Data are shown as the mean ± S.D. (n = 4), and the differences were significant at ∗P < 0.05 (Mann–Whitney U-test). F1, 1-day fasting; F2, 2-day fasting; S. intestine, small intestine; S. muscle, skeletal (rectus femoris) muscle. Highly atrophied (>31% weight decrease), moderately atrophied (18–19% weight decrease), and non-atrophied tissues (no significant change) are labeled in red, green, and black, respectively.
Fig. 4.
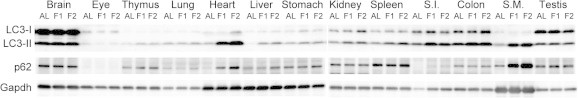
Western blot analysis of various mouse tissue homogenates for LC3b and p62 expression. Anti-LC3B antibody detected both LC3-I (∼18 kDa) and LC3-II (∼16 kDa) bands while anti-p62 and anti-GAPDH antibodies detected ∼62 kDa and ∼38 kDa bands, respectively. AL, ad libitum-fed; F1, 1-day fasting; F2, 2-day fasting; S.I., small intestine; S.M., skeletal (rectus femoris) muscle.
3.4. Amino acid (starvation) response
We examined the expression of five genes, Asns (Asparagine synthetase), Trib3 (Tribbles homolog 3), Herpud1 (Homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1), xCT (Sodium-independent aspartate/glutamate/cystine transporter; known as Slc7a11 [Solute carrier family 7, member 11]), and Chop (C/EBP homology protein; known as Ddit3 [DNA-damage inducible transcript 3]), as the amino acid response markers whose expressions could be regulated via ATF4-responsive C/EBP-ATF response elements (CARE) in their promoter regions [7,14]. Asns is an Asn-producing enzyme induced by the amino acid response as well as the unfolded protein response [14]; Trib3 is a regulator of various cellular processes such as proliferation, migration, invasion, and apoptosis [14,15]; Herpud1 is an endoplasmic reticulum (ER) stress-associated protein [16]; xCT is a transporter to uptake cystine and excrete Glu, thereby supplying Cys for reduced glutathione synthesis and thus cell survival [17]; and Chop is a transcriptional factor involved in ER stress-induced cell apoptosis [18]. The expression of Asns was upregulated upon fasting only in thymus > testis and brain (Fig. 5A) while that of Trib3 was upregulated in thymus, spleen > kidney, colon, and lung (Fig. 5B). In addition, the expression of Herpud1 was induced in thymus > small intestine and lung (Fig. 6A); that of xCT was upregulated only in thymus (Fig. 6B); and that of Chop was upregulated in thymus and testis (Fig. 6C).
Fig. 5.
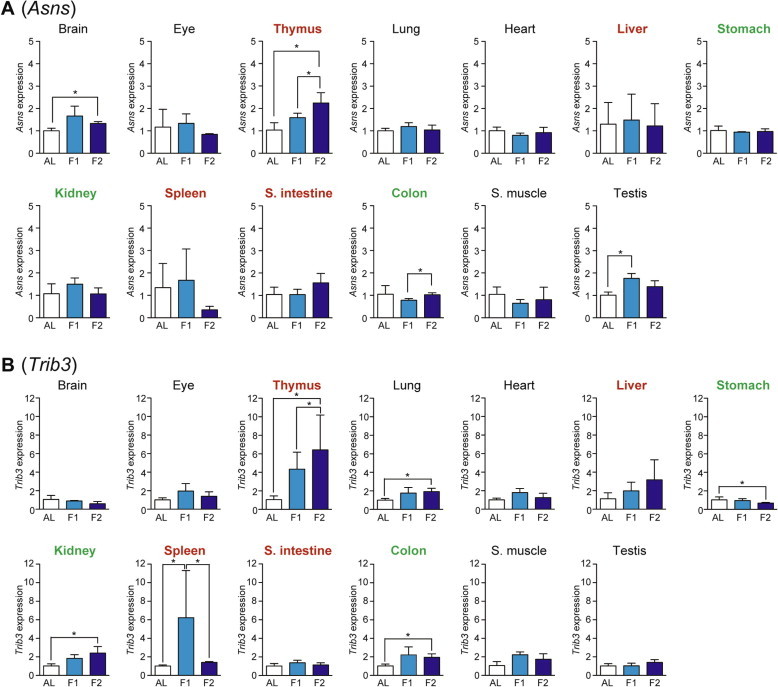
Fasting-induced tissue-specific expression changes in genes related to the amino acid response. The expression of Asns (A) and Trib3 (B) in various tissues was analyzed by qPCR and normalized to Hprt expression. The expression ratios in ad libitum-fed (AL) mice were set at 1. Data are shown as the mean ± S.D. (n = 4), and the differences were significant at ∗P < 0.05 (Mann–Whitney U-test). F1, 1-day fasting; F2, 2-day fasting; S. intestine, small intestine; S. muscle, skeletal (rectus femoris) muscle. Highly atrophied (>31% weight decrease), moderately atrophied (18–19% weight decrease), and non-atrophied tissues (no significant change) are labeled in red, green, and black, respectively.
Fig. 6.
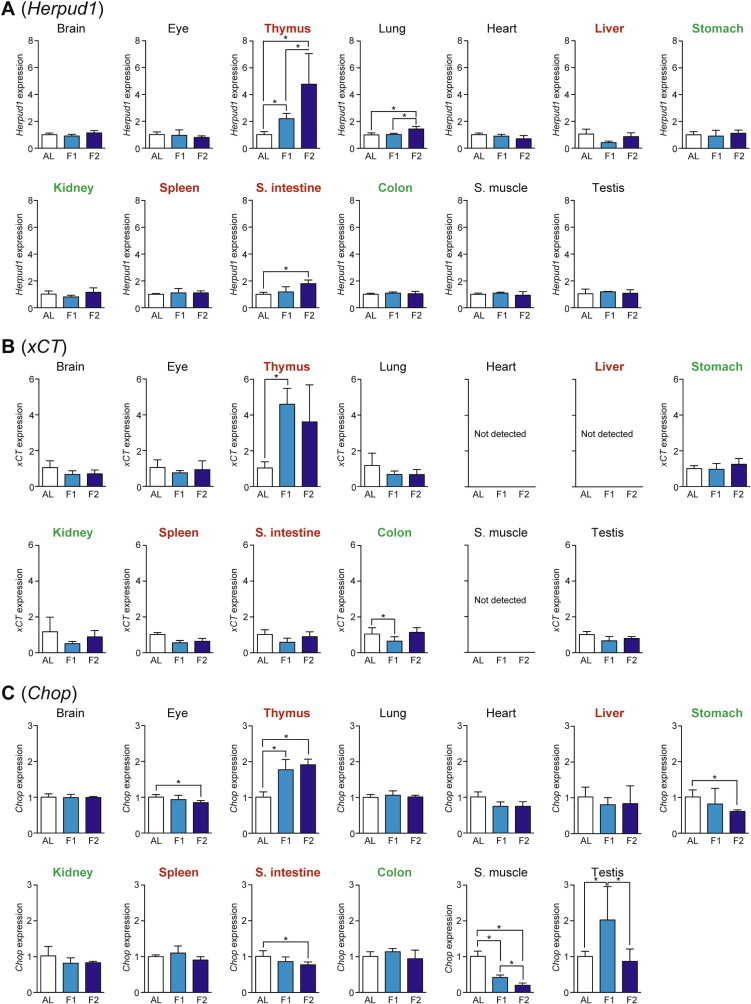
Fasting-induced tissue-specific expression changes in genes related to the amino acid response. The expression of Herpud1 (A), xCT (B), and Chop (C) in various tissues was analyzed by qPCR and normalized to Hprt expression. The expression ratios in ad libitum-fed (AL) mice were set at 1. Data are shown as the mean ± S.D. (n = 4), and the differences were significant at ∗P < 0.05 (Mann–Whitney U-test). F1, 1-day fasting; F2, 2-day fasting; S. intestine, small intestine; S. muscle, skeletal (rectus femoris) muscle. Highly atrophied (>31% weight decrease), moderately atrophied (18–19% weight decrease), and non-atrophied tissues (no significant change) are labeled in red, green, and black, respectively.
3.5. Nrf2 antioxidant system
The expression levels of HO-1 (Heme oxygenase-1) and Gsta1 (Glutathione S-transferase α1) were examined as representatives of Nrf2-regulated antioxidant defense/drug metabolism genes [19]. HO-1 expression was upregulated upon fasting in heart > thymus > skeletal muscle > kidney, spleen, and small intestine (Fig. 7A). Gsta1 expression was increased in kidney > thymus >> heart whereas it decreased in liver and skeletal muscle (Fig. 7B).
Fig. 7.
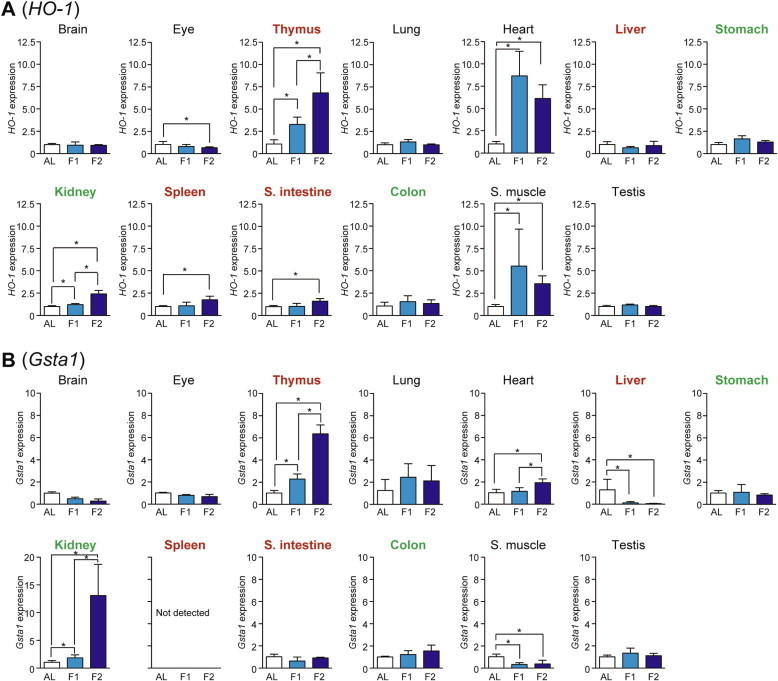
Fasting-induced tissue-specific expression changes in genes related to the Nrf2-mediated antioxidant system. The expression of HO-1 (A) and Gsta1 (B) in various tissues was analyzed by qPCR and normalized to Hprt expression. The expression ratios in ad libitum-fed (AL) mice were set at 1. Data are shown as the mean ± S.D. (n = 4), and the differences were significant at ∗P < 0.05 (Mann–Whitney U-test). F1, 1-day fasting; F2, 2-day fasting; S. intestine, small intestine; S. muscle, skeletal (rectus femoris) muscle. Highly atrophied (>31% weight decrease), moderately atrophied (18–19% weight decrease), and non-atrophied tissues (no significant change) are labeled in red, green, and black, respectively.
3.6. Amino acid transporters
Altered serum amino acid profiles could be a reflection of active amino acid import/export between tissues. In addition to xCT above, the expression levels of three major neutral/cationic amino acid transporter genes: Slc38a2 (Solute carrier family 38, member 2; also known as Snat2 [Sodium-coupled neutral amino acid transporter 2] [20], Slc7a5 (also known as Lat1 [L-type amino acid transporter 1]) [21], and Slc7a1 (also known as Cat1 [Cationic amino acid transporter 1]) [22] were examined. Slc38a2 expression (which could be activated by the amino acid response [14]) was increased upon fasting in eye and heart whereas it decreased in the colon (Fig. 8A). Slc7a5 expression was increased in colon but decreased in eye and kidney (Fig. 8B). Slc7a1 expression in skeletal muscle was increased at day 1 but then decreased at day 2, and that in stomach was decreased at day 2 (Fig. 8C).
Fig. 8.
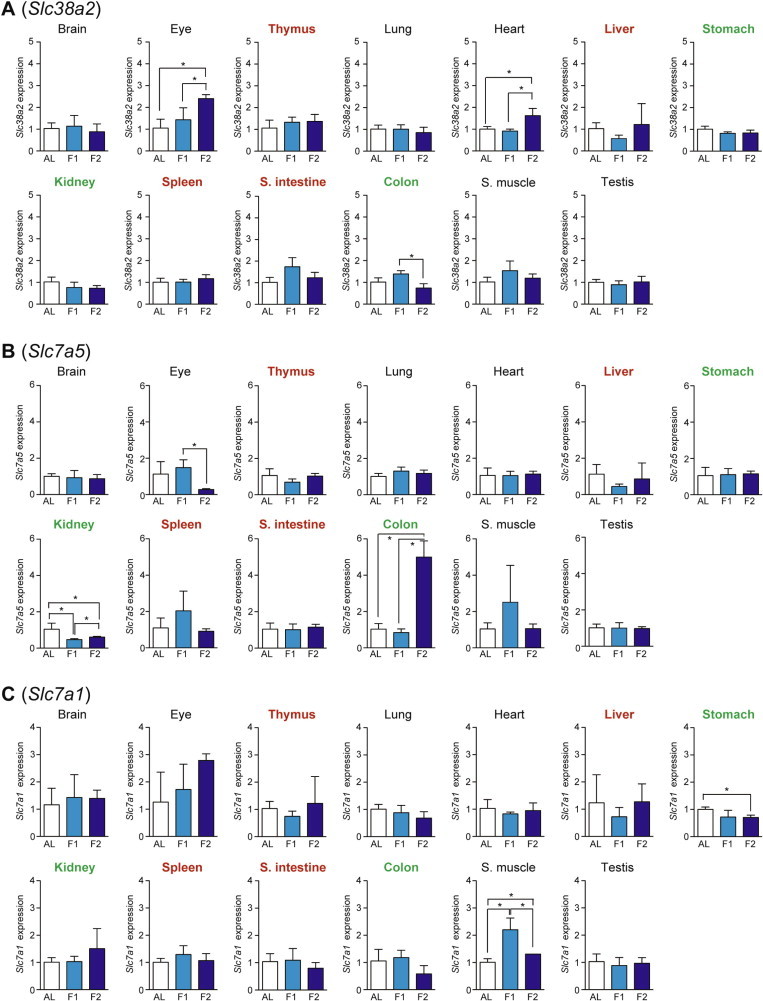
Fasting-induced tissue-specific expression changes in amino acid transporter genes. The expression of Slc38a2 (A), Slc7a5 (B), and Slc7a1 (C) in various tissues was analyzed by qPCR and normalized to Hprt expression. The expression ratios in ad libitum-fed (AL) mice were set at 1. Data are shown as the mean ± S.D. (n = 4), and the differences were significant at ∗P < 0.05 (Mann–Whitney U-test). F1, 1-day fasting; F2, 2-day fasting; S. intestine, small intestine; S. muscle, skeletal (rectus femoris) muscle. Highly atrophied (>31% weight decrease), moderately atrophied (18–19% weight decrease), and non-atrophied tissues (no significant change) are labeled in red, green, and black, respectively.
4. Discussion
Mammals survive (water-only) fasting by activating glycogenolysis, lipolysis (Fig. 1), ketogenesis (Table 3), and proteolysis in different tissues to maintain minimal blood glucose levels (∼100 mg/dL; Table 3) and indispensable energy production [1,23]. These responses are triggered, at least in part, by regulation of the levels of various hormones such as insulin, glucagon, growth hormone, insulin-like growth factor-1, glucocorticoids, epinephrine, triiodothyronine (T3), and thyroxine (T4) [1,23]; therefore, the proteolytic pathways upon fasting could be diverse depending on varying hormone sensitivity among different tissues. Here, we performed comprehensive analyses to gain an overview of tissue-specific adaptive responses upon fasting in a single/simple experimental paradigm; the results are summarized in Table 5. It should be noted that mice were deprived of food but had the opportunity to consume feces (coprophagia) and bedding (consists of cellulose, hemicellulose, and lignin; thus indigestible for mice) in this study; therefore, altered patterns of responses between 1- and 2-day fasting could be attributable to such consumption [6].
Table 5.
Summary of the tissue-specific adaptive responses to fasting in mice.
| Tissues |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Brain | Eye | Thymus | Lung | Heart | Liver | Stomach | Kidney | Spleen | S.I. | Colon | S.M. | Testis | |
| Weight loss (wet gram) | – | – | +++ | – | – | ++ | + | + | ++ | ++ | + | – | – |
| 1. Ubiquitin–proteasome system | |||||||||||||
| Atrogin-1 (MAFbx) | – | – | ++ | ++ (F1) | ++ | – | +(F1) | + | + | ++ | + | +++ (F1) | – |
| MuRF1 (Trim63) | – | – | +++ | ++ (F1) | ++ (F1) | ND | ++ (F1) | ND | ND | ND | ND | +++ (F1) | – |
| 2. Autophagy–lysosome system | |||||||||||||
| LC3b (Map1lc3b) | – | – | ++ | +(F1) | +++ (F1) | +(F1) | – | – | – | + | +(F1) | ++ (F1) | + (F1) |
| p62 (Sqstm1) | – | – | ++ | +(F1) | ++ (F1) | – | – | + | + | + | +(F1) | ++ (F1) | – |
| Lamp2 | – | – | ++ | – | – | down | down | – | – | – | – | – | – |
| 3. Amino acid response | |||||||||||||
| Asns | + | – | ++ | – | – | – | – | – | – | – | – | – | +(F1) |
| Trib3 | – | – | +++ | + | – | – | down | ++ | +++ (F1) | – | + | – | – |
| Herpud1 | – | – | +++ | + | – | – | – | – | – | + | – | – | – |
| xCT (Slc7a11) | – | – | +++ (F1) | – | ND | ND | – | – | – | – | down | ND | – |
| Chop (Ddit3) | – | down | + | – | – | – | down | – | – | down | – | down | ++ (F1) |
| 4. Nrf2 antioxidant system | |||||||||||||
| HO-1 | – | down | +++ | – | +++ (F1) | – | – | + | + | + | – | ++ (F1) | – |
| Gsta1 | – | – | +++ | – | + | down | – | +++ | ND | – | – | down | – |
| 5. Amino acid transporter | |||||||||||||
| Slc38a2 (Sat2) | – | ++ | – | – | + | – | – | – | – | – | down | – | – |
| Slc7a5 (Lat1) | – | down | – | – | – | – | – | down | – | – | +++ | – | – |
| Slc7a1 (Cat1) | – | – | – | – | – | – | down | – | – | – | – | ++ (F1) | – |
Systemic responses to 1- or 2-day (water-only) fasting were evaluated. Changes in wet tissue weights were categorized into – (no significant change), + (18–19% decrease), ++ (31–42% decrease), or +++ (54.7% decrease). Gene expression changes were categorized into – (no significant change), down (significant down-regulation), +(1 < X (fold-induction) < 3 for Atrogin-1, MuRF1, HO-1, and Gsta1, and 1 < X < 2 for others), ++ (3 < X < 6 for Atrogin-1, MuRF1, HO-1, and Gsta1, and 2 < X < 3 for others), and +++ (6 < X for Atrogin-1, MuRF1, HO-1, and Gsta1, and 3 < X for others); when the maximal responses were observed in 1-day fasted mice, “(F1)” was labeled. “down” represents significant down-regulation at day 1 or 2 in gene expression. S.I., small intestine; S.M., skeletal (rectus femoris) muscle; ND, not detected.
Previous studies mention that the ubiquitin–proteasome system is primarily responsible for increased skeletal muscle protein breakdown during starvation, whereas the autophagy–lysosome system is stimulated in most other tissues [23]. As ubiquitin–proteasome system markers, we examined the expression of Atrogin-1 and MuRF1, the two major skeletal muscle E3 ubiquitin ligases that are induced by a number of stressors (such as neural inactivity, malnutrition, oxidative stress, and glucocorticoids) and via a number of transcriptional factors (such as Foxo1/Foxo3a, NF-κB, C/EBPβ, and glucocorticoid receptor) [10]. Although the muscle loss upon 1- or 2-day fasting was not so apparent by X-ray CT analyses (compared with fat loss; Fig. 1) and weight measurements of rectus femoris muscles (Table 2), and serum levels of branched chain amino acids (Val, Leu, and Ile) that comprise ∼35% of the essential amino acids in muscle proteins and ∼40% of the amino acids required preformed by mammals [24] were not significantly changed (compared with most other amino acids; Table 4), Atrogin-1 and MuRF1 were upregulated not only in skeletal muscle [25] but also in thymus, heart, lung, and stomach (plus small intestine, colon, spleen, and kidney for Atrogin-1) (Fig. 2A and B). Moreover, both (macro) autophagy/lysosome system markers, LC3b and p62 [11,12], were induced in skeletal muscle (although their induction was not so great compared with Atrogin-1 and MuRF1) as well as in heart, thymus, lung, small intestine, and colon (Fig. 3A and B), and Lamp2 as a chaperone-mediated autophagy marker [13] was increased in thymus (Fig. 3C). Taken together, both ubiquitin–proteasome and autophagy–lysosome systems seemed to be activated, at least in thymus, lung, heart, and skeletal muscle (Table 5); importantly, these three tissues except thymus were all non-atrophied tissues (Table 2). A previous study mentioned that 0.5–1.5-day fasting-induced LC3-II accumulation in the liver was consistently apparent only in the presence of leupeptin (a lysosomal cysteine proteinase inhibitor) [26], but we could detect both Lc3b mRNA and LC3-II protein induction in the liver of 1 day fasted mice (Figs. 3A and 4). Liver autophagy may contribute to the maintenance of blood glucose levels (Table 3) and amino acid levels (Table 4) [26], although the autophagy–lysosome system might be more activated in heart and skeletal muscle (Fig. 4).
Next we examined the expression of five ATF4-regulated “amino acid response” genes (Asns, Trib3, Herpud1, xCT, and Chop [14]) because fasting induced a dramatic reduction in the serum levels of most proteinogenic amino acids (Table 4). General (amino acid) control depressible 2 (GCN2) kinase acts as an amino acid sensor by binding to any uncharged RNA, leading to the phosphorylation/activation of eukaryotic (translation) initiation factor (eIF) 2α and thus transcriptional activation of ATF4-regulated genes [14,27]. Therefore, a depletion at the cellular level of any individual amino acid could activate this system [7]. All five genes were markedly upregulated only in thymus, although slight but significant concomitant increases were found for Asns/Chop in testis and Trib3/Herpud1 in lung (Figs. 5 and 6). We previously demonstrated much more-marked Asns induction (30-folds at maximum) in skeletal muscle, liver, kidney, and small intestine of mice fed each essential amino acid-free diet for 2 weeks; the altered responses could be caused by altered serum levels of deprived amino acids [7]. In the state of 1- or 2-day fasting, the thymus may be the most sensitive tissue to amino acid starvation.
Fasting did not decrease serum total Cys levels but did decrease total glutathione levels (Table 4); therefore, the Nrf2-regulated system including drug metabolism/disposition and antioxidant defense [19,28] could give way upon fasting. Indeed the two major Nrf2-target genes, HO-1 and Gsta1, were upregulated in thymus, heart, and kidney (plus skeletal muscle, spleen, and small intestine for HO-1) (Fig. 7A and B). Such antioxidant responses may underlie cytoprotective effects conferred by fasting [1,3]. Indeed we observed that 2-day fasting of mice provides marked cardioprotection against ischemia/reperfusion injury [29] and thus, fasting-induced cytoprotection could be expected also in thymic and renal injuries. Furthermore, xCT, an aspartate/glutamate/cystine transporter gene, was induced upon fasting in thymus (Fig. 6B), and the three major amino acid transporter genes, Slc38a2, Slc7a5, and Slc7a5, were upregulated or downregulated in particular tissues (Fig. 8), suggesting the presence of controlled amino acid redistribution among tissues upon fasting. This may be reflected by changes in serum amino acid profile (Table 4).
In summary, single short-term fasting of mice conferred tissue-specific adaptive responses not only in atrophied tissues but also in non-atrophied tissues. Highly atrophied liver is a silent organ whereas non-atrophied lung, heart, and skeletal muscle are very active in this setting. Further investigation to clarify such responses may contribute to a better understanding of clinical benefits conferred by fasting, and our designed primer sets may be helpful for follow-on research.
Conflict of interest
There are no conflicts of interest to declare.
Acknowledgements
This study was supported by the Grants-in-Aid for Scientific Research (25460072 and 25220103) and the Program for Strategic Research Foundation at Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan; Keio University Program for the Advancement of Next Generation Research Projects and Keio Gijuku Fukuzawa Memorial Fund for the Advancement of Education and Research (to I.I.). I.I. conceived and designed the project; J.Y., S.K., A.M., T.N., and R.K. acquired the data; J.Y., S.K., and I.I. analyzed and interpreted the data; I.I. wrote the paper.
References
- 1.Longo V.D., Mattson M.P. Fasting: molecular mechanisms and clinical applications. Cell Metab. 2014;19:181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Praag H., Fleshner M., Schwartz M.W., Mattson M.P. Exercise, energy intake, glucose homeostasis, and the brain. J. Neurosci. 2014;34:15139–15149. doi: 10.1523/JNEUROSCI.2814-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robertson L.T., Mitchell J.R. Benefits of short-term dietary restriction in mammals. Exp. Gerontol. 2013;48:1043–1048. doi: 10.1016/j.exger.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verweij M., van Ginhoven T.M., Mitchell J.R., Sluiter W., van den Engel S., Roest H.P., Torabi E., Ijzermans J.N., Hoeijmakers J.H., de Bruin R.W. Preoperative fasting protects mice against hepatic ischemia/reperfusion injury: mechanisms and effects on liver regeneration. Liver Transpl. 2011;17:695–704. doi: 10.1002/lt.22243. [DOI] [PubMed] [Google Scholar]
- 5.Jongbloed F., de Bruin R.W., Pennings J.L., Payan-Gomez C., van den Engel S., van Oostrom C.T., de Bruin A., Hoeijmakers J.H., van Steeg H., JN I.J., Dolle M.E. Preoperative fasting protects against renal ischemia-reperfusion injury in aged and overweight mice. PLoS One. 2014;9:e100853. doi: 10.1371/journal.pone.0100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen T.L., Kiersgaard M.K., Sorensen D.B., Mikkelsen L.F. Fasting of mice: a review. Lab. Anim. 2013;47:225–240. doi: 10.1177/0023677213501659. [DOI] [PubMed] [Google Scholar]
- 7.Kamata S., Yamamoto J., Kamijo K., Ochiai T., Morita T., Yoshitomi Y., Hagiya Y., Kubota M., Ohkubo R., Kawaguchi M., Himi T., Kasahara T., Ishii I. Dietary deprivation of each essential amino acid induces differential systemic adaptive responses in mice. Mol. Nutr. Food Res. 2014;58:1309–1321. doi: 10.1002/mnfr.201300758. [DOI] [PubMed] [Google Scholar]
- 8.Mudd S.H., Levy H.L., Kraus J.P. Disorders of transsulfuration. In: Scriver C.R., Beaudet A.L., Sly W.S., Valle D., editors. The Metabolic and Molecular Basis of Inherited Disease. 8th ed. MaGraw-Hill; New York: 2001. pp. 2007–2056. [Google Scholar]
- 9.Koyama S., Hata S., Witt C.C., Ono Y., Lerche S., Ojima K., Chiba T., Doi N., Kitamura F., Tanaka K., Abe K., Witt S.H., Rybin V., Gasch A., Franz T., Labeit S., Sorimachi H. Muscle RING-finger protein-1 (MuRF1) as a connector of muscle energy metabolism and protein synthesis. J. Mol. Biol. 2008;376:1224–1236. doi: 10.1016/j.jmb.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 10.Bodine S.C., Baehr L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 2014;307:E469–E484. doi: 10.1152/ajpendo.00204.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid. Redox Signal. 2011;14:2201–2214. doi: 10.1089/ars.2010.3482. [DOI] [PubMed] [Google Scholar]
- 13.Majeski A.E., Dice J.F. Mechanisms of chaperone-mediated autophagy. Int. J. Biochem. Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 14.Kilberg M.S., Shan J., Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 2009;20:436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang N., Zhang W., Xu S., Lin H., Wang Z., Liu H., Fang Q., Li C., Peng L., Lou J. TRIB3 alters endoplasmic reticulum stress-induced beta-cell apoptosis via the NF-kappaB pathway. Metabolism. 2014;63:822–830. doi: 10.1016/j.metabol.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Shinozaki S., Chiba T., Kokame K., Miyata T., Kaneko E., Shimokado K. A deficiency of Herp, an endoplasmic reticulum stress protein, suppresses atherosclerosis in ApoE knockout mice by attenuating inflammatory responses. PLoS One. 2013;8:e75249. doi: 10.1371/journal.pone.0075249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewerenz J., Hewett S.J., Huang Y., Lambros M., Gout P.W., Kalivas P.W., Massie A., Smolders I., Methner A., Pergande M., Smith S.B., Ganapathy V., Maher P. The cystine/glutamate antiporter system x(c)(−) in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid. Redox Signal. 2013;18:522–555. doi: 10.1089/ars.2011.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Guo Y., Tang J., Jiang J., Chen Z. New insights into the roles of CHOP-induced apoptosis in ER stress. Acta Biochim. Biophys. Sinica. 2014;46:629–640. doi: 10.1093/abbs/gmu048. [DOI] [PubMed] [Google Scholar]
- 19.Ma Q. Role of Nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palii S.S., Thiaville M.M., Pan Y.X., Zhong C., Kilberg M.S. Characterization of the amino acid response element within the human sodium-coupled neutral amino acid transporter 2 (SNAT2) System A transporter gene. Biochem. J. 2006;395:517–527. doi: 10.1042/BJ20051867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mastroberardino L., Spindler B., Pfeiffer R., Skelly P.J., Loffing J., Shoemaker C.B., Verrey F. Amino-acid transport by heterodimers of 4F2hc/CD98 and members of a permease family. Nature. 1998;395:288–291. doi: 10.1038/26246. [DOI] [PubMed] [Google Scholar]
- 22.Closs E.I., Graf P., Habermeier A., Cunningham J.M., Forstermann U. Human cationic amino acid transporters hCAT-1, hCAT-2A, and hCAT-2B: three related carriers with distinct transport properties. Biochemistry. 1997;36:6462–6468. doi: 10.1021/bi962829p. [DOI] [PubMed] [Google Scholar]
- 23.Finn P.F., Dice J.F. Proteolytic and lipolytic responses to starvation. Nutrition. 2006;22:830–844. doi: 10.1016/j.nut.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Harper A.E., Miller R.H., Block K.P. Branched-chain amino acid metabolism. Annu. Rev. Nutr. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- 25.Sandri M. Autophagy in skeletal muscle. FEBS Lett. 2010;584:1411–1416. doi: 10.1016/j.febslet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- 26.Ezaki J., Matsumoto N., Takeda-Ezaki M., Komatsu M., Takahashi K., Hiraoka Y., Taka H., Fujimura T., Takehana K., Yoshida M., Iwata J., Tanida I., Furuya N., Zheng D.M., Tada N., Tanaka K., Kominami E., Ueno T. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gallinetti J., Harputlugil E., Mitchell J.R. Amino acid sensing in dietary-restriction-mediated longevity: roles of signal-transducing kinases GCN2 and TOR. Biochem. J. 2013;449:1–10. doi: 10.1042/BJ20121098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagiya Y., Kamata S., Mitsuoka S., Okada N., Yoshida S., Yamamoto J., Ohkubo R., Abiko Y., Yamada H., Akahoshi N., Kasahara T., Kumagai Y., Ishii I. Hemizygosity of transsulfuration genes confers increased vulnerability against acetaminophen-induced hepatotoxicity in mice. Toxicol. Appl. Pharmacol. 2015;282:195–206. doi: 10.1016/j.taap.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 29.Nakano S., Ishii I., Shinmura K., Tamaki K., Hishiki T., Akahoshi N., Ida T., Nakanishi T., Kamata S., Kumagai Y., Akaike T., Fukuda K., Sano M., Suematsu M. Hyperhomocysteinemia abrogates fasting-induced cardioprotection against ischemia/reperfusion by limiting bioavailability of hydrogen sulfide anions. J. Mol. Med. 2015 doi: 10.1007/s00109-015-1271-5. [DOI] [PubMed] [Google Scholar]


