Abstract
Genetic determinants appear to play a role in susceptibility to chronic diarrhea, but the genetic abnormalities involved have only been identified in a few conditions. The Na+/H+ exchanger 3 (NHE3) accounts for a large fraction of physiologic intestinal Na+ absorption. It is highly regulated through effects on its intracellular COOH-terminal regulatory domain. The impact of genetic variation in the NHE3 gene, such as single nucleotide polymorphisms (SNPs), on transporter activity remains unexplored. From a total of 458 SNPs identified in the entire NHE3 gene, we identified three nonsynonymous mutations (R474Q, V567M, and R799C), which were all in the protein's intracellular COOH-terminal domain. Here we evaluated whether these SNPs affect NHE3 activity by expressing them in a mammalian cell line that is null for all plasma membrane NHEs. These variants significantly reduced basal NHE3 transporter activity through a reduction in intrinsic NHE3 function in variant R474Q, abnormal trafficking in variant V567M, or defects in both intrinsic NHE3 function and trafficking in variant R799C. In addition, variants NHE3 R474Q and R799C failed to respond to acute dexamethasone stimulation, suggesting cells with these mutant proteins might be defective in NHE3 function during postprandial stimulation and perhaps under stressful conditions. Finally, variant R474Q was shown to exhibit an aberrant interaction with calcineurin B homologous protein (CHP), an NHE3 regulatory protein required for basal NHE3 activity. Taken together, these results demonstrate decreased transport activity in three SNPs of NHE3 and provide mechanistic insight into how these SNPs impact NHE3 function.
Keywords: NHE3, SNP, CHP, dexamethasone
chronic diarrhea remains a significant burden on the U.S. healthcare system (18). Abnormal function of intestinal ion transporters have long been known to be causative for the pathophysiology of diarrhea (15). Among apical membrane transport proteins, the Na+/H+ exchanger 3 (NHE3) (also known as solute carrier family 9 member 3; SLC9A3) is a major contributor to salt absorption in the gastrointestinal and renal systems, where it helps maintain body water and sodium homeostasis (3, 21, 39). Genetic evidence of its role in epithelial Na+ absorption includes that NHE3 knockout mice display modest diarrhea along with increased fluid in the small intestine and colon (17, 24). These mice also exhibit reduced fractional proximal renal reabsorption of Na+, resulting in significantly reduced blood pressure (37).
NHE3 is composed of two functionally distinct domains: 1) an NH2-terminal transmembrane domain that mediates Na+ and H+ ion exchange, and 2) a large cytoplasmic COOH-terminal domain that regulates transporter activity by affecting either NHE3 exocytosis or endocytosis (11, 22). The ∼377 amino-acid COOH terminus of NHE3 interacts both directly and indirectly with an array of regulatory proteins, including cytoskeletal proteins, kinases, phosphatases, scaffold proteins, and others, by which the regulation of NHE3 activity is achieved (11–13, 43). This regulation involves both increased and reduced NHE3 activity, as occurs sequentially after meals. Mutations in this region lead to dysregulation of NHE3 to physiological activators or inhibitors (27, 38, 44).
Recent studies have identified genetic variations in genes that account for individual susceptibility to chronic diarrhea (16, 42). Consequently, we searched the National Center for Biotechnology Information dbSNP database (www.ncbi.nlm.nih.gov/projects/SNP, 2008) and found a total of 458 SNPs in NHE3. All but nine of these SNPs are located in introns. Of the nine SNPs in exons, three are nonsynonymous mutations, each affecting different amino acid residues in the intracellular COOH terminus (Fig. 1 and Table 1). In the present study, we examined the effect of these three variants on NHE3 transporter activity under basal, inhibitory, and stimulatory conditions designed to mimic postprandial regulation of NHE3. We also evaluated the effect of these mutations on the interaction between NHE3 and calcineurin B homologous protein (CHP), an NHE3 binding protein that associates with the NHE3 COOH terminus under basal conditions and is necessary for NHE activity (10).
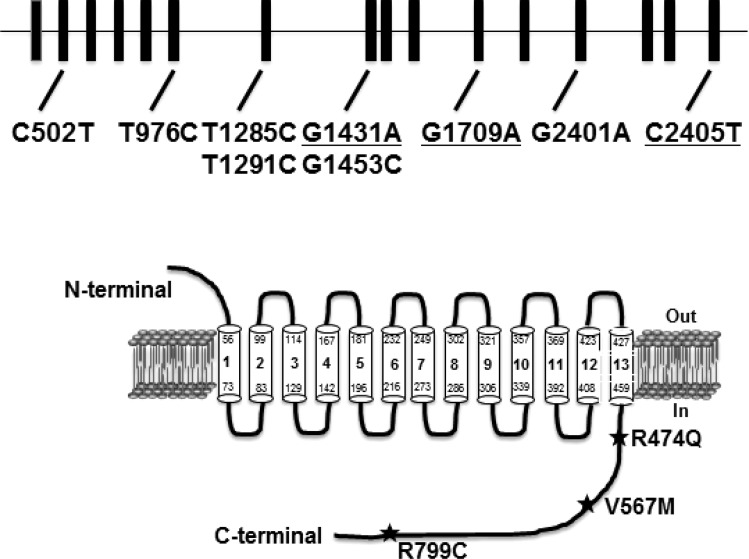
Fig. 1.
Identification of three nonsynonymous single nucleotide polymorphisms (SNPs) in the Na+/H+ exchanger 3 (NHE3) NHE3 gene. SNPs in the NHE3 gene were identified from the dbSNP(build 129) and Ensembl databases searched in 2008. Schematic diagram of the NHE3 gene and the locations of 9 SNPs in the exons are shown at top. Six silent mutations are in not underlined and three missense SNPs are underlined. All 3 missense SNPs, producing amino acid changes R474Q, V567M, and R799C, are located in the cytoplasmic tail of NHE3, as shown at bottom.
Table 1.
Summary of total 43 nonsynonymous SNPs at the COOH terminus of NHE3 present in the database
| NHE3 COOH Terminus | Nonsynonymous SNP |
|---|---|
| α0 (αα 458–471) | rs368029489 Arg471Trp; |
| Loop (aa472–474) | rs144153604 Glu472Asp; rs13154302 Arg474Gln |
| α1 (aa 475–494) | rs372558316 Gly481Arg |
| Loop (aa 495–503) | rs377525383 Gly496Arg |
| α2 (αα 504–513) | rs143487075 Asp505Asn |
| α3 (αα 518–560) | rs139467260 Arg522Gln; rs144147429 Gln525Arg; rs376396781 Arg528Gln; rs371820443 Glu537Lys; rs371187218 Ser561Tyr |
| α4 (αα 562–593) | rs368459613 Asp565Asn; rs41282627 Val567Met; rs 148343380 Thr573Lys; rs200347051 Ala581Ser; rs376013690 Val591Ala |
| α5 (αα 599–605) | rs115689317 Arg604Gln; rs371725135 Arg605Trp; rs146547322 Arg605Gln |
| Loop (aa 606–613) | rs145635595 Ala611Val |
| α6 (aa 614–637) | rs140444435 Thr619Met; rs202223784 Gln621Arg |
| Loop (aa 638–645) | rs372661265 Thr641Met; rs150973602 Thr643Met; rs377432139 Asp645Asn |
| α7 (aa 646–667) | rs145136022 Lys647Glu; rs373185294 Arg650Pro; rs199498162 Arg650Gly; rs377627891 Arg655Ly; rs144798475 Arg658Gln; rs201741190 Lys659Asn; rs199502995 Glu662Gln |
| α8 (aa 675–691) | |
| Loop (aa 692–749) | rs372062509 Asn696Ser; rs368610931 Ser703Gly; rs199817159 Gln706Arg; rs374244347 Glu712Lys; rs377080845 Asp719Glu; rs145183553 Thr720Ala |
| α9 (aa 750–752) | rs189017275 Gly751Glu |
| Loop (aa 753–806) | rs202198760 Ala762Asp; rs200138066 Gln790His; rs368047255 Pro792Ser; rs58623748 Cys799Arg |
Single nucleotide polymorphisms (SNPs) that are located near R474, V567, and R799 are highlighted by an underline. Database was accessed in May 2014.
NHE3, Na+/H+ exchanger 3.
MATERIALS AND METHODS
Materials.
All chemicals were purchased from Sigma (St. Louis, MO) except as follows: penicillin and streptomycin were from Whittaker MA Bioproducts (Walkersville, MD); culture media and G-418 (neomycin) were from GIBCO-BRL (Grand Island, NY); polyclonal anti-vesicular stomatitis virus (anti-VSVG) and anti-HA antibodies were from Covance Research Products (Princeton, NJ); mouse monoclonal anti-VSVG antibody was kindly provided by Thomas Kreis via Daniel Louvard (Curie Institute, Paris, France). The BCECF-AM [acetoxymethyl derivative of (2′7′)bis(2-carboxyethyl)-5,6-carboxyfluorescein] was from Invitrogen (Carlsbad, CA).
Cell culture, plasmid construction, and transfection.
PS120 fibroblasts do not express endogenous plasma membrane NHEs. We used these cells for stable expression of human NHE3 wild-type (26, 27) and mutants R474Q, R474A, V567M, and V567A, epitope-tagged with a COOH-terminal vesicular stomatitis virus glycoprotein (VSV-G) epitope (28). Rabbit NHE3 wild-type and mutant R799C and R799A with a triple-HA epitope tag at the NH2 terminus were also stably expressed in PS120 fibroblasts. All the NHE3 mutants were made using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The templates for mutagenesis were the pcDNA3.1/Neomycin+ vector (Invitrogen) containing human NHE3-VSVG or rabbit HA3-NHE3. NHE3 mutants were transfected into PS120 cells as described previously (28). G-418-resistant cells were selected by growth in 400 μg/ml and maintained in 200 μg/ml G-418. Clonal cell lines were isolated by limiting dilution and screened by Western blot for maximal expression. All PS120 cell lines were grown in DMEM supplemented with 25 mM NaHCO3, 10 mM HEPES, 50 U/ml penicillin, 50 μg/ml streptomycin, 400 μg/ml G418, and 10% fetal bovine serum in a 5% CO2-95% O2 incubator at 37°C.
Measurement of Na+/H+ exchange activity.
Na+/H+ exchange activity was determined fluorometrically using the intracellular pH-sensitive dye BCECF-AM (5 μM) in PS120 cells stably expressing wild-type NHE3 and mutants as described previously (28). Na+/H+ exchange activity was calculated as the product of the change in pHi (ΔpHi/dt) times the intracellular buffering capacity at each of multiple pHi values, as described previously (8). Kinetics of Na+/H+ exchange were analyzed by Hill plot using Origin (Microcal Software) to estimate NHE3 transport activity (Vmax) and K'(H+)i in individual experiments (28). K'(H+)i is a complex term composed of the effective concentration or two H+ binding/transport sites, interaction factors, and dissociation constants, as described previously (28). Means ± SE were determined from at least three experiments. In the experiments to determine the effects of cAMP, dexamethasone, and serum on NHE3 transporter activity, PS120 cells were first serum starved for different time periods (cAMP 3 h, serum 6 h, and dexamethasone 48 h). Cells were then incubated with 100 μM cAMP for 30 min, 10 μM dexamethasone for 2 h, or 10% dialyzed fetal bovine serum for 3 h before Na+/H+ exchange measurement.
Measurement of surface NHE3.
To measure surface levels of NHE3, PS120 cells stably expressing NHE3 wild-type, R474Q, V567M, or R799C were grown in 10-cm dishes to 90% confluency. Cells were incubated in serum-free medium for 3 h, followed by biotinylation assay with NHS-SS-biotin as described previously (22). Briefly, cells were washed twice in ice-cold PBS and once in borate buffer (154 mM NaCl, 10 mM boric acid, 7.2 mM KCl, and 1.8 mM CaCl2, pH 9.0). Plasma membrane proteins were biotinylated in 5 ml borate buffer containing 5 μg of sulfo-NHS-SS-biotin by gently shaking at 4°C for 30 min. Cells were then washed extensively with quenching buffer (120 mM NaCl and 20 mM Tris, pH 7.4) to remove excessive sulfo-NHS-SS -biotin, followed by two PBS rinses. Tissues were then scraped and lysed in 1 ml of N' buffer (60 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM KCl, 5 mM EDTA trisodium, 3 mM EGTA, 0.05% SDS, and 1% Triton X-100) and sonicated on ice. An aliquot of 80 μl lysate was saved for total NHE3 measurement. The total cell lysate (T) (0.8 ml) was incubated with streptavidin-agarose beads (50 μl of bed volume) to pull down the surface biotinylated proteins (S) that were eluted from the beads with 80 μl of Laemmli sample buffer (1). The supernatant, after incubation with streptavidin-agarose beads, was saved as the intracellular fraction (C). As indicated, each sample with several dilutions (μl) from total, surface, and intracellular fractions was subjected to 12.5% SDS-PAGE. Western blot assay was employed to quantify NHE3 protein by anti-VSV-G or anti-HA antibodies, and GAPDH was used as a loading control. Protein concentrations were determined (intensity units/ul) with an Odyssey Infrared Imaging System. Multiple volumes for each total, surface, and intracellular sample were plotted using linear regression, and the intensity of the signal was used to obtain a single value for each sample, which was expressed as intensity units per microliter. The results were presented as the percentage of total NHE3 on the surface by considering the dilution of each fraction.
Immunoprecipitation of NHE3 by CHP protein.
PS120 cells expressing NHE3 or its mutants were grown on 10-cm Petri dishes to 70–80% confluency, and cell lysates were prepared and incubated with agarose beads conjugated to anti-HA antibody. The agarose beads were then washed and bound proteins were eluted with Laemmli sample buffer. Protein samples were analyzed by SDS-PAGE and detected by Western blotting with anti-NHE3 and anti-CHP antibodies. Protein bands in the Western blots were quantified using an Odyssey Infrared Imaging System.
RESULTS
Effect of nonsynonymous SNPs on Na+/H+ exchange activity under basal conditions.
We searched the 2008 dbSNP(build 129) and Ensembl databases and found 458 SNPs for human NHE3, with 9 SNPs in exons and the rest in introns. Of these nine SNPs, six are synonymous silent substitutions giving the same amino acid sequence as wild type and three are missense mutations, R477Q, V567M, and R799C (Fig. 1 and Table 1). These three SNPs all affect amino acid residues in the cytoplasmic COOH-terminal domain of NHE3. To determine whether these affect NHE3 transporter activity, we introduced the same three point mutations and also separately replaced each amino acid with Ala, by site-directed mutagenesis into human and/or rabbit NHE3 and stably transfected the expression constructs into PS120 fibroblasts, a cell line that does not express any endogenous plasma membrane NHE. This cell line also lacks the NHE3 regulatory protein, NHERF2, which is required for many aspects of NHE3 activity (22). Accordingly, NHERF2 and each NHE3 variant were stably cotransfected into PS120 cells. Ala substitution tested the hypothesis that any mutation other than the usual polymorphic amino acid would similarly affect NHE3 activity. We monitored Na+/H+ exchange activity using BCECF/fluorometry (28). Basal NHE3 transport activity (Vmax) was reduced in all three NHE3 variants (Fig. 2A). In all cases, K'(H+)i was significantly increased compared with wild type (Fig. 2A, inset).
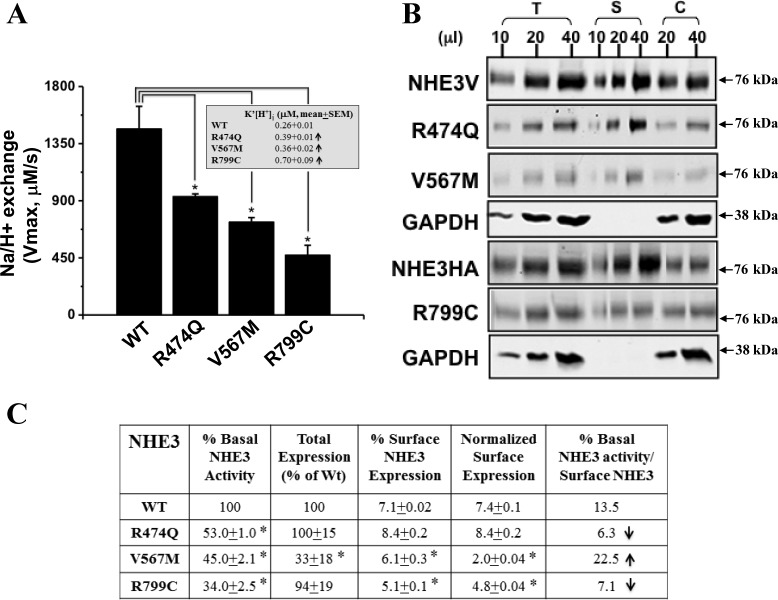
Fig. 2.
The 3 NHE3 variants have decreased basal Na+/H+ exchange activity. A: Na+/H+ transporter activity was measured in PS120/NHERF2 cells transfected with wild-type (WT) NHE3 or its mutants R474Q, V567M, and R799C. Data are presented as means ± SE from at least 3 independent experiments. *P < 0.05, comparison between wild-type NHE3 and the individual mutants (Student's t-test). Inset: K'(H+)i obtained by kinetic analysis of the same experiments used for the NHE3 transport activity (Vmax) calculations. K'(H+)i, ↑P < 0.05. B: surface expression of NHE3 [either epitope tagged with VSVG (NHE3V) or HA (NHE3HA)] and its polymorphic variants R474Q, V567M, and R799C was determined by cell surface biotinylation assay. NHE3 proteins in total lysates (T), surface fractions (S), and intracellular fractions (C) are labeled. A representative experiment, repeated 4 times is shown. Molecular mass standards are shown at right. GAPDH is only shown for WT. C: table shows results are means of 3–4 experiments ± SE. Column 1: relative Na+/H+ transporter activity in the variants after normalizing to WT NHE3 in each experiment. Column 2: total expression of each polymorphism normalized to WT NHE3. Column 3: percentage of total NHE3 on the plasma membrane. Column 4: normalized surface NHE3 per cell, which was obtained as the product of the normalized surface NHE3 (column 2) times the percent surface expression of each construct (column 3). Rounding causes some variation. In columns 1-4, *P < 0.05. Column 5 is column 1/column 4; however, since experiments in columns 1 and 4 did not entirely overlap, statistical evaluation is not provided.
The effect of nonsynonymous SNPs on surface expression of NHE3.
Previous studies have shown that NHE3 resides in both the plasma membrane and endosomal compartments and trafficks between these pools by exocytosis and endocytosis under both basal and acutely regulated conditions (1, 6, 9, 23, 26, 29, 40, 41). To determine whether the decreased basal NHE3 activity in these NHE3 mutations was due to reduced total or surface expression of NHE3, we performed cell surface biotinylation assays to measure the total amount of NHE3 protein expressed in the cells and on the plasma membrane surface (Fig. 2B). There were reduced levels of NHE3 in the surface fraction in the V567M and R799C mutants but not in R474Q (Fig. 2, B and C). Normalization of NHE3 in the surface fraction to total NHE3 compared with wild-type total set as 100% revealed that the percentage of surface expression of NHE3 was reduced in both the V567M and R799C mutants, whereas R474Q had a surface expression level similar to that of wild-type NHE3 (Fig. 2C).
Impaired intrinsic transporter activity in R474Q and R799C variants.
The intrinsic activities of the wild-type and mutant NHE3 molecules were compared by normalizing NHE3 activity (Vmax) to the relative amount of plasma membrane NHE3 (Fig. 2C) again normalized to that of wild-type NHE3. There was a significant reduction in the transporter activity/surface expression in variants R474Q and R799C, indicating that there are defects in intrinsic protein function (turnover number of Na+ and H+). In contrast, the intrinsic activity/surface molecule of variant V567M was increased compared with wild type (Fig. 2C, 5th column).
Alanine replacement at the polymorphic sites impairs NHE3 activity.
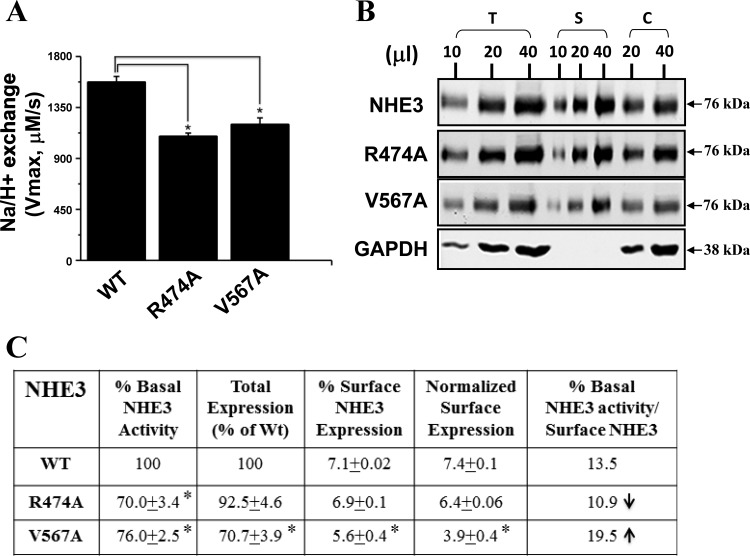
Whether the altered Na+/H+ exchange activity observed in the three NHE3 SNPs was due to substitution by a particular amino acid residue used or to the specific location of the polymorphic sites was further determined. Mutations at these three SNP sites were engineered, converting each mutated amino acid into alanine. The NHE3 mutant R474A displayed reduced transporter activity without a change in its surface expression. The V567A variant exhibited reduced transport activity and surface expression. Therefore, these two mutants behaved similarly to the corresponding SNP variants (compare Figs. 2 and 3). The R799A mutant appeared to be unstable with too low protein expression to perform the assays (data not shown); therefore, this alanine mutant was excluded from further study.
Fig. 3.
Alanine substitutions at two polymorphic sites of NHE3 also decreased basal NHE3 activity. Na+/H+ basal activity was measured in PS120/NHERF2 cells stably expressing WT, R474A, or V567A. Data are presented as means ± SE from at least 3 independent experiments. *P < 0.05, comparison between WT NHE3 and R474A and V567A (Student's t-test). B: surface expression of NHE3 and the mutants R474A and V567A determined by surface biotinylation assay. Studies as described in the legend of Fig 2B. Representative experiment is shown that was repeated at least 3 times. C: table shows results of 4–6 experiments and statistical evaluation as described in Fig. 2C. ↑P < 0.05. *P < 0.05 for columns 1–4.
Altered response to dexamethasone stimulation but normal serum simulated and cAMP inhibited activity in the NHE3 SNPs.
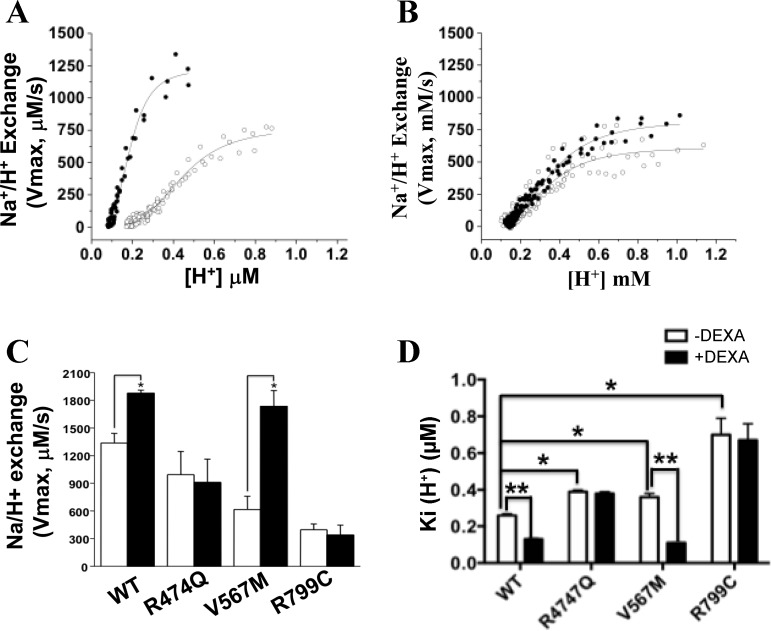
Previous studies have shown that dexamethasone acutely (short times of exposure) stimulates NHE3 activity through the activation of SGK1 (40, 41, 45) without altering NHE3 transcription. We evaluated the effect of dexamethesone (10 μM, 4 h) on NHE3 activity in PS120 cells stably cotransfected with NHERF2 and NHE3 mutants. Dexamethasone increased NHE3 Vmax and reduced K'(H+)i in wild-type NHE3 (Fig. 4, A, C, and D) and NHE3 V567M but had no effect on either Vmax or K'(H+)i of R474Q (Fig. 4, B, C, and D) and R799C (Fig. 4, C and D). Wild-type NHE3 activity was inhibited by cAMP (33–35) and stimulated by 10% dialyzed serum (27). All three variants were similarly inhibited by 100 μM 8-Br-cAMP and stimulated by serum to similar extents as wild-type NHE3 (data not shown).
Fig. 4.
Loss of response to dexamethasone stimulation in NHE3 variants. NHE3 activity was measured in PS120 cells stably expressing WT (A) and R474Q (B) in the presence (●) or absence (○) of dexamethasone (Dexa). C: data are presented as means ± SE of Vmax (C) and K'(H+)i (D) for NHE3 activity measured in PS12/NHERF2 cells stably expressing WT, R474Q, V567M, and R799C in the presence (filled bars) and absence (open bars) of dexamethasone from at least 3 independent experiments. *P < 0.05, comparison between controls and dexamethasone-treated groups (paired Student's t-test).
NHE3 binding to CHP is altered in some NHE3 SNPs.
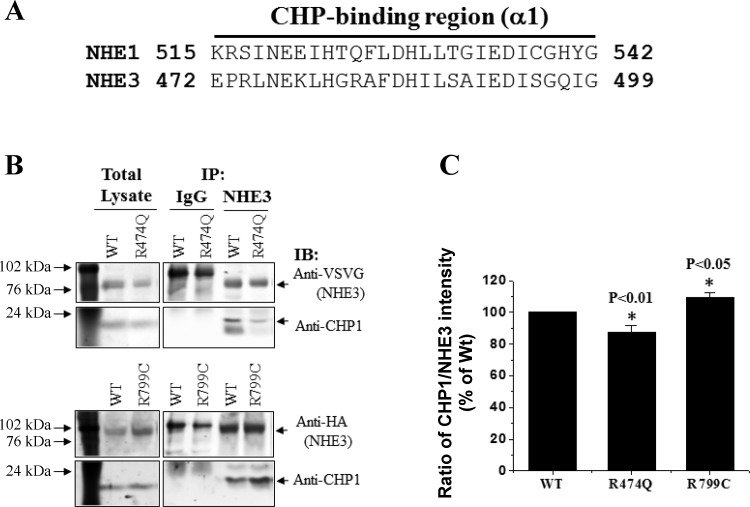
The structure for human NHE3 has not been solved. However, the NH2-terminal domain of other members of the NHE superfamily (NHE1 and Nha2) has been predicted based on homology to the bacterial Na+/H+ antiporter NhaA, which has been crystalized (22, 2). The bacterial Na+/H+ antiporter has a very short COOH-terminal domain. The only NHE COOH-terminal domain for which reliable structural information is available is the beginning of the NHE1 COOH terminus that was crystalized binding to CHP2 (2). By sequence homology, the NHE3 domain that binds CHP is predicted to include amino acid 473–499 and has 90% amino acid similarity with the CHP binding domain of NHE1 (amino acid 516–542) (Fig. 5A). The polymorphic NHE3-R474Q is in this area. Given that R474 maps to the putative NHE3 binding CHP domain, we postulated that a subtle amino acid substitution (e.g., R474Q) in this region could cause a conformational change in NHE3 and thereby alter its association with CHP. To test this hypothesis, PS120 cells were stably transfected with Flag-NHERF2 and HA-tagged wild-type NHE3 or NHE3-R474Q or R799C. HA antibody was used to pull down NHE3 protein, and anti-CHP1 antibody was used to detect the associated CHP1 by Western blot. Wild-type NHE3 and the NHE3 polymorphisms R474Q and R799C were coimmunoprecipitated by CHP1 (Fig. 5B). However, there was a significant decrease in coimmunoprecipitation of CHP by R474Q compared with wild type (0.88 ± 0.04; P < 0.01; Fig. 5, B and C), indicating that changing amino acid R474 to Q alters the interaction between NHE3 and CHP. Surprisingly, the binding of R799C to CHP was increased (1.09 ± 0.03) by ∼10% (P < 0.05; Fig. 5, B and C).
Fig. 5.
Altered binding of NHE3 variants to calcineurin B homologous protein 1 (CHP1). A: sequence alignments of the CHP-binding region in NHE1 and NHE3. B: Coimmunoprecipitation of CHP1 with WT NHE3, R474Q, or R799C illustrated by Western blot analysis. Molecular mass standards are at left. IB, immunoblot; IP, immunoprecipitation, C: quantification of binding of CHP1 to NHE3 and its variants. Results normalized to WT set as 100 for each experiment. *P < 0.05, comparison between wild-type NHE3 and R474Q or R799C (paired Student's t-test, n = 3).
DISCUSSION
The present study identified and characterized three missense SNPs, which affect residues in the intracellular COOH-terminal domain of NHE3. The effects of these polymorphisms were investigated on NHE3 transporter activity under both basal and acutely regulated conditions, as well as the ability of the mutant proteins to bind the NHE3 interacting protein CHP. These three variants all have significantly reduced basal NHE3 activity, although the compromised function was identified as being due to different mechanisms. This included reduced intrinsic NHE3 transport protein function (R474Q), abnormal trafficking or membrane stability (V567M), or defects in both intrinsic function and trafficking/stability (R799C). R474Q and R799C also failed to respond to acute stimulation with dexamethasone, while responding normally to inhibition by cAMP and stimulation by serum. These results suggest that there could be a defect in NHE3 function during some postprandial stimulation and under stress conditions in patients expressing these SNPs. In addition, we demonstrated that the R474Q variant had reduced interaction with CHP, providing a mechanism by which the compromised function might occur since CHP binding has been shown to be necessary for Na+/H+ exchange activity by the homologous NHE1 protein (34, 35).
SNPs in the NHE3 gene.
We focused on three nonsynonymous SNPs that result in change in three amino acids within the cytoplasmic, COOH-terminal domain of NHE3. However, we cannot conclude that other SNPs in the NHE3 gene are less important in the setting of NHE3 activity. For instance, SNPs could alter NHE3 activity by influencing expression of the NHE3 gene by altering transcription and/or translation of NHE3 or affecting mRNA stability or microRNA effects. The SNP database searched included all SNPs identified up to 2008. Since that time, many new SNPs have been identified in the exons of NHE3, including 40 additional nonsynonymous SNPs that affect residues in the NHE3 COOH-terminal domain. These are summarized in Table 1. Notably, two SNPs affect residues close to R474, seven affect residues near V567, and two affect residues near R799. Whether these SNPs impact NHE3 activity in a similar way to the SNPs characterized in the present study is not known. Our targeted sequencing of the three SNPs in DNA samples from six patients with congenital Na+ diarrhea did not detect mutations at these residues (data not shown), which suggests that other mutations in NHE3 (and/or in other genes) might be contributing to that form of diarrhea.
SNPs and NHE3 exchanger activity.
The three characterized SNPs affect residues in the COOH-terminal regulatory domain of NHE3. This domain is necessary for the physiologic function of NHE3 that includes inhibition in the immediate postprandial state, presumably to increase luminal water to spread the digestive enzymes over the absorptive surface, followed by stimulation later in the digestive period, presumably to prevent dehydration related to eating. This regulation appears to occur via the COOH terminus acting as a scaffold on which large, multiprotein regulatory complexes form (11, 13, 22). Models of the secondary structure of the NHE3 COOH terminus predict that the NH2-terminal component of the intracellular domain is highly structured and contains multiple α-helices, while the COOH-terminal half is largely disordered under basal conditions (11, 20). The R474 and V567 polymorphisms are present in the NHE3 COOH-terminal domains that take part in the formation or stabilization of these signaling complexes. R474Q impairs intrinsic transporter activity under basal conditions, prevents dexamethasone stimulation of NHE3 activity, and reduces CHP binding to NHE3. These are predicted consequences of disrupting what we named the stimulatory regulatory complex that forms on the NHE3 COOH terminus involving amino acids ∼474–554 (10). This complex involves CHP binding and direct ezrin binding to NHE3. CHP is a two EF-hand domain containing protein with three isoforms (19, 31, 34, 35). The structure of the CHP-NHE association has been solved for NHE1 with the CHP binding domain of NHE1 being α-helical and fitting into the hydrophobic cleft separating the NH2 and COOH termini of CHP, which is felt to stabilize this domain of NHE3. A CHP binding defective mutant of NHE3 has reduced basal activity and is unable to alter NHE3 activity in response to growth factors and osmotic stress (35). The CHP biding domain is conserved in other NHE isoforms (34) and as shown in Fig. 5A, this area is highly homologous for NHE3. There is 90% amino acid similarity between NHE1 (amino acids 516–542) and NHE3 (amino acids 473–499). Based on the analysis of the cocrystal structure of CHP2 and NHE1 (2, 35) and sequence alignment (Fig. 5A), we predict that the Arg residue at amino acid 474 in NHE3 may be positioned at the entry of the hydrophobic cleft, where it would be expected to help retain and stabilize the long stretch of the NHE3 α-helix in the CHP cleft. Since the R474Q polymorphism reduced CHP coprecipitation with NHE3, it is likely that the disrupted CHP binding explains the phenotype we described of reduced basal NHE3 activity. However, since only a 13% reduction of CHP binding was demonstrated, cause and effect has not been established. In addition, this stimulatory signaling complex involves direct binding of ezrin to NHE3 amino acids 517, 521, and 528, which requires phosphorylation of amino acids 516 and 527 (human NHE3) by Akt and GSK-3, respectively, as well as CHP binding (38). This ezrin binding to NHE3 is required not only for basal NHE3 activity but for multiple examples of stimulated NHE3 activity, and it is possible that ezrin binding is disrupted by this polymorphism to explain the failure of dexamethasone to stimulate NHE3.
The V567M polymorphism has a different effect on NHE3 activity than the R474Q, reducing NHE3 activity by decreasing the percentage of transporter on the plasma membrane as well as having less made although the function of each NHE3 molecule on the plasma membrane had full or increased Vmax activity but with an increased K'(H+)i. This suggests that this phenotype is due to abnormal NHE3 trafficking or stability on the membrane but with some altered turnover number. Due to the level of expression being variable among clones from the same construct in NHE3 studies in general, we are reluctant to attribute a role for altered expression in patients with these polymorphisms. The amino acid V567 is just outside the beginning of what we have called the stimulatory/inhibitor regulatory complex (11) and is predicted to help stabilize this part of the NHE3 COOH terminus. This NHE3 complex involves the direct binding of two kinases, one of which stimulates basal NHE3 activity (CK2) (36) while the other inhibits basal NHE3 activity (CaM kinase II) (1), as well as the area that the NHERF family of multi-PDZ domain proteins binds. The NHERFs form a large percentage of the NHE3 complexes and fix it to the plasma membrane in a dynamic manner that is necessary for regulation by trafficking (7). Whether this V567M polymorphism disrupts this NHE3 regulatory complex is not known, although we did not find that V567M altered NHERF2 binding (data not shown).
R799 lies in the downstream part of the NHE3 COOH terminus that appears nonstructured under basal conditions (11, 20). While this part of NHE3 binds megalin and PP2A and is the domain phosphorylated by both CK2 and CaM kinase II (7, 47), how it contributes to NHE3 function is not known. The observation that R799C increases CHP binding to NHE3 (Fig. 5, B and C) demonstrates that there is an interaction among seemingly distant parts of the COOH terminus and that understanding the structure of the COOH terminus, probably in the presence of the components of the signaling complexes, will be needed to fully understand how regulation of NHE3 occurs. The K'(H+)i was increased for all three polymorphisms, which supports that this complex characteristic of NHE3 activity is influenced by the regulatory domain as well as by the transport domain. Given that the K'(H+)i was affected by changes in different parts of the regulatory domain along with different mechanisms explaining the changes in NHE3 function, conclusions cannot be made about how these changes in the NHE3 COOH terminus alter K'(H+)i.
Altered response to dexamethasone.
Dexamethasone has been shown to acutely stimulate NHE3 activity through increased surface expression of NHE3 (40, 41, 45). Both R474Q and R799C, which exhibited decreased intrinsic activity, lost the response to dexamethasone, whereas V567M, which displayed impaired membrane trafficking or stability, maintained a normal stimulatory response. This suggests that short-term dexamethasone exposure may influence NHE3 activity via an additional unidentified mechanism. cAMP is a key second messenger that mediates the regulatory effect of hormones and growth factors on NHE3 (5, 46, 47). The normal extent of cAMP related inhibition of NHE3 that was present in all these polymorphisms indicates that the contribution to diarrhea of the reduced NHE3-related Na absorption is likely to occur as in the population with wild-type NHE3, while the compensatory increased Na+ absorption due to NHE3 is likely to be compromised in some patients with these polymorphisms.
Potential clinical significance of SNPs with reduced NHE3 activity.
Although we have identified three SNPs that have reduced NHE3 activity, it is not known if individuals bearing these SNPs have a phenotype related to abnormal bowel function. It would be predicted that this could take several forms that include an increased susceptibility to or increased severity of diarrhea, for instance that associated with a high glucocorticoid environment, such as inflammatory bowel disease, acute infectious diarrhea or even irritable bowel syndrome-diarrhea predominant, and/or reduced response to drugs that target stimulation of NHE3 to treat diarrhea or target inhibition of NHE3 to treat constipation and perhaps treat hypertension by reducing total body Na+, as recently suggested (30). In our separate studies, severely reduced NHE3 activity in several cases of congenital Na+ diarrhea was shown to be due to nonfunctional NHE3 mutations in the NH2-terminal transport domain in both alleles of NHE3 presenting as an autosomal recessive condition (unpublished observations, Janecke et al.). Thus in searching for clinical relevance of the functionally compromised polymorphisms, the population at risk is likely to be that in which both alleles are affected.
While the three polymorphisms studied have variable allele frequency, studies in specific populations with increased prevalence offer the best chance of understanding whether phenotypes exist as consequences of altered NHE3 activity from these SNPs. For instance, the genotype frequencies for 474R/474Q and 567V/567M are 0.02 and 0.22, respectively (14, 33). In contrast, the R799C polymorphism is distributed widely in European, African, and Asian populations (33); in fact, the European populations have genotype frequencies of 0.014 for 799R/799R, 0.215 for 799C/799R, and 0.771 for 799C/799C alleles, whereas the Asian-Han populations have much higher genotype frequencies for 799R/799R (0.302) and 799C/799R (0.514) but less frequency for 799C/799C (0.278), likely conferring reduced NHE3 activity (14, 33). Needing to be examined in these populations is the effects of the presence of a Cys rather than an Arg at amino acid position 799 on intestinal Na+ absorption and predisposition to acute and chronic diarrhea.
These and other nonsynonymous polymorphisms of NHE3 exist in variable gene frequencies among populations and since they have variable transport activities should be evaluated for possible contributions to diseases associated with abnormal water and electrolyte homeostasis.
GRANTS
This work was supported, in whole or in part, by National Institute of Diabetes and Digestive and Kidney Diseases Grants R01-DK-26523, R01-DK-61765, P01-DK-072084, T32-DK-07632, and P30-DK-089502 (The NIDDK Hopkins Digestive Diseases Basic and Translational Research Core Center; all to M. Donowitz) and K08-DK-088950 and R03-DK-099566 (both to X. C Zhu).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: X.C.Z., R.S., M.C., T.-E.C., C.M.T., B.C., and M.D. conception and design of research; X.C.Z., R.S., M.C., T.-E.C., and B.C. performed experiments; X.C.Z., R.S., J.R.H., M.C., C.M.T., B.C., and M.D. analyzed data; X.C.Z., R.S., J.R.H., T.-E.C., C.M.T., B.C., and M.D. interpreted results of experiments; X.C.Z. and M.D. prepared figures; X.C.Z. and M.D. drafted manuscript; X.C.Z., R.S., and M.D. edited and revised manuscript; X.C.Z., R.S., J.R.H., M.C., T.-E.C., C.M.T., B.C., and M.D. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Sandra Guggino for critical suggestions on the dexamethasone experiment and Denise Cheshner for help with cell culture. We thank Dr. Vladimir Yarov-Yarovoy for modeling the NHE3 COOH terminus.
REFERENCES
- 1.Akhter S, Cavet ME, Tse CM, Donowitz M. C-terminal domains of Na(+)/H(+) exchanger isoform 3 are involved in the basal and serum-stimulated membrane trafficking of the exchanger. Biochemistry 39: 1990–2000, 2000. [DOI] [PubMed] [Google Scholar]
- 2.Ammar YB, Takeda S, Hisamitsu T, Mori H, Wakabayashi S. Crystal structure of CHP2 complexed with NHE1-cytosolic region and an implication for pH regulation. EMBO J 25: 2315–2325, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amemiya M, Loffing J, Lotscher M, Kaissling B, Alpern RJ, Moe OW. Expression of NHE-3 in the apical membrane of rat renal proximal tubule and thick ascending limb. Kidney Int 48: 1206–1215, 1995. [DOI] [PubMed] [Google Scholar]
- 4.Brant SR, Yun CH, Donowitz M, Tse CM. Cloning, tissue distribution, and functional analysis of the human Na+/N+ exchanger isoform, NHE3. Am J Physiol Cell Physiol 269: C198–C206, 1995. [DOI] [PubMed] [Google Scholar]
- 5.Cabado AG, Yu FH, Kapus A, Lukacs G, Grinstein S, Orlowski J. Distinct structural domains confer cAMP sensitivity and ATP dependence to the Na+/H+ exchanger NHE3 isoform. J Biol Chem 271: 3590–3599, 1996. [DOI] [PubMed] [Google Scholar]
- 6.Cha B, Tse M, Yun C, Kovbasnjuk O, Mohan S, Hubbard A, Arpin M, Donowitz M. The NHE3 juxtamembrane cytoplasmic domain directly binds ezrin: dual role in NHE3 trafficking and mobility in the brush border. Mol Biol Cell 17: 2661–2673, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cha B, Chen T, Sarker R, Yang J, Raben D, Tse CM, Kovbasnjuk I, Donowitz M. Lysophosphatidic acid stimulation of NHE3 exocytosis in polarized epithelial cells occurs with release from NHERF2 via ERK-PLC-PKCδ signaling. Am J Physiol Cell Physiol 307: C55–C65, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen T, Kocinsky HS, Cha B, Murtazina R, Yang J, Tse CM, Singh V, Cole R, Aronson PS, de Jonge H, Sarker R, Donowtiz M. Cyclic GMP kinase (cGKII) inhibits NHE3 by altering its trafficking and phosphorylating NHE3 at three required sites: identification of a multifunctional phosphorylation site. J Biol Chem 290: 1952–1965, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi JW, Lee-Kwon W, Jeon ES, Kang YJ, Kawano K, Kim HS, Suh PG, Donowitz M, Kim JH. Lysophosphatidic acid induces exocytic trafficking of Na(+)/H(+) exchanger 3 by E3KARP-dependent activation of phospholipase C. Biochim Biophys Acta 1683: 59–68, 2004. [DOI] [PubMed] [Google Scholar]
- 10.Di Sole F, Babich V, Moe OW. The calcineurin homologous protein-1 increases Na(+)/H(+)-exchanger 3 trafficking via ezrin phosphorylation. J Am Soc Nephrol 20: 1776–1786, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donowitz M, Mohan S, Zhu CX, Chen TE, Lin R, Cha B, Zachos NC, Murtazina R, Sarker R, Li X. NHE3 regulatory complexes. J Exp Biol 212: 1638–1646, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donowitz M, Cha B, Zachos NC, Brett CL, Sharma A, Tse CM, Li X. NHERF family and NHE3 regulation. J Physiol 567: 3–11, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donowitz M, Li X. Regulatory binding partners and complexes of NHE3. Physiol Rev 87: 825–872, 2007. [DOI] [PubMed] [Google Scholar]
- 14.Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP), Seattle, WA: (Online). http://evs.gs.washington.edu/EVS/ [December2014]. [Google Scholar]
- 15.Field M. Intestinal ion transport and the pathophysiology of diarrhea. J Clin Invest 111: 931–943, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiskerstrand T, Arshad N, Haukanes BI, Tronstad RR, Pham KD, Johansson S, Havik B, Tonder SL, Levy SE, Brackman D, Boman H, Biswas KH, Apold J, Hovdenak N, Visweswariah SS, Knappskog PM. Familial diarrhea syndrome caused by an activating GUCY2C mutation. N Engl J Med 366: 1586–1595, 2012. [DOI] [PubMed] [Google Scholar]
- 17.Gawenis LR, Stien X, Shull GE, Schultheis PJ, Woo AL, Walker NM, Clarke LL. Intestinal NaCl transport in NHE2 and NHE3 knockout mice. Am J Physiol Gastrointest Liver Physiol 282: G776–G784, 2002. [DOI] [PubMed] [Google Scholar]
- 18.Guerrant RL, DeBoer MD, Moore SR, Scharf RJ, Lima AA. The impoverished gut–a triple burden of diarrhoea, stunting and chronic disease. Nat Rev Gastroenterol Hepatol 10: 220–229, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez-Ford C, Levay K, Gomes AV, Perera EM, Som T, Kim YM, Benovic JL, Berkovitz GD, Slepak VZ. Characterization of tescalcin, a novel EF-hand protein with a single Ca2+-binding site: metal-binding properties, localization in tissues and cells, and effect on calcineurin. Biochemistry 42: 14553–14565, 2003. [DOI] [PubMed] [Google Scholar]
- 20.Hendus-Altenburger R, Kragelund BB, Pedersen SF. Structural dynamics and regulation of the mammalian SLC9A family of Na+/H+ exchangers. Curr Top Membr 73: 9–148, 2014. [DOI] [PubMed] [Google Scholar]
- 21.Hoogerwerf WA, Tsao SC, Devuyst O, Levine SA, Yun CH, Yip JW, Cohen ME, Wilson PD, Lazenby AJ, Tse CM, Donowitz M. NHE2 and NHE3 are human and rabbit intestinal brush-border proteins. Am J Physiol Gastrointest Liver Physiol 270: G29–G41, 1996. [DOI] [PubMed] [Google Scholar]
- 22.Hunte C, Screpanti E, Venturi M, Rimon A, Padan E, Michel H. Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH. Nature 435: 1197–1202, 2005. [DOI] [PubMed] [Google Scholar]
- 23.Kim JH, Lee-Kwon W, Park JB, Ryu SH, Yun CH, Donowitz M. Ca2+-dependent inhibition of Na+/H+ exchanger 3 (NHE3) requires an NHE3–E3KARP-alpha-actinin-4 complex for oligomerization and endocytosis. J Biol Chem 277: 23714–23724, 2002. [DOI] [PubMed] [Google Scholar]
- 24.Ledoussal C, Woo AL, Miller ML, Shull GE. Loss of the NHE2 Na+/H+ exchanger has no apparent effect on diarrheal state of NHE3-deficient mice. Am J Physiol Gastrointest Liver Physiol 281: G1385–G1396, 2001. [DOI] [PubMed] [Google Scholar]
- 25.Lee C, Kang HJ, von Ballmoos C, Newstead S, Uzdavinys P, Dotson DL, Iwata S, Beckstein O, Cameron AD, Drew D. A two-domain elevator mechanism for sodium/proton antiport. Nature 501: 573–577, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee-Kwon W, Kawano K, Choi JW, Kim JH, Donowitz M. Lysophosphatidic acid stimulates brush border Na+/H+ exchanger 3 (NHE3) activity by increasing its exocytosis by an NHE3 kinase A regulatory protein-dependent mechanism. J Biol Chem 278: 16494–16501, 2003. [DOI] [PubMed] [Google Scholar]
- 27.Levine SA, Nath SK, Yun CH, Yip JW, Montrose M, Donowitz M, Tse CM. Separate C-terminal domains of the epithelial specific brush border Na+/H+ exchanger isoform NHE3 are involved in stimulation and inhibition by protein kinases/growth factors. J Biol Chem 270: 13716–13725, 1995. [DOI] [PubMed] [Google Scholar]
- 28.Levine SA, Montrose MH, Tse CM, Donowitz M. Kinetics and regulation of three cloned mammalian Na+/H+ exchangers stably expressed in a fibroblast cell line. J Biol Chem 268: 25527–25535, 1993. [PubMed] [Google Scholar]
- 29.Li X, Galli T, Leu S, Wade JB, Weinman EJ, Leung G, Cheong A, Louvard D, Donowitz M. Na+-H+ exchanger 3 (NHE3) is present in lipid rafts in the rabbit ileal brush border: a role for rafts in trafficking and rapid stimulation of NHE3. J Physiol 537: 537–552, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linz D, Wirth K, Linz W, Heuer HO, Frick W, Hofmeister A, Heinelt U, Arndt P, Schwahn U, Bohm M, Ruetten H. Antihypertensive and laxative effects by pharmacological inhibition of sodium-proton-exchanger subtype 3-mediated sodium absorption in the gut. Hypertension 60: 1560–1567, 2012. [DOI] [PubMed] [Google Scholar]
- 31.Mohan S, Tse CM, Gabelli SB, Sarker R, Cha B, Fahie K, Nadella M Zachos NC, Tu-Sekine B, Raben D, Amzel LM, Donowitz M. NHE3 activity is dependent on direct phophoinositide binding at the N-terminus of its intracellular cytosolic region. J Biol Chem 285: 34566–34578, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naoe Y, Arita K, Hashimoto H, Kanazawa H, Sato M, Shimizu T. Structural characterization of calcineurin B homologous protein 1. J Biol Chem 280: 32372–32378, 2005. [DOI] [PubMed] [Google Scholar]
- 33.National Center for Biotechnology Information. dbSNP Short Genetic Variations (Online). http://www.ncbi.nlm.nih.gov/projects/SNP [December2014].
- 34.Pang T, Su X, Wakabayashi S, Shigekawa M. Calcineurin homologous protein as an essential cofactor for Na+/H+ exchangers. J Biol Chem 276: 17367–17372, 2001. [DOI] [PubMed] [Google Scholar]
- 35.Pang T, Hisamitsu T, Mori H, Shigekawa M, Wakabayashi S. Role of calcineurin B homologous protein in pH regulation by the Na+/H+ exchanger 1: tightly bound Ca2+ ions as important structural elements. Biochemistry 43: 3628–3636, 2004. [DOI] [PubMed] [Google Scholar]
- 36.Sarker R, Gronborg M, Cha B, Moran S, Chen Y, Pandey A, Litchfield D, Donowitz M, Li X. Casein kinase 2 binds to the C terminus of Na+/H+ exchanger 3 (NHE3) and stimulates NHE3 basal activity by phosphorylating a separate site in NHE3. Mol Biol Cell 19: 389–3870, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultheis PJ, Clarke LL, Meneton P, Miller ML, Soleimani M, Gawenis LR, Riddle TM, Duffy JJ, Doetschman T, Wang T, Giebisch G, Aronson PS, Lorenz JN, Shull GE. Renal and intestinal absorptive defects in mice lacking the NHE3 Na+/H+ exchanger. Nat Genet 19: 282–285, 1998. [DOI] [PubMed] [Google Scholar]
- 38.Singh V, Lin R, Yang J, Cha B, Sarker R, Tse CM, Donowitz M. AKT and GSK-3 are necessary for direct ezrin binding to NHE3 as part of a C-terminal stimulatory complex: role of a novel Ser-rich NHE3 C-terminal motif in NHE3 activity and trafficking. J Biol Chem 289: 5449–5461, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vallon V, Schwark JR, Richter K, Hropot M. Role of Na+/H+ exchanger NHE3 in nephron function: micropuncture studies with S3226, an inhibitor of NHE3. Am J Physiol Renal Physiol 278: F375–F379, 2000. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Sun H, Lang F, Yun CC. Activation of NHE3 by dexamethasone requires phosphorylation of NHE3 at Ser663 by SGK1. Am J Physiol Cell Physiol 289: C802–C810, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang D, Zhang H, Lang F, Yun CC. Acute activation of NHE3 by dexamethasone correlates with activation of SGK1 and requires a functional glucocorticoid receptor. Am J Physiol Cell Physiol 292: C396–C404, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wedenoja S, Pekansaari E, Hoglund P, Makela S, Holmberg C, Kere J. Update on SLC26A3 mutations in congenital chloride diarrhea. Hum Mutat 32: 715–722, 2011. [DOI] [PubMed] [Google Scholar]
- 43.Yang J, Singh V, Chen TE, Sarker R, Xiong L, Cha B, Jin S, Li X, Tse CM, Zachos NC, Donowitz M. NHERF2/NHERF3 protein heterodimerization and macrocomplex formation are required for the inhibition of NHE3 activity by carbachol. J Biol Chem 289: 20039–20053, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yun CH, Tse CM, Donowitz M. Chimeric Na+/H+ exchangers: an epithelial membrane-bound N-terminal domain requires an epithelial cytoplasmic C-terminal domain for regulation by protein kinases. Proc Natl Acad Sci USA 92: 10723–10727, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yun CC, Chen Y, Lang F. Glucocorticoid activation of Na(+)/H(+) exchanger isoform 3 revisited. The roles of SGK1 and NHERF2. J Biol Chem 277: 7676–7683, 2002. [DOI] [PubMed] [Google Scholar]
- 46.Zhao H, Wiederkehr MR, Fan L, Collazo RL, Crowder LA, Moe OW. Acute inhibition of Na/H exchanger NHE-3 by cAMP. Role of protein kinase A and NHE-3 phosphoserines 552 and 605. J Biol Chem 274: 3978–3987, 1999. [DOI] [PubMed] [Google Scholar]
- 47.Zizak M, Lamprecht G, Steplock D, Tariq N, Shenolikar S, Donowitz M, Yun CH, Weinman EJ. cAMP-induced phosphorylation and inhibition of Na(+)/H(+) exchanger 3 (NHE3) are dependent on the presence but not the phosphorylation of NHE regulatory factor. J Biol Chem 274: 24753–24758, 1999. [DOI] [PubMed] [Google Scholar]